Abstract
Bacillus subtilis 168 is resistant to phenolic acids by expression of an inducible enzyme, the phenolic acid decarboxylase (PadC), that decarboxylates these acids into less toxic vinyl derivatives. In the phenolic acid stress response (PASR), the repressor of padC, PadR, is inactivated by these acids. Inactivation of PadR is followed by a strong expression of padC. To elucidate the functional interaction between PadR and the padC promoter, we performed (i) footprinting assays to identify the region protected by PadR, (ii) electrophoretic mobility shift assays (EMSAs) with a modified padC promoter protected region to determine the interacting sequences, and (iii) random mutagenesis of padR to identify amino acid residues essential for the function of PadR. We identified an important consensus dyad sequence called IR1-2 (ATGT-8N-ACAT) overlapping a second dyad element (GTGT-8N-ACAT) that we named dIR1-2bis. The entire dIR1-2bis/IR1-2 sequence permits binding of two PadR dimers in EMSAs, which may be observed for bacteria grown under noninduced conditions where the padC promoter is completely repressed. Three groups of modified PadRs giving a PASR phenotype were characterized in vivo. The DNA sequences of certain mutant padR alleles indicate that important residues are all located in the region containing the coiled-coil leucine zipper domain that is involved in dimerization. These substitutions reduce the affinity of PadR binding to the padC promoter. Of particular interest are residue L128, located at the center of the putative coiled-coil leucine zipper domain, and residue E97, which is conserved among all PadRs.
INTRODUCTION
Phenolic acids (also termed substituted hydroxycinnamic acids) are naturally abundant plant compounds with important roles as lignin-related aromatic acids. These acids can be released by cinnamonyl esterase activities, which are expressed by various microorganisms (10, 12, 27) and in their free form induce a specific chemical stress response in microorganisms. Certain bacteria, such as the probiotic organism Lactobacillus plantarum (6, 8, 15), Pediococcus pentosaceus (7), and Bacillus subtilis (9, 29, 32), are resistant to the toxicity of phenolic acids, such as ferulic, p-coumaric, and caffeic acids. This resistance is due to the rapid induction of the padA or padC gene, which encodes a phenolic acid decarboxylase (PadA or PadC) that can rapidly degrade these antimicrobial acids into less toxic vinyl derivatives (6). This resistance mechanism is termed the phenolic acid stress response (PASR) (32). In B. subtilis 168, the expression of padC, which is cotranscribed with upstream noncharacterized and supposedly nonfunctional yveFG genes (29, 32) (Fig. 1), is controlled by a negative transcriptional regulator (PadR) that was identified as the first member of what is now a large family of transcriptional regulators (Pflam PF03551) (7, 14). This family, which is subdivided into two distinct subfamilies (17), consists of more than 2,800 entries in GenBank for completed genomes or running sequencing projects. PF03551 belongs to the gluconate operon repressor (GntR) superfamily, whose members possess an alpha C-terminal core. To date, the function is known for only a few members of the PadR family, which have been shown to play a major role in the biology of their host bacteria. Among these members, (i) AphA from Vibrio cholerae is a quorum sensing-regulated activator that initiates the virulence cascade and is a repressor of penicillin amidase activity (pva gene) (18, 19, 20, 23), (ii) LadR from Listeria monocytogenes negatively regulates the expression of the multidrug efflux pump MdrL (17), (iii) LstR is required for effective thermal resistance (35), and (iv) LmrR from Lactococcus lactis regulates the production of LmrCD, a major multidrug ABC transporter (1, 25). Crystal structures of two PadR-like proteins, AphA (11) and Pex (5), revealed a protein structure containing a conserved N-terminal winged helix-turn-helix (WHTH) that acts as the DNA-binding motif (4). This protein architecture is similar to that of the repressor MarR, which controls antibiotic resistance (2), and further shows the existence of a highly divergent C-terminal domain involved in dimerization. Since to our knowledge the existence of a putative PASR has not been investigated in these species, the biochemical characteristics of initially described PadRs have not been studied. In the PASR, PadR binds to the padC promoter to repress the expression of padC in the absence of phenolic acids (32), but the site of interaction and amino acid residues involved in the functionality of PadR are unknown.
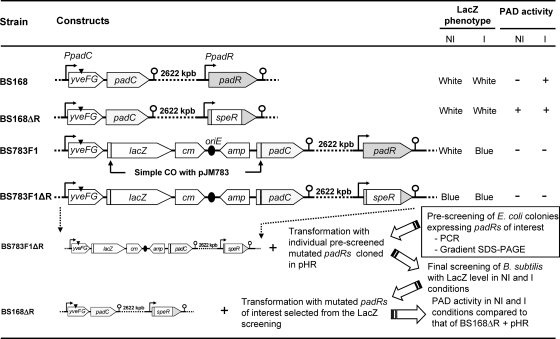
Fig. 1.
