Abstract
The Escherichia coli flagellar master regulator, FlhD4C2, binds to the promoter regions of flagellar class II genes, yet, despite extensive analysis of the FlhD4C2-regulated promoter region, a detailed consensus sequence has not emerged. We used in vitro and in vivo experimental approaches to determine the nucleotides in the class II promoter, fliAp, required for the binding and function of FlhD4C2. FlhD4C2 protects 48 bp (positions −76 to −29 relative to the σ70-dependent transcriptional start site) in the fliA promoter. We divided the 48-bp footprint region into 5 sections to determine the requirement of each DNA segment for the binding and function of FlhD4C2. Results from an in vitro binding competition assay between the wild-type FlhD4C2-protected fragment and DNA fragments possessing mutations in one section of the 48-bp protected region showed that only one-third of the 48 bp protected by FlhD4C2 is required for FlhD4C2 binding and fliA promoter activity. This in vitro binding result was also seen in vivo with fliA promoter-lacZ fusions carrying the same mutations. Only seven bases (A12, A15, T34, A36, T37, A44, and T45) are absolutely required for the promoter activity. Moreover, A12, A15, T34, T37, and T45 within the 7 bases are highly specific to fliA promoter activity, and those bases form an asymmetric recognition site for FlhD4C2. The implications of the asymmetry of the FlhD4C2 binding site and its potential impact on FlhD4C2 are discussed.
INTRODUCTION
The bacterial flagellum is an external helical filament whose rotation is responsible for swimming motility. Over 60 genes belong to the flagellar regulon of Escherichia coli, and these are grouped into transcriptional classes (class I, class II, and class III) comprising a three-level, hierarchical regulatory cascade (24, 27, 33). At the top of the hierarchy is the sole member of class I operons, the flhDC operon, which encodes the master transcriptional regulator, FlhD4C2, of the flagellar regulon. Together with σ70, FlhD4C2 activates the transcription of class II promoters, including those of fliA, flgM, and genes for hook-basal body assembly. fliA encodes σ28, a flagellar gene-specific σ factor that is responsible for initiating the transcription of class III promoters (31, 32). Some operons have both class II and class III promoters, including fliA and flgM, encoding the major checkpoint for flagellum biosynthesis. In the early stages of flagellar synthesis, FlgM, the anti-σ28 factor, binds σ28 to inhibit the transcription of class III promoters until the flagellar hook-basal body complex is complete. Subsequently, FlgM is secreted through the flagellar type III secretion system to release free σ28 that associates with RNA polymerase (RNAP) to initiate transcription from class III promoters (9, 10, 19). Other class III genes encode flagellar filament components, flagellar basal bodies, and the chemotaxis system. The flhDC operon is regulated at the transcriptional level by many environmental factors via global regulators, such as CRP (catabolite repression) (53), OmpR (osmolarity) (50), H-NS (DNA topology) (5, 53), LrhA (28), RcsAB (16), HdfR (23), and QseBC (quorum sensing) (55). CsrA regulates the expression of the flhDC operon at the posttranscriptional level (54, 65). The activity and stability of FlhDC are regulated by FliZ, FliT, and FliD, the downstream gene products in the flagellar regulon, by YdiV, an EAL-like protein acting as an anti-FlhD4C2 factor, by ClpXP, an ATP-dependent protease, and by DnaK, a protein chaperone (1, 45, 46, 58, 59, 62, 67). Microarray studies suggest that FlhDC also functions as a global regulator involved in carbon metabolism and anaerobic respiration (38, 39).
Typically, bacterial transcriptional regulators use a helix-turn-helix (HTH) motif to bind DNA and commonly form homodimers (e.g., λ cI, λ cro, trp repressor, CRP, NtrC, MarR, and FadR) (2, 22, 36, 40, 42, 43, 49, 60, 64). Some transcriptional regulators bind their target sites as monomers (e.g., members of the AraC family and MotA from bacteriophage T4) (11, 17), or as homomultimers (e.g., LacI, H-NS, and the LysR family) (4, 6, 34, 52). Heteromeric transcriptional activators are unusual in bacteria but have been reported, such as the dimer of integration host factor, IHF-αβ, the dimer of the regulator of capsular biosynthesis, RcsBA, and dimers of the nucleoprotein H-NS with another nucleoprotein, Hha or StpA (15, 57). FlhDC is a unique bacterial transcriptional regulator composed of heterologous subunits, FlhD (13 kDa) and FlhC (22 kDa), that form a functional heterohexamer complex, FlhD4C2 (63).
Due to the formation of a large complex, FlhD4C2 protects a large nucleotide “footprint” in the promoters that it regulates. DNase I footprinting analyses have shown that the FlhD4C2 complex binds to 48 to 50 bp in the promoter regions of the fliA, flhB, and fliL operons of Escherichia coli and to 46 to 59 bp in the promoter regions of the flhB, fliA, and flgA-flgB operons of Proteus mirabilis (13, 32). The FlhD4C2 binding site (from approximately positions −30 to −80 relative to the transcriptional start site) overlaps the σ70-specific −35 element (Fig. 1A). The C-terminal domain of the α-subunit (α-CTD) of RNA polymerase is required for FlhD4C2-mediated transcription (30) and presumably binds to the upstream region of the FlhD4C2 binding site.
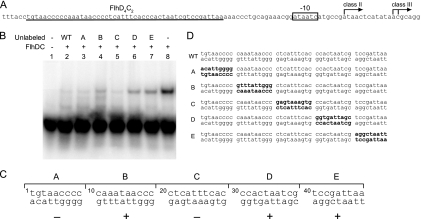
Fig. 1.
Map of the in vitro binding region on the FlhD4C2 footprint sequence. (A) Structure of the fliA promoter. Transcriptional start sites of class II and class III (arrows), class II promoter-dependent −10 element (box), and FlhD4C2 protected region (underlined) are illustrated. (B) A 500-fold excess of unlabeled fragments (WT, A, B, C, D, and E) competed with radioactively labeled WT fragment for FlhD4C2 complex binding. As controls, lane 1 contains the radioactively labeled probe DNA only and lane 8 contains both the radioactively labeled probe and the FlhD4C2 complex but no competitor DNA fragments (+, present; -, absent). (C) The effects of mutations in each section on FlhD4C2 binding (+, positive; -, negative). (D) Sequences of DNA fragments used for the in vitro binding competition assay, including the wild-type FlhD4C2 footprint sequence in the fliA promoter and derivative sequences with complementary mutations (shown in bold) in sections A, B, C, D, and E.
In electrophoretic mobility shift assays (EMSAs), FlhD does not have DNA binding activity by itself, but reconstituted FlhC (from Proteus mirabilis) does, and binding is enhanced in the FlhD4C2 complex (12, 32). The FlhD4C2 complex binds potentially to DNA by a helix-turn-helix structure and the region near a zinc binding area in the FlhC protomers (Y.-Y. Lee and P. Matsumura, unpublished data). However, results from photo-cross-linking, alanine scanning mutagenesis, and heparin affinity of protein variants suggest that FlhD also contributes to DNA binding through Ser-82, Arg-83, and Val-84, located in the C terminus of the protein (7, 13).
While sequence alignment of the FlhD4C2 footprint fragments does not produce a compelling consensus sequence, a consensus sequence of FlhD4C2 binding nevertheless has been generated using different approaches. Kutsukake and coworkers found σ70-dependent −10 consensus elements in the promoters of the fliA, flgA, flgB, and fliD genes from Salmonella enterica serovar Typhimurium, but no evident −35 consensus elements; instead, they observed a sequence element, TTATTCC, centered at −42 of these class II promoters (21, 26, 37). In subsequent studies, the same group found another weakly conserved sequence element, GCAATAA, present 15 to 19 bp upstream of the TTATTCC element in flagellar class II promoters. The two elements make an imperfect palindromic sequence motif, and deletion of the GCAATAA half-site eliminates promoter activity (20). An informatics study of FlhD4C2 binding sequences was done by the Hughes group at Cambridge (England), who aligned the promoter regions of 12 flagellar class II genes from E. coli (fliA, fliL, and flhB), S. Typhimurium (fliA, fliL, flhB, fliE, and fliF), and P. mirabilis (fliA, flhB, and flgA-flgB). From this study, they proposed a consensus “FlhDC box”: an imperfect inverted repeat of TNAA(C/T)G(C/G)N2–3AAATA(A/G)CG in each half-site separated by a 10- to 12-nucleotide spacer (13). This FlhDC box is found adjacent to the putative −35 elements in the promoters of flagellar class II genes and some nonflagellar genes (56). Significantly, empirical data that correlate the informatic consensus sequence and the function of FlhD4C2 have not been reported.
In this report, we studied the fliA promoter as an example of a FlhDC-regulated promoter and determined the regions where FlhD4C2 binds in the FlhD4C2 footprint. Unlike previous efforts, we randomized the sequence in the FlhD4C2 footprint on the fliA promoter to experimentally build, using both in vitro and in vivo approaches, a minimal DNA sequence required for FlhD4C2 binding and activity. Our approaches allowed us to build a sequence of the FlhD4C2 binding site with biological significance that can lead to a greater understanding of the functional mechanisms of complex and global regulators, such as FlhD4C2. The resulting sequence not only shares common bases and features with the informatically derived consensus sequence but also provides higher resolution and specificity than previous approaches. Importantly, we found that only 7 bases in the sequence are absolutely required for FlhD4C2 binding, and they form a unique asymmetric site.
MATERIALS AND METHODS
Bacteria strains, plasmids construction, and growth conditions.
The fliA-deficient nonmotile E. coli strain YK4104 (25), a derivative of YK410 [F− araD139 Δ(argF-lac)169 λ− pyrC46 Δ(fruK-yeiR)725(fruA25) thi gyrA-0(NalR) relA1 thyA0 rpsL150(strR) rbsR22 deoC1 his] (25) was used for all in vivo tests. E. coli strain XL1-Blue (Strategene) was used for routine plasmid manipulation. E. coli strain BL21(DE3) was used to overexpress the FlhDC complex. pXL11, bearing the fusion of the fliA promoter (210 bp inserted into the EcoRI and the BamHI sites) with lacZ in pRS528 (51), was used as a transcriptional reporter of fliA promoter activity. pXL11 derivatives were constructed to have the fliA promoter bearing mutations on the FlhD4C2 footprint. Derivative fliA promoter fragments were produced by sequential PCR using a single or a pair (sequence complementary to each other) of mutagenic primers (see Table S1 in the supplemental material) and two flanking primers, 5′fliAFP (CGGGATCCATCCGGCAACATAAA) and 3′fliAFP_rev (CGTCGCCGCTTTCATCGGTT). 5′fliAFP contains a BamHI recognition site (sequence underlined), and 3′fliAFP_rev matches the opposite strand sequence that is downstream and adjacent to an EcoRI recognition site into which the right end of the fliA promoter fragment is cloned. Each step of amplification was done using pXL11 or the derivatives as a template, PCR SuperMix HiFi (Invitrogen) as the DNA polymerase mix, and the following thermocycler program: 94°C for 3 min; 25 cycles of 94°C for 45 s, 58°C for 30 s, and 72°C for 15 s; 72°C for 5 min. pXL27, bearing the flhDC operon in pT7-7, was used to overexpress the FlhDC complex (32).
Bacteria were grown in Luria-Bertani (LB) broth (Bacto tryptone, 10 g liter−1; yeast extract, 5 g liter−1; sodium chloride, 10 g liter−1) (35) at 37°C. LB was supplemented with 20 g liter−1 thiamine and 20 g liter−1 thymine when growing YK4104 to improve the growth, as they supplement the inability of the strain to synthesize thiamine and thymine de novo due to thi and thyA0 mutations (3).
FlhD4C2 complex overexpression and purification.
Recombinant native FlhD4C2 complex was expressed by 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) induction at 37°C using mid-exponential-phase cultures. Overexpression of IPTG-induced FlhD4C2 proteins was examined in whole-cell extracts using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Cells were harvested by centrifugation after 3 h of induction and incubation and resuspended in 50 mM Tris (pH 7.9), resulting in a concentration of 1 g of cells in 3 ml of 50 mM Tris (pH 7.9). Cells were disrupted by sonication, and the resulting lysate was passed through a HiTrap heparin HP column (Amersham Bioscience) using an EconoPump system (Bio-Rad). The protein complex was eluted in 50 mM Tris (pH 7.9) with a gradient of 0 to 1.4 M NaCl, which separated overexpressed FlhD4C2 complex from other proteins with nonspecific binding. The protein composition was confirmed by SDS-PAGE followed by Coomassie brilliant blue staining. Fractions containing only bands corresponding to the sizes of FlhD and FlhC proteins were collected for further use.
EMSA.
The DNA probes used for EMSA were made by two methods. Oligonucleotides (48 bp in length) were suspended in water to a final concentration of 1 mg ml−1. Five picomoles of the respective oligonucleotide was labeled in 10 μl with 1 μl of [γ-32P]ATP (6,000 Ci mmol−1; 10 μCi ml−1) using T4 polynucleotide kinase (Invitrogen) at 37°C for 20 min, and then complementary labeled oligonucleotides were combined and annealed at room temperature for 20 min. After separation using a 10% (wt/vol) polyacrylamide gel in Tris-borate-EDTA (TBE) running buffer (100 mM Tris-HCl, 90 mM boric acid, 1 mM EDTA; pH 8.4) by a “crush and soak” method, the double-stranded 32P-labeled DNA was purified and resuspended in TE (pH 8.0) (47). The 210-bp DNA fragment of the fliA promoter was obtained by PCR using the flanking primers 5′fliAFP and 3′fliAFP_rev following the program described above.
Twenty microliters of each binding reaction mixture containing 1× binding buffer (10 mM Tris [pH 7.9], 100 mM potassium chloride, 5 mM EDTA [pH 8.0], and 5% glycerol), DNA probe, and FlhD4C2 as required was incubated at 30°C for 20 min. For the binding competition assay, 50 ng FlhD4C2, 2 ng 32P-labeled DNA probe, and 1 μg unlabeled DNA fragments (as required) were added. For binding sequence selection, 147 ng FlhD4C2 and 44 ng 210-bp DNA probe were used. Products from the binding reaction mixture were separated on 5% (wt/vol) polyacrylamide gels buffered with TBE at 8 mA or 50 to 80 V for 90 min. The 32P-labeled DNA fragments and the PCR-generated DNA fragments, respectively, were visualized by autoradiography and UV light illumination of the ethidium bromide (EtBr)-stained DNA.
SAAB assay.