Construction and phenotypes of B. subtilis 168 mutant strains. This includes the strategy used to screen for mutant padR genes obtained by complementation of a padR mutant strain (BS783F1ΔR). The black triangle shown above yveFG indicates the stop codon (32). NI, noninduced; I, induced by 1 mM ferulic acid. An absence of PAD activity in the induced fusion strain 783F1 results from the absence of the promoter region upstream of the padC gene.
In this work, we identify for the first time the necessary sequences of the padC promoter involved in the interaction with PadR. We also identify single amino acid substitutions that modify the function of PadR. Results from electrophoretic mobility shift assays (EMSAs) with native and modified padC promoters and modified PadR were supported by in vivo experiments using wild-type B. subtilis 168 and a padR mutant strain.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli, B. subtilis 168, and corresponding mutant strains were grown aerobically in Erlenmeyer flasks on a rotary shaker in Luria-Bertani (LB) medium at 37°C. For selection and growth, antibiotics were used at the following concentrations: erythromycin, 100 μg/ml for E. coli and 5 μg/ml for B. subtilis 168; chloramphenicol, 5 μg/ml for B. subtilis 168; ampicillin, 200 μg/ml; and kanamycin, 50 μg/ml for E. coli.
Table 1.
Plasmids and bacterial strains
| Plasmid or strain | Genotype and/or relevant feature(s) | Source or reference |
|---|---|---|
| Plasmids | ||
| pET28a+ | Kanr; vector for overexpression of His-tagged proteins using T7 bacteriophage promoter | Novagen |
| pER | pET28a+ containing padR between BspHI and XhoI sites to overproduce PadR with His6 tag | 32 |
| pERM (1-16) | pET28a+ containing modified padR genes | This work (Fig. 5B) |
| pJM783 | Ampr Cmr; integrative vector used to construct the lacZ transcriptional fusion F1 | 30 |
| pHT315 | Eryr Ampr | 3 |
| pHR | pHT315 containing padR between XbaI and PstI sites | This work (Fig. 4A) |
| pHRM (1-16) | pHR containing modified padR genes between BseRI and PstI sites | This work (Fig. 4A) |
| Strains | ||
| B. subtilis strains | ||
| 168 | trpC2 | Institut Pasteur, France |
| 168 ΔpadR | trpC2 Specr ΔpadR mutant of BS168 | 32 |
| 783F1 | trpC2 Ampr Cmr; carries the F1 padC::lacZ fusion | 32 |
| 783F1 ΔpadR | trpC2 Specr ΔpadR Ampr Cmr; contains the F1 padC::lacZ fusion | This work (Fig. 1) |
| 783F1 ΔpadR/pHRM | trpC2 Specr ΔpadR Ampr Cmr; contains the F1 padC::lacZ fusion and pHRM plasmid | This work (Fig. 1) |
| 168 ΔpadR/pHRM | trpC2 Specr ΔpadR mutant containing pHRM plasmid | This work (Fig. 4B) |
| E. coli strains | ||
| TG1 | supE hsdΔ5thi Δ(lac-proAB) F′ traD36 proAB+lacIqlacZΔM15 | Invitrogen |
| TG1/pHR | Ampr; carries plasmid pHR | This work (Fig. 4A) |
| TG1/pHRM | Ampr; carries plasmid pHRM | This work (Fig. 4A) |
| BL21(DE3) Star | pThsdSB(rB− mB−) gal dcm (DE3) | Invitrogen |
| BL21/pER | BL21(DE3) Star carrying plasmid pER | This work (Fig. 5B) |
| BL21/pERM (1-16) | BL21(DE3) Star carrying plasmid pERM | This work (Fig. 5B) |
DNA extraction, PCR amplification, sequencing, bacterial transformation, and bioinformatic analysis.
Standard molecular procedures were used as described by Sambrook et al. (31). Genomic DNA was extracted as described previously (32). PCR amplifications were performed in 50-μl reaction mixtures, using 0.1 unit of Platinum high-fidelity Taq DNA polymerase (Invitrogen) in a thermocycler (Bio-Rad) with the primers (Eurogentec) listed in Table 2. PCR products and digested DNA fragments were purified using either a QIAquick PCR purification kit or a QIAgel agarose gel extraction kit (Qiagen). E. coli was transformed by electroporation as described by Dower et al. (13). B. subtilis 168 was transformed with linear plasmid DNA or chromosomal DNA by using a two-step nutrient downshift as previously described (28). DNA sequences (Cogenic) were analyzed using Bio-Edit software.
Table 2.