The selection and amplification of binding sequence (SAAB) method is also known as cyclic amplification and selection of targets (CAST) and systematic evolution of ligands by exponential enrichment (SELEX). As used in this study, the SAAB method was as follows. A primer pair complementary to each other and containing random sequences in the middle of each oligonucleotide was used to construct a fliA promoter segment with degenerate sequence in the target region. The degenerate fliA promoter was used in EMSAs, and DNAs contained within the gel were stained with EtBr and visualized following exposure to 320-nm UV light. DNA bands that had shifted mobility were purified from the polyacrylamide gel by using a QIAEX II gel extraction kit (Qiagen) and eluted in 20 μl of water. The fragments were amplified by PCR following the program described above. This procedure was repeated up to 7 times, and the sequence of the PCR product from each cycle was determined with an ABI 3730 DNA analyzer (University of Illinois at Chicago Research Resources Center DNA sequencing facility).
β-Galactosidase assay.
β-Galactosidase activity expressed from each promoter-lacZ transcriptional fusion was determined in Miller units (35) as follows. Cells were harvested from exponential-phase cultures (optical density at 600 nm [OD600], 0.3 to 0.7), and 0.1 ml of the culture was added to a reaction mixture containing 0.9 ml of Z buffer (8.517 g Na2HPO4, 5.5 g NaH2PO4·H2O, 0.75 g KCl, 0.246 g MgSO4·7H2O, 2.7 ml β-mercaptoethanol in 1,000 ml of distilled water), 60 μl of chloroform, and 40 μl of 0.1% SDS. The mixture was vortexed for 10 s, and the assay was started by adding 200 μl of 4 mg ml−1 o-nitrophenyl-β-d-galactopyranoside (dissolved in Z buffer) and stopped by adding 0.5 ml of 1 M Na2CO3 once a yellow color had developed (OD420, 0.4 to 0.6). The reaction mixture was incubated at 28°C. β-Galactosidase activity was determined using the following formula: (1,000 × OD420 of the reaction mixture after the assay terminated)/(OD600 of the culture suspension × 0.1 × reaction time, in minutes). Samples were measured from three independent experiments in triplicate.
Data calculation.
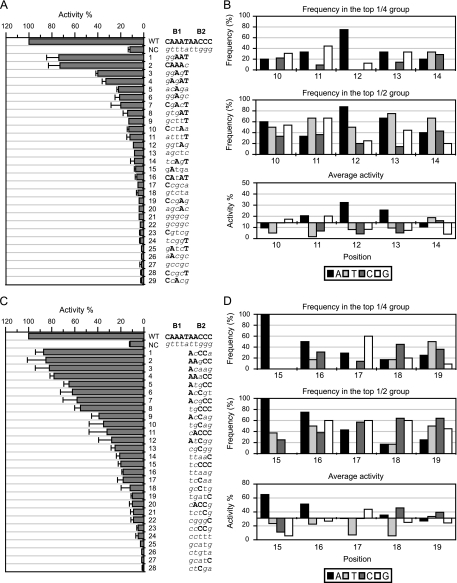
The frequency of each base at each position in the high-activity groups (defined as the top one-fourth [top 1/4] and top one-half [top 1/2] highest activities after screening, as shown below in Fig. 4 to 6) was calculated as described below. The frequency was determined as the ratio of the number of times a particular base appeared at a specific position in the defined group (top 1/4 or 1/2 activities) out of the number of times that base appeared at that specific position in all samples. For example, 29 samples had been screened in section B1, so samples 1 to 7 are in the top 1/4 activities group. A12 appeared 6 times in the top 1/4 group and 8 times in all samples (see Fig. 4A). Therefore, the frequency of A12 in the top 1/4 group is 75%.
Fig. 4.
Activities of the fliA promoter with sequence alterations in sections B1 and B2. (A and B) Section B1; (C and D) section B2. (A and C) The percent activity (normalized to the wild type, set as 100%) is listed in decreasing order, with the corresponding nucleotide sequence. The wild-type sequence is shown in bold uppercase letters, and the mutant sequence is shown in italic lowercase letters. NC, negative control (possesses complementary mutations in the whole B section [positions 10 to 19]). The promoter activity was followed by measuring β-galactosidase activity in three independent experiments. Error bars represent standard deviations of the means. (B and D) The frequency of each base at each position in the top 1/4 (upper graphs) and the top 1/2 (middle graphs) high activity groups and the average activity contributed by each base at each position (bottom graphs). In the bottom graphs of panels B and D, the x axis crosses the y axis at the mean value of the activity of all samples for section B1 and B2, respectively.
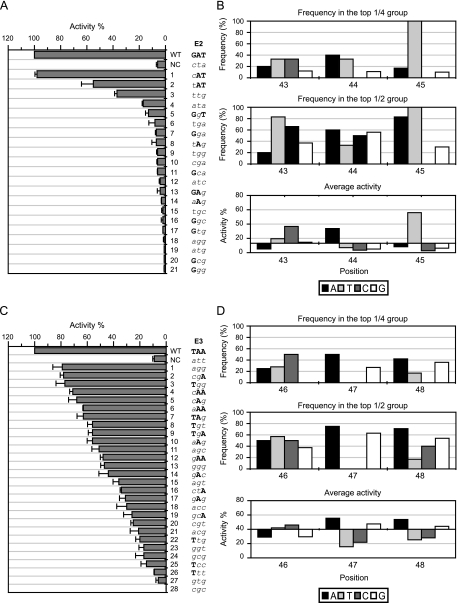
Fig. 6.
Activities of the fliA promoter with sequence alterations in sections E2 and E3. (A and B) Section E2; (C and D) section E3. (A and C) The percent activity (normalized to the wild type, set as 100%) is listed in decreasing order, with the corresponding nucleotide sequence. The wild-type sequence is shown in bold uppercase letters, and the mutant sequence is shown in italic lowercase letters. NC, negative control (possesses complementary mutations in section E2 [positions 43 to 45] and section E3 [positions 46 to 48], respectively). The promoter activity was followed by measuring β-galactosidase activity in three independent experiments. Error bars represent standard deviations of the means. (B and D) The frequency of each base at each position in the top 1/4 (upper panels) and the top 1/2 (middle panels) high activity groups and the average activity contributed by each base at each position (bottom panel). In the bottom graphs of panels B and D, the x axis crosses the y axis at the mean value of the activity of all samples for sections E2 and E3, respectively.
The mean activity of each base at each position was calculated by taking the mean value of the activities from the samples with a particular base appearing at a specific position. For example, average activity of A12 is the mean value of the activities of samples possessing A at position 12.
RESULTS
Mapping the binding regions of the FlhD4C2 complex on the fliA promoter.
We started this study by mapping the regions required for FlhD4C2 complex binding on the reported FlhD4C2 footprint region in the fliA promoter. The FlhD4C2 complex protects a 48-bp region of the fliA promoter from −76 to −29 relative to the σ70-dependent (class II) transcriptional start site and −88 to −41 relative to the σ28-dependent (class III) transcriptional start site, respectively (31, 32) (Fig. 1A). The 48-bp DNA fragment found by footprinting is sufficient for FlhD4C2 complex binding, as shown in Fig. 1B (lane 8). To define further the required bases, we divided the 48-bp fragment into 5 sections, called A to E (Fig. 1C). Section A contained bases 1 to 9; B contained bases 10 to 19; C contained bases 20 to 29; D contained bases 30 to 39; E contained bases 40 to 48. The five 48-bp DNA fragments (A, B, C, D, and E) were synthesized with complementary mutations, i.e., strands were “flipped over” in each corresponding section, as shown in Fig. 1D. The wild-type fragment, along with fliA fragments A, B, C, D, and E, were used in in vitro binding competition assays in which unlabeled fliA fragments A to E competed against labeled wild-type fliA fragment (WT in Fig. 1). The advantages in making these types of mutations are that (i) they are the same length (an important consideration in binding competition assays), (ii) every section is kept in the same position relative to the others, (iii) every base is mutated within the particular section of interest, and (iv) this prevents purine or pyrimidine bias in the mutated sequence. With this approach, the better the unlabeled fragments compete, the less significant the mutated sections are to FlhD4C2 binding, and thus a shifted band in the competition indicates that the mutations in the unlabeled DNA section are important to FlhD4C2 binding. As can be observed in Fig. 1B, binding of the FlhD4C2 complex to labeled wild-type fliA DNA without unlabeled competitor DNA caused an upward shift in band electrophoretic mobility (lane 8). Unlabeled wild-type fliA (Fig. 1B, lane 2) outcompeted the binding of radioactively labeled fliA DNA to FlhD4C2. Mutant fragments A and C had the same capability as unlabeled wild-type fliA to outcompete radioactively labeled wild-type fliA fragment in binding to the FlhD4C2 complex (Fig. 1B, compare lane 2, the control, to lanes 3 and 5 containing fragments A and C, respectively). In contrast, mutated fragments B, D, and E did not have the same capabilities as the mutant fragments A and C to compete with binding of wild-type fliA. These data indicate that sections B, D, and E of the FlhD4C2 footprint in the fliA promoter contribute strongly to FlhD4C2 binding, while sections A and C do not.
We next asked whether the bases of fliA required for FlhD4C2 binding are also required for FlhD4C2 activation of transcription from the fliA promoter. To address this, a series of fliA promoter (210 bp)-lacZ transcriptional fusions were made using plasmid pRS528 (see Materials and Methods). The FlhD4C2 footprint sequence was replaced by each of the respective 48-bp fragments (Fig. 1C, sections A to E) used in the previous binding competition assay. Since fliA contains both class II and class III promoters, a set of plasmids bearing mutated fliA promoter-lacZ fusions was introduced in YK4104, a nonmotile E. coli strain with a fliA-deficient background, to avoid false-positive expression from the class III fliA promoter. Promoter activity was measured indirectly as β-galactosidase activity normalized against the wild-type activity. Figure 2 shows the results from these experiments. Almost 80% of the activity was retained when either section A or section C was mutated. However, transcription was reduced to 12% when section B was mutated, and promoter activity was abolished when either section D or section E was mutated (Fig. 2A). In addition, knocking out flhDC (YK410 flhDC::kan) abolished transcription of the wild-type fliA promoter (17 units versus 20 units from a flhDC strain containing pRS528 [empty vector control]). This result indicates that the reduction of transcription resulting from the mutated sections is due to the action of FlhD4C2 specifically and is not caused by an alteration in the basal level of transcription from the fliA promoter. These results suggest that the footprint region required for FlhD4C2 binding is also required for the promoter activity, i.e., transcription of fliA.
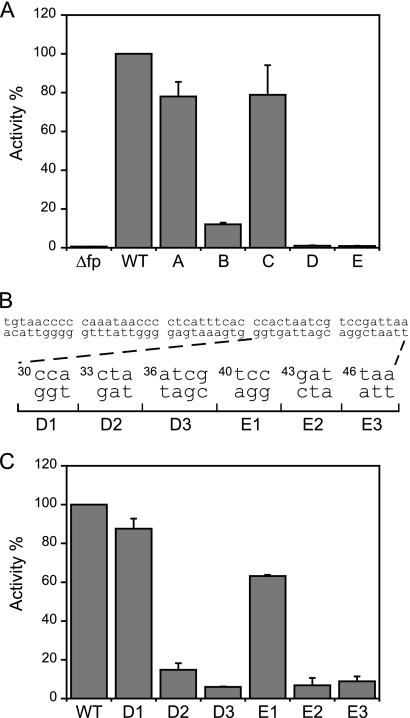
Fig. 2.
Regions on the FlhD4C2 footprint required for fliA promoter activity. (A) The activity of the fliA promoter (normalized to the WT fliA promoter activity), compared with those after deletion of the FlhD4C2 footprint sequence (Δfp) and of the mutated FlhD4C2 footprint sequences A, B, C, D, and E. (B) Dissection of fragments D and E. (C) The percentages of activity of the fliA promoter containing mutations in sections D1, D2, D3, E1, E2, and E3. Promoter activity was followed by measuring the β-galactosidase activity in three independent experiments. The error bars represent standard deviations of the means.
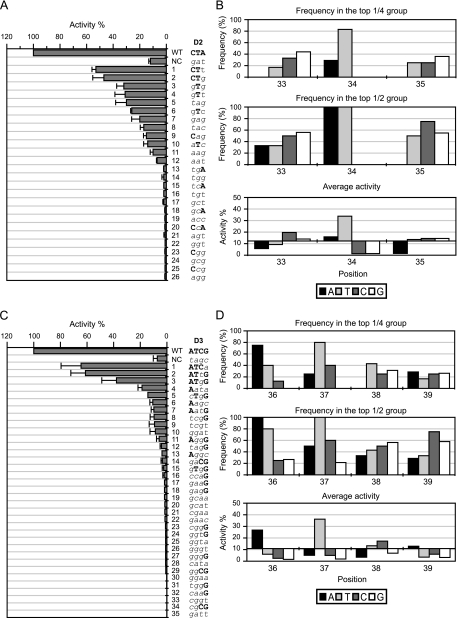
We further dissected sections D and E (10 and 9 bp, respectively) into 6 parts: D1, position 30 to 32; D2, position 33 to 35; D3, position 36 to 39; E1, position 40 to 42; E2, position 43 to 45; E3, position 46 to 48. β-Galactosidase activities of these and complementary mutations in each subsegment were measured (Fig. 2B). The results indicated that only D2/D3 and E2/E3 are required for FlhD4C2 function (Fig. 2C). Combining the in vitro (Fig. 1) and in vivo (Fig. 2) results, we found that FlhD4C2 binding to and activation of the fliA promoter requires sections B, D (specifically D2/D3), and E (specifically E2/E3) of the FlhD4C2-protected region of fliA.
Determination of DNA sequence for FlhD4C2 binding.
We employed a method (a selection and amplification of binding sequence or SAAB assay [see Materials and Methods]) to select and amplify random mutations in fliA segments that enhance FlhD4C2 binding. Briefly, we amplified fragments of the 210-bp fliA promoter carrying random sequences in sections B, C, D2/D3, or E2/E3. Mixtures of the probe DNA and the purified FlhD4C2 complex were used in an EMSA, and the FlhD4C2-bound DNA was isolated from the gel and amplified by PCR. These amplified DNAs became new probes for subsequent FlhD4C2 binding in EMSAs.
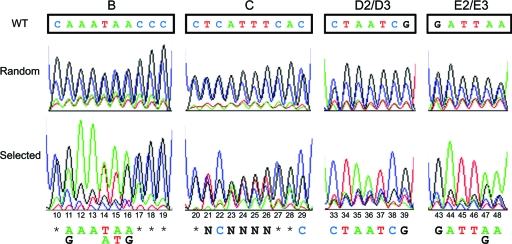
In the early cycles of the SAAB assay selection, randomization of the bases in sections B, D2/D3, or E2/E3 resulted in a low degree of shifted DNA, but in subsequent cycles, we observed an enrichment of selected sequences that optimized binding to FlhD4C2. The efficiency of the SAAB selection was examined by sequencing the amplified DNA mixture after each cycle, and the resulting sequence chromatograms were used to show the frequency of distribution of each nucleotide position in the mix of resulting randomized DNAs. Specific optimized binding sequences were obtained in 5 cycles of SAAB for section B, in 4 cycles for section D2/D3, and in 3 cycles for section E2/E3. SAAB failed to identify a specific sequence for section C, despite extending the assay to 7 cycles (Fig. 3). The results obtained through SAAB indicate that fragment C is not essential, which agrees with our results obtained from binding assays and fliA transcriptional fusions. As shown in Fig. 3, the sequence ultimately selected by SAAB for optimized binding of FlhD4C2 to the fliA promoter is (A/g)AA(T/A)(A/t)(A/g) at positions 11 to 16 of section B, CTAATCG at positions 33 to 39 of section D2/D3, and GATT(A/g)A at positions 43 to 48 of section E2/E3. The sequence distribution of the three segments is very similar to the wild-type fliA sequence and consists of mostly As and Ts. This result is not surprising, since the wild-type fliA promoter sequence is optimal for properly regulated transcription activation, but it does highlight the importance of A/T base pairs as binding targets of FlhD4C2.