Primers used for this study
| Primer use and name | Sequence (5′→3′)a | Site created/modified |
|---|---|---|
| Cloning of padR into pET28a+ | ||
| BSR1 | GACTCATGAGAGTATTAAAATACGCC | BspHI |
| BSR2 | GCTCTCGAGATCCTTATCTATCATAG | XhoI |
| Cloning of padR into pHT315 | ||
| BSR3 | TACGTCTAGAGACAGGATTATGTACTGACT | XbaI |
| BSR4 | AAGCTGCAGGATCGACATTGAA | PstI |
| Random mutagenesis of padR by error-prone PCR | ||
| BSR5 | ATGCTGCAGATTATCGCTAACGGTGCC | PstI |
| BSR101 | ATGAGAGTATTAAAATACGCC | BseRI (native) |
| Production of padC::lacZ fusions | ||
| BSDF1 | CCAGAATTCACGGCAAGTCAGCAAGCCGT | EcoRI |
| BSDFR | TCAGGATCCGATAAAGTTTTCCATCTTACAC | BamHI |
| Sequencing of padR mutants | ||
| BSR6 | TCGGATACCTTCTGACAA | |
| Probes for DNA binding | ||
| BSD1 | CAAAGCTAGCTTCAGACAAGG | |
| BSD2 | CACTTTAACACCATTGCAG | |
| BSD4 | ATGTAACTATTTACATGTTCAC | |
| BSD5 | GCAATGGTGTTAAAGTGAACATGT | |
| BSD5IR1 (forward) | GCAATGGTGTTAAAGTGAACΔAAATAGTTACATGATTTTTTC | ΔIR1 (ATGT) |
| BSD5IR2 (forward) | GCAATGGTGTTAAAGTGAACATGTAAATAGTTΔGATTTTTTCTGAAGGTGAGGTG | ΔIR2 (ACAT) |
| BSD5IR12 (forward) | GCAATGGTGTTAAAGTGAACΔAAATAGTTΔGATTTTTTC | ΔIR1, ΔIR2 |
| BSD6 | ACATGTTCACTTTAACACCATTGC | |
| BSD8 (reverse) | GAATCATCTCAGTCCCAGGCTTG |
Underlined nucleotides correspond to restriction sites for the enzymes given in the right column.
DNase I footprinting assays.
DNase I protection assays were performed as previously described (22). Briefly, a 234-bp probe (P1), encompassing positions −97 to +137 relative to the transcription start site of the padC promoter (see Fig. 3), was produced by PCR amplification with primer BSD1 or BSD8 (Table 2) previously labeled with T4 polynucleotide kinase (Invitrogen) in the presence of [γ-32P]ATP (Perkin-Elmer). The probe-PadR protein binding reactions were conducted under the conditions for EMSA (see below), with 0.1 to 10 nM purified PadR, as previously described (32). DNase I digestions were optimized. Briefly, 100-μl reaction mixtures included radioactive DNA probes incubated for 1 min at 37°C with 0.5 U of DNase I. DNase I digestion was stopped by addition of a solution containing 200 mM NaCl, 20 mM EDTA, 10 g/liter SDS, and 10 mg/ml salmon sperm DNA. DNA products were purified by ethanol precipitation and resuspended in 10 μl of TE (10 mM Tris-HCl, 1 mM EDTA) buffer. Samples were mixed 1:1 with gel sequencing loading buffer and then resolved in a denaturing 6% (wt/vol) polyacrylamide-urea electrophoresis gel and visualized by autoradiography. Sequencing reactions were performed with corresponding primers for each strand, using a CycleReader DNA sequencing kit (MBI-Fermentas). These reactions were run adjacent to their respective DNase I footprinting samples.
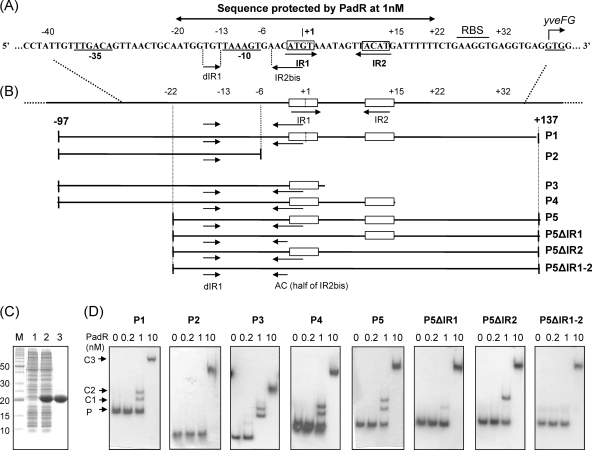
Fig. 3.