Fig. 3.
Chromatograms of SAAB results. The sequencing chromatograms of the original degenerated probe sequences (random [upper panel]) and of the sequence from SAAB selection (selected [lower panel]) are shown. The wild-type sequence of the corresponding regions (WT [top boxes]) and the selected nucleotide sequence (bottom) are displayed. N, any base; *, no selectivity. The numbers beneath the chromatograms indicate the relative position in the footprint. The abundance of Gs and Cs in the “random” (upper panel) chromatograms is due to an artifact of the oligonucleotide synthesis.
Identification of bases required for FlhD4C2-activated fliA transcription.
In order to determine which bases in the FlhD4C2 binding sites (sections B, D2/D3, and E2/E3) are required for fliA promoter activity, β-galactosidase activity was measured in a population containing derivative fliA promoter-lacZ fusions with randomly mutated bases in sections B1 and B2. The population of degenerated sequence of 5 bp was 45 (1,024), and the number of samples screened in sections B1 and B2 was, respectively, 29 and 28, which was 3% of the population. As is shown in Fig. 4A and C, the screened samples covered a range of activities from high to low that was considered representative of the whole population. The results indicated that As at positions 12 and 15, in sections B1 and B2, respectively, are positively correlated with high fliA transcription, while other wild-type bases in these sections are widely distributed throughout the sequences in all samples (Fig. 4A and C). The frequency at which a particular base appeared at a specific position in the top 1/4 and top 1/2 activity groups was analyzed, and the mean activity was calculated based on these subsets of data (see Materials and Methods). Since every base in the sequence of each section contributes to the activity; the mean activity contributed by an individual base at each position can be calculated. As shown in Fig. 4B, no specific base appeared 100% of the time at any single position in the top 1/2 activity group of section B1. However, in the top 1/4 activity group, an A at position 12 was found 75% of the time, and it possessed the highest average activity (32%; 2.3-fold above the average of all samples) (Fig. 4B, lower panel). When bases in section B2 were examined, a 100% frequency of occurrence of an A at position 15 was found in both high-activity groups (100% probability in both), with a notable mean activity (65%; 2.1-fold above the average of all samples) (Fig. 4D). Similarly, an A at position 13, an A at position 16, and a C at position 18 each had notable mean activities, but the frequencies of appearance of each of these bases were not distinguishable from the background.
Similar results occurred when sections D2, D3, and E2 were examined (Fig. 5 and 6). As shown in Fig. 5A and C and 6A, a T at position 34 (section D2), an A and a T at positions 36 and 37 (section D3), respectively, and an A and a T at positions 44 and 45 (section E2), respectively, were each positively correlated with high fliA transcription. T34 occurred with 83% frequency in the top 1/4 high activity group and produced significant activity (2.7-fold above the average of all samples in section D2) (Fig. 5B, top and bottom panels). A36 and T37 occurred at 75% and 80% frequencies, respectively, in the top 1/4 high activity group and had significant activities (2.5-fold and 3.3-fold above the averages of all samples, respectively, in section D3) (Fig. 5D, top and bottom panels). A44 and T45 both had significant activities in section E2 (2.6-fold and 4.2-fold above the averages of all samples, respectively), and T45 occurred at 100% frequency in both the top 1/2 and the 1/4 high activity groups (Fig. 6B).
Fig. 5.
Activities of the fliA promoter with sequence alterations in sections D2 and D3. (A and B) Section D2; (C and D) section D3. (A and C) The percent activity (normalized to the wild type, set as 100%) is listed in decreasing order, with the corresponding nucleotide sequence. The wild-type sequence is shown in bold uppercase letters, and the mutant sequence is shown in italic lowercase letters. NC, negative control (possesses complementary mutations in section D2 [positions 33 to 35] and section D3 [positions 36 to 39], respectively). The promoter activity was followed by measuring β-galactosidase activity in three independent experiments. Error bars represent standard deviations of the means. (B and D) The frequency of each base at each position in the top 1/4 (upper graphs) and the top 1/2 (middle graphs) high activity groups and the average activity contributed by each base at each position (bottom graphs). In the bottom graphs of panels B and D, the x axis crosses the y axis at the mean value of the activity of all samples for sections D2 and D3, respectively.
Section E3 (Fig. 6C) exhibited a different pattern of base selection from the other sections. First, the mean activity decreased at a lower rate and most of the samples retained substantial activity, and second, the wild-type bases at all three positions spread out evenly among the screened samples (Fig. 6C). These observations suggest that there is no predominant position in the E3 group at positions 46 to 48, despite the evidence from other methods (Fig. 2) that indicates its importance. The lack of one or more predominant bases was also observed in the frequency and the mean activity data (Fig. 6D). However, a striking observation was made when the frequencies of appearance of purines versus pyrimidines (frequency of appearance among the sample numbers in the group) at these three positions were calculated: the presence of purine (A or G) was dominant at positions 47 and 48 and correlated with high activity. A similar preference for either purines or pyrimidines at position 46 was not as obvious (Table 1).
Table 1.
Frequencies of purines and pyrimidines appearing at positions 46, 47, and 48
| Activity group | Frequency (%) of basea at position: |
|||||
|---|---|---|---|---|---|---|
| 46 |
47 |
48 |
||||
| Ra | Ya | R | Y | R | Y | |
| Top 1/4 | 29 | 71 | 100 | 0 | 100 | 0 |
| Top 1/2 | 50 | 50 | 100 | 0 | 79 | 21 |
| Total | 54 | 46 | 68 | 32 | 64 | 36 |
R, purine; Y, pyrimidine.
Specificity of the important bases for FlhD4C2-activated fliA promoter activity.
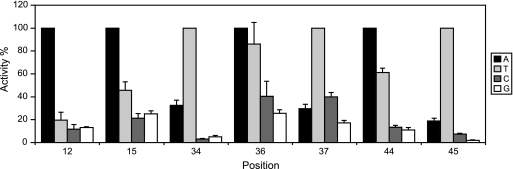
To measure the specificity of the sequence identified from the promoter activity screening (Fig. 4 to 6), single mutations were introduced directly at positions 12, 15, 34, 36, 37, 44, and 45 to change the sequence to each of the other three bases. Most of the mutations reduced the activity to 40% or less, but there were two exceptions: A→T36 and A→T44 (Fig. 7). These changes resulted in 80% and 60% mean activity, respectively. Taken as a whole, the results demonstrated that A12, A15, T34, T37, and T45 are highly specific for FlhD4C2-dependent fliA promoter activity and that positions 36 and 44 are A/T specific. Significantly, these results showed that these seven bases form an asymmetric FlhD4C2 site on fliA, making this a unique feature of this regulatory protein complex.
Fig. 7.