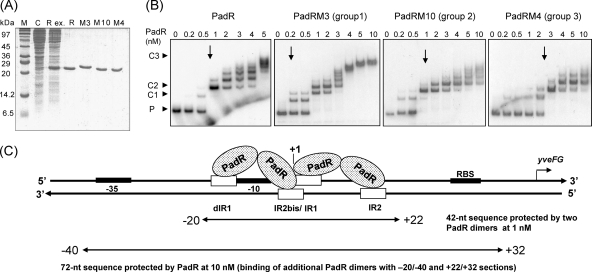
EMSA for PadR with native and modified padC promoters. (A) padC promoter DNA sequence indicating nt positions from the +1 transcription start site, −35 and −10 boxes, and IR1-IR2 and dIR1-2bis, the inverted repeat sequences that form the dIR1-2bis/IR1-2 pattern. The start codon (GTG) of yveFG, a nonfunctional gene cotranscribed with padC (32), is indicated. (B) Map of native (P1) and modified (P2 to P5 and P5 derivatives) padC promoters used in EMSA. (C) SDS-PAGE of protein extracts containing purified PadR used in EMSA. Lane M, molecular mass standard; lanes 1 and 2, crude extracts from E. coli BL21 and E. coli BL21/pER, respectively; lane 3, purified PadR. (D) EMSAs with native and modified padC promoters (0.2 nM). PadR was used in EMSAs at concentrations ranging from 0 to 10 nM. A concentration of 1 nM PadR gave specific binding (C1 and C2 complexes), while 10 nM PadR gave unspecific binding (C3 complex). P, unbound probe. For panel D, all PAGE experiments were run under the same conditions, and the fact that the probes and the complexes in the different panels are not aligned horizontally results from the different sizes of the probes (P1, 234 bp; P2, 91 bp; P3, 102 bp; P4, 112 bp; P5, 159 bp; P5ΔIR1 and P5ΔIR2, 155 bp; and P5ΔIR1-2, 151 bp).
EMSA.
The 234-bp padC promoter DNA probe P1 used for DNase I footprinting assays was also used for EMSA. Matching experiments were performed with modified padC promoters generated with the appropriate primers listed in Table 2. PCR products were purified using a QIAquick PCR purification kit. Standard EMSA was performed as described previously (32). Briefly, purified native or modified PadR was incubated for 20 min at 28°C in 15 μl binding buffer containing 0.2 nM DNA probe, 10 mM Tris-HCl, pH 7.8, 5% (vol/vol) glycerol, 0.2 mM EDTA, 2.5 mM MgCl2, 50 mM KCl, 2 mM dithiothreitol (DTT), 2.5 μg/ml bovine serum albumin (BSA), and 2.5 μg/ml salmon sperm DNA.
The samples were resolved in 5% (wt/vol) polyacrylamide gels and analyzed by autoradiography. To test the effects of phenolic acids on PadR-padC promoter DNA binding, PadR was preincubated with 1 mM p-coumaric, ferulic, or o-coumaric acid in 15 μl of binding buffer for 5 min at room temperature. o-Coumaric acid, an isomer of p-coumaric acid unable to induce the expression of padC (32), was used as a negative control at 1 mM. The probe was then added to the above mixture and incubated for 20 min at 28°C before loading onto a polyacrylamide gel.
Production of modified padC promoter probes.
All probes used in this study were PCR amplified from B. subtilis 168 genomic DNA by use of three different primers: an internal primer carrying the desired modification and two external primers (Table 2). The external primers were previously labeled with T4 polynucleotide kinase (Invitrogen) in the presence of [γ-32P]ATP (Perkin-Elmer). The PCR consisted of two steps. First, the internal primer was used together with the appropriate external primer to introduce the modification, and second, the resulting product was used as a primer together with the second external primer (Table 2) to obtain the final product. Nonlabeled corresponding modified sequences were verified by sequencing (Cogenic).
Random mutagenesis of padR by error-prone PCR and screening for modified padR genes of interest.
Random mutagenesis of padR was carried out in vitro with a GeneMorph II random mutagenesis kit (Stratagene). PCR amplification was performed in a 50-μl reaction mixture containing 100 ng of genomic DNA extracted from B. subtilis 168 cells, 2.5 U of Mutazyme II DNA polymerase, a 40 mM deoxynucleoside triphosphate (dNTP) mix, and 250 ng/μl (each) of BSR5 and BSR101 primers (Table 2). The reaction conditions were designed to achieve 0 to 4 mutations per kb of DNA. To avoid any modification to the unknown promoter region of padR and to facilitate cloning, the native BseRI restriction site was used with the PstI restriction site to replace the native padR gene with the mutated padR genes. Error-prone PCR products were purified, digested by the BseRI and PstI enzymes, and cloned into plasmid pHR. After ligation, the reaction mixtures containing the resulting pHRM plasmids were transformed into E. coli TG1. About 5,000 colonies were obtained. A three-step procedure was used to efficiently screen padR mutant genes of interest (Fig. 1). Two hundred colonies were analyzed individually by PCR amplification to verify the presence of a padR amplicon with the expected size. Positive colonies were used in a second step consisting of gradient SDS-polyacrylamide gels to select cultures producing a modified PadR protein (PadRM). pHRM plasmids from these cultures were transformed individually into the B. subtilis 783F1 ΔpadR reporter strain (see below) to screen modified padR genes of interest with the LacZ phenotype. Modified padR genes of interest were sequenced to identify mutated nucleotides. These plasmids were then transformed into a B. subtilis ΔpadR strain. Comparisons of strains with plasmids to control strains were made by analysis of the LacZ phenotype and phenolic acid decarboxylase (PAD) activities.
Construction of B. subtilis 168 reporter strains to screen modified padR genes of interest.
B. subtilis strain 783F1 ΔpadR (Fig. 1) was obtained by transforming strain 783F1 with chromosomal DNA from a B. subtilis ΔpadR strain (32) and selecting LacZ+ colonies (blue) on LB agar plates containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) supplemented with spectinomycin. B. subtilis 783F1 ΔpadR did not display PAD activity with (inducing [I] conditions) or without (noninducing [NI] conditions) ferulic acid. The absence of PAD activity is due to the separation of the functional padC promoter from the padC gene by integration of a pJM783 vector containing a truncated, nonfunctional padC promoter (32). The constitutive LacZ+ phenotype results from the deletion of padR by replacement with the speR cassette. The wild-type padR gene in plasmid pHR was replaced by different modified padR genes to generate the pHRM plasmids. These plasmids were transformed into strain 783F1 ΔpadR (Fig. 1). The transformation mixture was poured onto LB agar plates supplemented with erythromycin and X-Gal and with (I conditions) or without (NI conditions) ferulic acid. The selection of colonies was based on LacZ reporter phenotypes, which we were able to classify into five discernible intensities ranging in color from blue to white. As a reference, we used strain 783F1 ΔpadR. For instance, 783F1 ΔpadR yields a deep-blue LacZ reporter phenotype, or LacZ 5; this strain complemented with native padR on LB medium without ferulic acid yields a white LacZ phenotype, or LacZ 0; and this strain on LB medium with ferulic acid yields a lighter blue color, or LacZ 4. LacZ level 2 and 3 colonies were colonies initially formed with at least two contiguous bacteria. However, after reisolation, these displayed LacZ levels 0 and 2 and then levels 2 and 4, respectively.
Overexpression and purification of native PadRs and PadRMs.
Expression and purification of PadR proteins were performed as described previously (32). The PadR and modified PadR (PadRM) coding regions were PCR amplified with the BSR1 and BSR2 primers to replace the TAA stop codon with an XhoI restriction site. The amplified DNA fragment was cloned into the pET28a+ vector by NcoI and XhoI digestion, generating the plasmids pER and pERM. Expression of pER and pERM in E. coli BL21(DE3) cells induced with IPTG (isopropyl-β-d-thiogalactopyranoside) produced PadR fusion proteins containing a His6 tag at the C-terminal end. Recombinant PadR and PadRM proteins were purified from E. coli BL21(DE3) cell extracts by elution from a 0.5-ml nickel-nitrilotriacetic acid (Ni-NTA) column (Qiagen) according to the manufacturer's guidelines. Purified protein samples were stored at −25°C.
Cell extracts, assay for PAD activity, and protein gel electrophoresis.
Wild-type and mutant B. subtilis 168 strains grown in LB medium were harvested and disrupted using a Z Plus series cell disrupter (Constant system) (15). PAD activity in cell extracts was measured by monitoring the kinetics of absorption peaks by UV spectrophotometry (6). Protein concentrations were determined using a Bio-Rad protein assay kit with BSA as the standard. Protein extracts were routinely resolved in a 12% (wt/vol) denaturing SDS-PAGE gel as previously described (14). To verify PadRM protein expression in crude extracts from recombinant E. coli strains, 8 to 15% (wt/vol) gradient SDS-PAGE (18-cm by 18-cm gel) was run for 5 h at 110 V.
RESULTS
Identification of PadR binding site for the padC promoter.
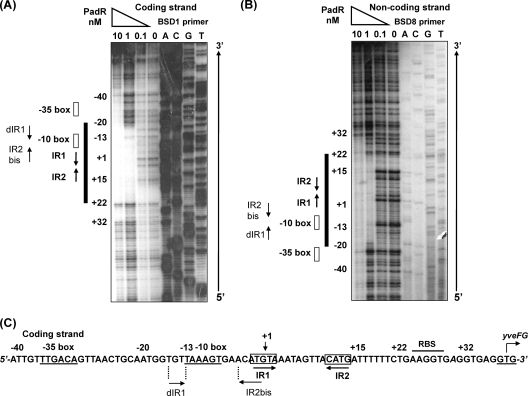
In the absence of phenolic acid (nonstress, noninduced conditions), PadR binds to the padC (yveFG-padC) promoter, thereby repressing its expression. Our previous studies showed a consensus inverted repeat DNA sequence (ATGT-8N-ACAT) in the padC (yveFG-padC) promoter that is found in the promoters of padC or padA genes in bacteria displaying the PASR (32) and has a suspected interaction with PadR (32). To identify the binding sites of PadR for the padC promoter, DNase I footprinting experiments were performed. For these assays, protection of a radioactively labeled 234-bp fragment extending from nucleotides (nt) −97 to +137 relative to the +1 start of transcription of the padC promoter was observed in the presence of different concentrations of purified PadR (Fig. 2). Consistent results were obtained for the two DNA strands (Fig. 2A and B). A PadR concentration of 0.1 nM did not result in detectable protection; however, we did observe DNA protection at a PadR concentration of 1 nM. At this concentration of PadR, previous gel shift assays demonstrated two specific shifted bands for the probe in EMSAs (32). This 42-bp region includes the previously supposed consensus 16-bp inverted repeat ATGT-8N-ACAT (IR1-2) (Fig. 2C). In addition to the IR1-2 sequence, there exists a degenerate IR1-2 sequence (GTGT-8N-ACAT), named dIR1-2bis, that contains an ACAT pattern overlapping the AT nucleotides (ATGT) in the IR1 region (Fig. 2C). Like the IR1 region, the dIR1 region is followed by a stretch of 3 adenosines. The dIR1-2bis/IR1-2 pattern (which includes nucleotides at positions −15 to +15 relative to the padC +1 start of transcription) resided within the region protected by PadR at 1 nM (from position −20 to position +22) (Fig. 3A). When 10 nM PadR was used, a concentration where a single shift was found by EMSA (32), this protected region could be extended upstream to encompass the −10 and −35 promoter boxes and to roughly 20 nt downstream of the dIR1-2bis region (Fig. 2C).