Activity of mutations at A12, A15, T34, A36, T37, A44, and T45 in the fliA promoter. β-Galactosidase activities of samples with single mutations at position 12, 15, 34, 36, 37, 44, and 45 were measured in three independent experiments. Error bars represent standard deviations of the means. The percentage of activity of each sample (normalized to the wild type) is shown. The 100% activity level was given to the wild type at each position.
DISCUSSION
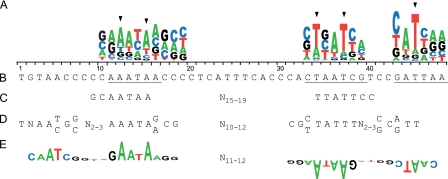
In this study, we refined the FlhD4C2 consensus sequence based on the functional activity of each respective base comprising the footprint on the fliA promoter. The data demonstrated that only 7 bases are required for FlhD4C2 binding and, surprisingly, the binding site is asymmetric. We used the fliA promoter, a flagellar class II promoter bearing a determined FlhD4C2-protected sequence, as an experimental model and found that only 3/5 of the footprint region in this promoter was required for FlhD4C2 binding. More significantly, the results from a SAAB selection further showed that FlhD4C2 binds to A/T base pairs, and only A12, A15, T34, A36, T37, A44, and T45 are required, with A12, A15, T34, T37, and T45 being highly specific for the function of the FlhD4C2 complex. The resulting consensus sequence resulting from these results is ANNAN18TN(A/T)TN6(A/T)T.
The consensus sequence proposed is not the first for the FlhD4C2 binding site. Other groups have also proposed consensus sequences for FlhD4C2 recognition, but what makes the current results noteworthy is that we used a single FlhDC-regulated promoter, fliA, and experimentally determined which bases were important for recognition and function of the protein complex. In earlier reports, Ikebe et al. proposed a weakly conserved, imperfectly inverted sequence motif, GCAATAAN15–19TTATTCC, for FlhD4C2 binding to flagellar class II promoters of S. Typhimurium (21). These binding half-sites are similar but not homologous to the sequences of sections B and D in the fliA promoter of E. coli identified herein (Fig. 8), and furthermore, our results showed that the sequence adjacent to the downstream half-site found by Ikebe et al. (our section E) is also required for FlhD4C2 function. In another study, Claret and Hughes proposed a large imperfectly palindromic motif as the FlhD4C2 binding consensus sequence, of which each half-site is a weakly conserved inverted repeat, TNAA(C/T)G(C/G)N2–3AAATA(A/G)CG, separated by a 10- to 12-nucleotide spacer (Fig. 8) (13). Although this sequence is present in all E. coli flagellar class II promoters (56), our results indicated that only 7 bases of it are required for FlhD4C2 function. Recently, Wozniak and Hughes combined in silico and genetics tools to construct a sequence for flagellar class II (FlhD4C2-specific) promoters. Our results agreed with their findings that bases equivalent to A12, A15, T34, T37, A44, and T45 in the present study are highly preferred in the sequence alignments and have strong impact on the fliA promoter of S. Typhimurium (Fig. 8) (66). The significant difference is that the approach used by Wozniak and Hughes relied on the conservation of nucleotides backed up with genetic mutagenesis, while our approach selected directly and without bias for base changes that improve the function (DNA binding and transcription), thereby emphasizing function rather than nucleotide sequence conservation.
Fig. 8.
The FlhD4C2 binding sequence. (A) Positional dependence of nucleotides required for FlhD4C2-dependent fliA promoter activity. The height of each sequence logo corresponds to the average activity contributed by each base at each position. Arrowheads indicate the bases with high specificity to fliA promoter activity. (B) The sequence of the FlhD4C2 footprint in the E. coli fliA promoter (32). Underlined bases represent those selected from SAAB. (C) The consensus sequence of class II flagellar promoters from S. Typhimurium (21). (D) The consensus FlhD4C2 binding sequence proposed from informatic alignment of 12 class II promoters from E. coli (fliA, flhB, and fliL), S. Typhimurium (fliA, flhB, fliL, flgA, fliE, and fliF), and P. mirabilis (flhB, fliA, and flgA-flgB) (13). (E) The FlhD4C2 consensus binding sequence (inverted half-sites separated by 11 to 12 bp) as proposed by Wozniak and Hughes. The heights of logos represent the degree of conservation of a given base at a given position within alignment, i.e., a larger letter indicates greater conservation of that nucleotide (66).
The current results revealed a surprising nature of FlhD4C2: it binds to an asymmetric site defined by 7 bases in the fliA promoter, which does not require the first 10 bp (section A) of the FlhD4C2 footprint sequence. In this context, it is noteworthy that FlhD4C2 produces a large footprint (∼50 bp) and bends the DNA that it binds to by 111° (7, 63). In most bacterial transcriptional regulatory proteins, a large DNA footprint (>30 bp) usually results from binding of multiple identical subunits, which in turn cause the target DNA to bend (6, 8, 34, 52). Examples are regulatory proteins in the LysR family (34, 48), H-NS (29, 61), and Lrp (41). The size of such regulatory proteins is relatively small (LysR family, ∼30 kDa; H-NS, 16 kDa; Lrp, 15 kDa) compared to the length of their protected DNA fragments. Importantly, binding sites with large DNA footprints frequently result from multiple symmetric contacts. There are exceptions, e.g., IHF is a special case that causes asymmetric DNA binding (14, 68).
IHF, a heteromeric regulator in E. coli, shares similar characteristics with FlhD4C2 in both its structure and DNA binding. IHF (i) folds into a symmetric heteromeric complex, (ii) protects large DNA fragments (32 to 48 bp) from DNase I footprinting with DNA bending, and (iii) has an asymmetric consensus binding sequence (i.e., WATCAANNNNTTR, located at the 3′-half of its footprint region) (14, 18, 68). IHF is a heterodimer consisting of two homologous (30% similarity) subunits, IHF-α and IHF-β (44). Despite the differences in their primary sequences, the architectures of IHF-α and IHF-β are very similar, and they form a symmetric heterodimer complex. The IHF-DNA crystal structure indicates that the large footprint is due to DNA wrapping around the regulator with two sharp bends in minor grooves. However, the structure shows that, in fact, the DNA binding residues of IHF contact mostly the phosphodiester backbone rather than bases of the DNA. This implies that IHF recognizes DNA conformation (specifically, the minor groove) rather than the sequence specificity of the DNA (44). While the crystal structure of FlhD4C2 has not been completely determined, the current data suggest that the FlhD4C2 complex has dyad symmetry (7, 63). The lack of data about FlhD4C2 structure makes understanding how the DNA binding sites fit into the geometry of the FlhD4C2 complex difficult. Therefore, it remains unknown how a symmetrically folded FlhD4C2 complex recognizes an asymmetric binding site.
Prior to the current study, much of what we knew about the FlhD4C2 binding site was derived from bioinformatic approaches, e.g., sequence alignments, frequency statistics, and matrix searches of genomes that were applied to identify a consensus binding sequence. Although conserved sequences have been found in homologous class II flagellar promoters from different genomes, the former attempts to identify the FlhDC binding sequence lacked biological significance and activity, as was determined in the current study. The current results revealed that bases in the consensus sequence are not equally important to FlhDC activity, in contrast to informatically derived results that assumed the frequency of finding a nucleotide within sequence entries from different databases reflected the physiological importance of that nucleotide. Thus, the current results provide important new experimental data that further define the FlhD4C2 binding site consensus sequence and its physiologically relevant bases. The currently defined sequence may serve as the primary elements for FlhD4C2 regulation, and the remaining nucleotides in FlhD4C2 regulatory promoters may work to fine-tune transcription.
Supplementary Material
ACKNOWLEDGMENTS
We thank Alan McLachlan for suggesting we divide the binding site into sections and for his advice on the binding competition assay. We also thank Andres Campos for assistance with FlhD4C2 complex purification, along with helpful discussions and comments on the manuscript, and Peggy O'Neill for critical reading of the manuscript.
This work was supported by a National Institutes of Health award (GM 062044) to P.M. and a National Science Foundation award (MCB-0919820) to R.B.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 17 June 2011.
REFERENCES
- 1. Aldridge C., et al. 2010. The interaction dynamics of a negative feedback loop regulates flagellar number in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 78:1416–1430 [DOI] [PubMed] [Google Scholar]
- 2. Alekshun M. N., Levy S. B., Mealy T. R., Seaton B. A., Head J. F. 2001. The crystal structure of MarR, a regulator of multiple antibiotic resistance, at 2.3 Å resolution. Nat. Struct. Biol. 8:710–714 [DOI] [PubMed] [Google Scholar]
- 3. Barker C. S., Pruss B. M., Matsumura P. 2004. Increased motility of Escherichia coli by insertion sequence element integration into the regulatory region of the flhD operon. J. Bacteriol. 186:7529–7537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bell C. E., Lewis M. 2001. The Lac repressor: a second generation of structural and functional studies. Curr. Opin. Struct. Biol. 11:19–25 [DOI] [PubMed] [Google Scholar]
- 5. Bertin P., et al. 1994. The H-NS protein is involved in the biogenesis of flagella in Escherichia coli. J. Bacteriol. 176:5537–5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brinkman A. B., Ettema T. J., de Vos W. M., van der Oost J. 2003. The Lrp family of transcriptional regulators. Mol. Microbiol. 48:287–294 [DOI] [PubMed] [Google Scholar]
- 7. Campos A., Matsumura P. 2001. Extensive alanine scanning reveals protein-protein and protein-DNA interaction surfaces in the global regulator FlhD from Escherichia coli. Mol. Microbiol. 39:581–594 [DOI] [PubMed] [Google Scholar]
- 8. Ceschini S., et al. 2000. Multimeric self-assembly equilibria involving the histone-like protein H-NS. A thermodynamic study. J. Biol. Chem. 275:729–734 [DOI] [PubMed] [Google Scholar]
- 9. Chadsey M. S., Hughes K. T. 2001. A multipartite interaction between Salmonella transcription factor σ28 and its anti-sigma factor FlgM: implications for σ28 holoenzyme destabilization through stepwise binding. J. Mol. Biol. 306:915–929 [DOI] [PubMed] [Google Scholar]
- 10. Chadsey M. S., Karlinsey J. E., Hughes K. T. 1998. The flagellar anti-sigma factor FlgM actively dissociates Salmonella typhimurium σ28 RNA polymerase holoenzyme. Genes Dev. 12:3123–3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cicero M. P., Alexander K. A., Kreuzer K. N. 1998. The MotA transcriptional activator of bacteriophage T4 binds to its specific DNA site as a monomer. Biochemistry 37:4977–4984 [DOI] [PubMed] [Google Scholar]
- 12. Claret L., Hughes C. 2000. Functions of the subunits in the FlhD2C2 transcriptional master regulator of bacterial flagellum biogenesis and swarming. J. Mol. Biol. 303:467–478 [DOI] [PubMed] [Google Scholar]
- 13. Claret L., Hughes C. 2002. Interaction of the atypical prokaryotic transcription activator FlhD2C2 with early promoters of the flagellar gene hierarchy. J. Mol. Biol. 321:185–199 [DOI] [PubMed] [Google Scholar]
- 14. Craig N. L., Nash H. A. 1984. E. coli integration host factor binds to specific sites in DNA. Cell 39:707–716 [DOI] [PubMed] [Google Scholar]
- 15. Fang F. C., Rimsky S. 2008. New insights into transcriptional regulation by H-NS. Curr. Opin. Microbiol. 11:113–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Francez-Charlot A., et al. 2003. RcsCDB His-Asp phosphorelay system negatively regulates the flhDC operon in Escherichia coli. Mol. Microbiol. 49:823–832 [DOI] [PubMed] [Google Scholar]
- 17. Gallegos M. T., Schleif R., Bairoch A., Hofmann K., Ramos J. L. 1997. Arac/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61:393–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Goodrich J. A., Schwartz M. L., McClure W. R. 1990. Searching for and predicting the activity of sites for DNA binding proteins: compilation and analysis of the binding sites for Escherichia coli integration host factor (IHF). Nucleic Acids Res. 18:4993–5000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hughes K. T., Gillen K. L., Semon M. J., Karlinsey J. E. 1993. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science 262:1277–1280 [DOI] [PubMed] [Google Scholar]
- 20. Ikebe T., Iyoda S., Kutsukake K. 1999. Promoter analysis of the class 2 flagellar operons of Salmonella. Genes Genet. Syst. 74:179–183 [DOI] [PubMed] [Google Scholar]
- 21. Ikebe T., Iyoda S., Kutsukake K. 1999. Structure and expression of the fliA operon of Salmonella typhimurium. Microbiology 145:1389–1396 [DOI] [PubMed] [Google Scholar]
- 22. Joachimiak A., Kelley R. L., Gunsalus R. P., Yanofsky C., Sigler P. B. 1983. Purification and characterization of trp aporepressor. Proc. Natl. Acad. Sci. U. S. A. 80:668–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ko M., Park C. 2000. H-NS-dependent regulation of flagellar synthesis is mediated by a LysR family protein. J. Bacteriol. 182:4670–4672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Komeda Y. 1982. Fusions of flagellar operons to lactose genes on a mu lac bacteriophage. J. Bacteriol. 150:16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Komeda Y., Kutsukake K., Iino T. 1980. Definition of additional flagellar genes in Escherichia coli K12. Genetics 94:277–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kutsukake K., Ide N. 1995. Transcriptional analysis of the flgK and fliD operons of Salmonella typhimurium which encode flagellar hook-associated proteins. Mol. Gen. Genet. 247:275–281 [DOI] [PubMed] [Google Scholar]
- 27. Kutsukake K., Ohya Y., Iino T. 1990. Transcriptional analysis of the flagellar regulon of Salmonella typhimurium. J. Bacteriol. 172:741–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lehnen D., et al. 2002. LrhA as a new transcriptional key regulator of flagella, motility and chemotaxis genes in Escherichia coli. Mol. Microbiol. 45:521–532 [DOI] [PubMed] [Google Scholar]
- 29. Lithgow J. K., Haider F., Roberts I. S., Green J. 2007. Alternate SlyA and H-NS nucleoprotein complexes control hlyE expression in Escherichia coli K-12. Mol. Microbiol. 66:685–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu X., Fujita N., Ishihama A., Matsumura P. 1995. The C-terminal region of the alpha subunit of Escherichia coli RNA polymerase is required for transcriptional activation of the flagellar level II operons by the FlhD/FlhC complex. J. Bacteriol. 177:5186–5188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu X., Matsumura P. 1996. Differential regulation of multiple overlapping promoters in flagellar class II operons in Escherichia coli. Mol. Microbiol. 21:613–620 [DOI] [PubMed] [Google Scholar]
- 32. Liu X., Matsumura P. 1994. The FlhD/FlhC complex, a transcriptional activator of the Escherichia coli flagellar class II operons. J. Bacteriol. 176:7345–7351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Macnab R. M. 1996. Flagella and motility, p. 123–145 In Neidhardt F. C., et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed American Society for Microbiology, Washington, DC [Google Scholar]