Fig. 2.
padC (yveFG-padC) promoter DNA protection with PadR. Purified PadR was incubated with a 234-bp single-end-labeled padC promoter DNA probe. The reaction mixtures contained a 5′-[γ-32P]ATP-labeled coding strand (BSD1 primer) (A) or noncoding strand (BSD8 primer) (B) that was digested by DNase I. The reaction mixtures were run adjacent to corresponding DNA sequencing reaction mixtures (ACGT). (C) padC promoter coding strand sequence, with nt positions, +1 transcription start site, and −35 and −10 boxes for the padC promoter operon indicated, as well as the GTG start codon of yveFG (32). RBS, ribosome binding site. IR1, IR2, dIR1, and IR2bis are inverted repeat sequences for PadR.
EMSA with PadR and mutated padC promoters.
Since we showed that the sequence containing the dIR1-2bis/IR1-2 pattern resided within the region protected by PadR (Fig. 2), our next objective was to determine the functional specificity of this binding. PCR was used to generate a probe containing the entire 234-bp native promoter and seven other mutated promoter probes (Fig. 3B). These probes were then tested for binding to PadR (Fig. 3C and D). With the P1 fragment, which contains the entire promoter region and 1 nM PadR, two specific complexes, C1 and C2, were found, accounting for two operator sites. Nevertheless, C3, a high-molecular-mass complex, was found with this probe at 10 nM PadR. This binding may be accounted for by additional lower-affinity or less specific bindings of PadR proteins to the region extending upstream of the −35 box (Fig. 2). This finding corroborates the results obtained after footprinting assays with 10 nM PadR, where we found that the protected region extends to the −40 position (Fig. 2). This suggests the existence of a possible additional, less specific binding site(s) of PadR, even though the IR1-IR2 pattern (or a degenerate version) is not observed within this sequence.
The presence of two inverted repeat dyads in the promoter and of two complexes, C1 and C2, in the presence of 1 nM PadR corresponds to a modified overlapping binding model, as described for the Fur repressor binding site involved in iron uptake in E. coli (21). These findings are reinforced by the fact that we demonstrated that in B. subtilis 168, PadR easily forms dimers at low concentrations of the cross-linking reagent glutaraldehyde (0.1% [vol/vol]) (see the supplemental material). Moreover, the 42-nt length of the padC promoter protected region with 1 nM PadR is sufficient to bind two PadR dimers. This is supported by many studies that report DNA regions protected by one dimer repressor that extend over 19 to 27 nt (16, 21). Binding of PadR at 1 nM to the P1, P4, and P5 probes, which contained the entire dIR1-2/IR1-2 sequence, produced C1 and C2 complexes. Using the P3 fragment, which contained only dIR1-2bis/IR1 and not the IR2 sequence, C1 and C2 complexes were detected, suggesting that this region is sufficient to bind to two PadR molecules. The low-intensity C1 band observed with the P5ΔIR1 probe probably resulted from binding to the IR2 sequence, since this C1 complex was not observed with the P5ΔIR1-2 probe.
Moreover, the C2 complex did not form when the sequences encompassing IR1 and half of IR2bis (AC) (probes P2, P5ΔIR1, and P5ΔIR1-2) were absent. Experiments with the P2 probe indicated that the dIR1 (GTGT) sequence, in addition to any upstream sequences, was not capable of binding to PadR at 1 nM but produced the C3 complex in the presence of 10 nM PadR. On the other hand, C1 and C2 complexes were present with the P4 fragment (the probe containing dIR1-2bis/IR1-2), indicating that the sequence downstream of IR2 is not necessary for C1 and C2 complex formation. However, this sequence might produce a lower-affinity or less specific binding participating in the formation of the less specific C3 complex, since it was partially protected at 10 nM PadR. Taken together, these findings indicate that the dIR1-2bis/IR1 sequence is mainly responsible for the interaction with PadR. A C2 complex was detected with the P5ΔIR2 probe, indicating that this probe is able to bind in vitro to two dimers of PadR, as does the P1 probe containing the IR2 sequence. It is likely that the IR1 sequence is sufficient to bind in vitro to a PadR dimer. Further comparison of these EMSA results with those obtained with P5ΔIR1 and P5ΔIR1-2 fragments indicates that IR2 (probe P5ΔIR1) contributes to PadR binding. Indeed, use of IR2 produced a significant amount of C1 complex, corresponding to binding of one PadR dimer. The C1 complex was not observed when the P5ΔIR1-2 probe was incubated with IR2-null sequences.