- 34. Maddocks S. E., Oyston P. C. 2008. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology 154:3609–3623 [DOI] [PubMed] [Google Scholar]
- 35. Miller J. H. 1992. A short Course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 36. Notka F., Linde H. J., Dankesreiter A., Niller H. H., Lehn N. 2002. A C-terminal 18 amino acid deletion in MarR in a clinical isolate of Escherichia coli reduces MarR binding properties and increases the MIC of ciprofloxacin. J. Antimicrob. Chemother. 49:41–47 [DOI] [PubMed] [Google Scholar]
- 37. Ohnishi K., Kutsukake K., Suzuki H., Iino T. 1990. Gene fliA encodes an alternative sigma factor specific for flagellar operons in Salmonella typhimurium. Mol. Gen. Genet. 221:139–147 [DOI] [PubMed] [Google Scholar]
- 38. Pruss B. M., et al. 2003. FlhD/FlhC is a regulator of anaerobic respiration and the Entner-Doudoroff pathway through induction of the methyl-accepting chemotaxis protein Aer. J. Bacteriol. 185:534–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pruss B. M., Liu X., Hendrickson W., Matsumura P. 2001. FlhD/FlhC-regulated promoters analyzed by gene array and lacZ gene fusions. FEMS Microbiol. Lett. 197:91–97 [DOI] [PubMed] [Google Scholar]
- 40. Ptashne M., et al. 1980. How the lambda repressor and cro work. Cell 19:1–11 [DOI] [PubMed] [Google Scholar]
- 41. Pul U., Wurm R., Wagner R. 2007. The role of LRP and H-NS in transcription regulation: involvement of synergism, allostery and macromolecular crowding. J. Mol. Biol. 366:900–915 [DOI] [PubMed] [Google Scholar]
- 42. Raman N., Black P. N., DiRusso C. C. 1997. Characterization of the fatty acid-responsive transcription factor FadR. Biochemical and genetic analyses of the native conformation and functional domains. J. Biol. Chem. 272:30645–30650 [DOI] [PubMed] [Google Scholar]
- 43. Reitzer L. J., Magasanik B. 1983. Isolation of the nitrogen assimilation regulator NR(I), the product of the glnG gene of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 80:5554–5558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rice P. A., Yang S., Mizuuchi K., Nash H. A. 1996. Crystal structure of an IHF-DNA complex: a protein-induced DNA U-turn. Cell 87:1295–1306 [DOI] [PubMed] [Google Scholar]
- 45. Saini S., Brown J. D., Aldridge P. D., Rao C. V. 2008. FliZ is a posttranslational activator of FlhD4C2-dependent flagellar gene expression. J. Bacteriol. 190:4979–4988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Saini S., et al. 2010. FliZ induces a kinetic switch in flagellar gene expression. J. Bacteriol. 192:6477–6481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sambrook J., Russell D. W. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 48. Schell M. A. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47:597–626 [DOI] [PubMed] [Google Scholar]
- 49. Schevitz R. W., Otwinowski Z., Joachimiak A., Lawson C. L., Sigler P. B. 1985. The three-dimensional structure of trp repressor. Nature 317:782–786 [DOI] [PubMed] [Google Scholar]
- 50. Shin S., Park C. 1995. Modulation of flagellar expression in Escherichia coli by acetyl phosphate and the osmoregulator OmpR. J. Bacteriol. 177:4696–4702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Simons R. W., Houman F., Kleckner N. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85–96 [DOI] [PubMed] [Google Scholar]
- 52. Smyth C. P., et al. 2000. Oligomerization of the chromatin-structuring protein H-NS. Mol. Microbiol. 36:962–972 [DOI] [PubMed] [Google Scholar]
- 53. Soutourina O., et al. 1999. Multiple control of flagellum biosynthesis in Escherichia coli: role of H-NS protein and the cyclic AMP-catabolite activator protein complex in transcription of the flhDC master operon. J. Bacteriol. 181:7500–7508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Soutourina O. A., Bertin P. N. 2003. Regulation cascade of flagellar expression in Gram-negative bacteria. FEMS Microbiol. Rev. 27:505–523 [DOI] [PubMed] [Google Scholar]
- 55. Sperandio V., Torres A. G., Kaper J. B. 2002. Quorum sensing Escherichia coli regulators B and C (QseBC): a novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in E. coli. Mol. Microbiol. 43:809–821 [DOI] [PubMed] [Google Scholar]
- 56. Stafford G. P., Ogi T., Hughes C. 2005. Binding and transcriptional activation of non-flagellar genes by the Escherichia coli flagellar master regulator FlhD2C2. Microbiology 151:1779–1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Stout V., Torres-Cabassa A., Maurizi M. R., Gutnick D., Gottesman S. 1991. RcsA, an unstable positive regulator of capsular polysaccharide synthesis. J. Bacteriol. 173:1738–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Takaya A., Matsui M., Tomoyasu T., Kaya M., Yamamoto T. 2006. The DnaK chaperone machinery converts the native FlhD2C2 hetero-tetramer into a functional transcriptional regulator of flagellar regulon expression in Salmonella. Mol. Microbiol. 59:1327–1340 [DOI] [PubMed] [Google Scholar]
- 59. Tomoyasu T., Takaya A., Isogai E., Yamamoto T. 2003. Turnover of FlhD and FlhC, master regulator proteins for Salmonella flagellum biogenesis, by the ATP-dependent ClpXP protease. Mol. Microbiol. 48:443–452 [DOI] [PubMed] [Google Scholar]
- 60. van Aalten D. M., DiRusso C. C., Knudsen J. 2001. The structural basis of acyl coenzyme A-dependent regulation of the transcription factor FadR. EMBO J. 20:2041–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. van Ulsen P., Hillebrand M., Zulianello L., van de Putte P., Goosen N. 1996. Integration host factor alleviates the H-NS-mediated repression of the early promoter of bacteriophage Mu. Mol. Microbiol. 21:567–578 [DOI] [PubMed] [Google Scholar]
- 62. Wada T., et al. 2011. EAL domain protein YdiV acts as an anti-FlhD4C2 factor responsible for nutritional control of the flagellar regulon in Salmonella enterica Serovar Typhimurium. J. Bacteriol. 193:1600–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wang S., Fleming R. T., Westbrook E. M., Matsumura P., McKay D. B. 2006. Structure of the Escherichia coli FlhDC complex, a prokaryotic heteromeric regulator of transcription. J. Mol. Biol. 355:798–808 [DOI] [PubMed] [Google Scholar]
- 64. Weber I. T., Steitz T. A. 1987. Structure of a complex of catabolite gene activator protein and cyclic AMP refined at 2.5 Å resolution. J. Mol. Biol. 198:311–326 [DOI] [PubMed] [Google Scholar]
- 65. Wei B. L., et al. 2001. Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol. Microbiol. 40:245–256 [DOI] [PubMed] [Google Scholar]
- 66. Wozniak C. E., Hughes K. T. 2008. Genetic dissection of the consensus sequence for the class 2 and class 3 flagellar promoters. J. Mol. Biol. 379:936–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yamamoto S., Kutsukake K. 2006. FliT acts as an anti-FlhD2C2 factor in the transcriptional control of the flagellar regulon in Salmonella enterica serovar typhimurium. J. Bacteriol. 188:6703–6708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yang C. C., Nash H. A. 1989. The interaction of E. coli IHF protein with its specific binding sites. Cell 57:869–880 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.