Identification of single amino acid substitutions modifying the functionality of PadR.
Random mutagenesis of padR and construction of an appropriate recipient and reporter B. subtilis strain, strain 783F1 ΔpadR, were performed to identify which amino acid residue(s) is important for PadR structure-function. To screen modified padR genes of interest, we employed the strategy schematized in Fig. 1. Two hundred colonies, from 5,000 originally isolated after transformation of E. coli, were analyzed individually by PCR amplification to verify the presence of a padR amplicon of the expected size. Seventy amplicons showed the expected size. Protein extracts from these clones were analyzed by SDS-PAGE. Among them, only 32 strains exhibited a PadR protein with the expected size. Plasmid DNAs from these strains were transformed into strain 783F1 ΔpadR. Transformed colonies were selected on LB–X-Gal medium with (I conditions) and without (NI conditions) ferulic acid by comparing their LacZ phenotype color intensities (from white [LacZ 0] to deep blue [LacZ 5]) to those of control strains 783F1 and 783F1 ΔpadR as described above. Mutant colonies with a LacZ phenotype different from that of 783F1 ΔpadR complemented with the native padR gene on LB medium, with or without ferulic acid, were selected. The corresponding mutant padR genes were then sequenced and translated to identify amino acid residue changes. Eleven putative mutants initially displayed a LacZ color phenotype of 3 to 5. During their propagation, many changed their LacZ color phenotype to level 4, which was obtained with wild-type PadR. Further sequence investigations showed that these putative mutants contained native padR, and therefore these clones were excluded from further analysis. Two mutants displaying a stable LacZ level of 5 under I or NI conditions had double mutations, at residues 46 to 128 and residues 61 to 128. These were not analyzed further. The remaining 19 mutants (M clones) were classified into three groups (Fig. 4B). Group 1 consisted of five mutants giving white colonies (LacZ 0) under NI conditions and a LacZ level of 2 under I conditions (which was lower than that of control strain 783F1). Sequencing of padR revealed modifications at residues 113, 138, and 149 for clones M15, M3, and M5, respectively. Group 2 contained six PadRMs giving white colonies (LacZ 0) under NI conditions and blue colonies (LacZ 5) under I conditions. Sequencing of padR revealed modifications at residues 145, 63, and 100 for clones M8, M10, and M13, respectively. Group 3 contained eight PadRMs with a LacZ level of 5 under both NI and I conditions, which is equivalent to the level for 783F1 ΔpadR (without padR). Sequencing revealed mutations at residues 85, 97, and 128 for clones M14, M16, and M4, respectively. Substitutions of nine different residues were found to modify the function of PadR.
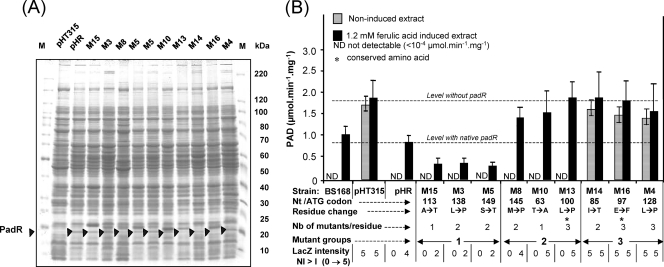
Fig. 4.
(A) SDS-PAGE analysis of modified PadR (black triangles) expressed in E. coli strains. (B) PAD activities (μmol min−1 mg−1; data are averages for three experiments) in mutant strains complemented with native padR (pHR) or modified padR genes (pHRM). Cultures were induced or not with 1 mM ferulic acid for 20 min before harvesting and disrupting the cells. B. subtilis 168, wild-type strain; pHT315, 168 ΔpadR mutant transformed with pHT315 (without padR); pHR, 168 ΔpadR mutant complemented with native padR; M15 to M4, 168 ΔpadR mutants complemented with modified padR. These mutants form three groups based on PAD activity compared to that with native padR (pHR).
The plasmids containing the 9 modified padR genes were transformed into the recipient ΔpadR strain with a functional padC gene (Fig. 1) in order to compare PAD activities (under I and NI conditions) with those observed for native PadR (Fig. 4B). For all PadRMs, the LacZ phenotype correlated well with the PAD activity. Group 1 mutants displayed no detectable activity under NI conditions and a PAD activity 2- to 2.5-fold lower than that of the pHR strain under I conditions. These results show that modified PadRMs maintained their repressor function but were not completely inactivated by ferulic acid. Repressor function was also retained in group 2 mutants, but PAD activity in induced cultures was increased 2-fold compared to that observed in controls (Fig. 4B). Finally, group 3 mutants under NI conditions displayed a PAD activity fairly identical to that obtained under I conditions. These findings indicate that PadRMs of this group are not functional. We then verified that the His-tagged native PadR protein, whose gene was cloned into vector pHT315 and expressed in strain 168 ΔpadR, displayed the same function as the native nontagged PadR protein (data not shown), demonstrating that the phenotypes of mutants were the consequences of the single amino acid changes. Interesting results were obtained for two lysine-to-proline substitutions, in M4 (position 128) and M3 (position 138), both of which affect the coiled-coil motif. The M4 substitution (which affects the middle of the predicted coiled-coil motif) produced a nonfunctional PadRM4, while the M3 mutation (which affects the tail end of the motif) led to a reduction of PAD activity only under induced conditions. These results demonstrate that replacement of a lysine residue at the center of a coiled-coil motif by a proline leads to a high degree of disorder of the secondary structure of PadR, while the same substitution in the extremity of the coiled-coil motif has less of an influence on PadR protein structure and function.
Interaction of PadRMs with native padC promoters.
To further characterize PadR-padC sites of interaction, one modified padR gene representative of each of our three groups was cloned into the vector pET28a+ and expressed in E. coli BL21 to produce the corresponding PadRMs (Fig. 5A). The levels of interaction between these purified PadRMs at different concentrations were analyzed by EMSA with the padC promoter probes at 0.2 nM (Fig. 5B). The padC promoter required concentrations ranging from 0.5 (not tested in the experiments shown in Fig. 3D) to 1 nM native PadR to yield complexes C1 and C2. C1 and C2 correspond to binding of the probe to one and two PadR dimers, respectively. Increasing the concentration of PadR from 2 to 5 nM produced new complexes with higher molecular weights that could correspond to additional bound PadR dimers, probably to the upstream region of the −35 box and possibly to the downstream region of IR2. This point was previously discussed in the analysis of the C3 complex and was incorporated into our model (Fig. 5C). In this case, the C3 complex corresponds to the saturation of the probe by PadR dimers, which was observed when incubation mixtures included 5 nM (Fig. 5B) and 10 nM (Fig. 3D) PadR. For PadRM3 (group 1), C1 and C2 complexes were observed at 0.2 nM, and saturation of the probe was obtained at approximately 4 nM. These results indicate that the PadRM3 group displays a higher affinity for the probe and corroborate the LacZ phenotype and PAD activity of these mutants as shown in Fig. 4B. For PadRM10 (group 2), C1 and C2 complexes were obtained at 0.5 to 1 nM, similar to the results obtained for native PadR. These findings may explain the absence of detectable PAD activity with this group of PadR mutants under NI conditions, identical to the case for native PadR. Nevertheless, increasing the concentration of PadRM10 did not produce complexes of significantly higher molecular mass, and C3 was not detected in the presence of 10 nM PadR. These results reflect a reduced capacity of PadRM10 to bind to the padC promoter and correlate with a higher PAD activity under ferulic acid-induced conditions (Fig. 4B). For PadRM4 (group 3), the C1 complex was detected by EMSA at 0.2 to 0.5 nM PadR, but the C2 complex was detected only when the PadR concentration was higher than 2 nM. Additionally, as observed with PadRM10, increasing the concentration to 10 nM PadRM4 did not produce the C3 complex. Taken together, these findings indicate that the lysine-to-proline substitution at position 128 in PadRM4 reduces the capacity to produce the C2 complex and causes an inability to produce the C3 complex. This substitution, which occurs at the center of the putative coiled-coil motif, may modify the dimer structure and alter the recognition of at least one of the two binding sites of the padC promoter. This correlates with the inability of PadRM4 to repress the expression of padC under noninduced conditions.
Fig. 5.
(A) SDS-PAGE of native (R) and modified (M3, M10, and M4) His6-tagged PadRs produced in E. coli and used in EMSA. M, molecular mass standard; C, control E. coli strain with pET28a+; Rex, crude extract of native PadR. The same types of extracts were produced to purify the native protein and M3, M10, and M4, shown in the corresponding lanes. (B) EMSAs of native and modified PadRs (M3, M10, and M4), representing the three groups based on PAD activity with the native padC promoter probe at 0.2 nM (see Fig. 4). P, promoter DNA probe without PadR. C1 and C2 indicate specific complexes formed between the DNA probe and PadR. C3 results from additional low-affinity or less specific binding. Arrows indicate the concentrations at which the specific complexes C1 and C2 are observed. (C) Model for PadR dimer binding to padC (yveFG) promoter. The protected sequence was deduced from the results presented in Fig. 2. One dimer binds to dIR1 and the complementary sequence of IR2bis, and the second dimer binds to IR1 and the complementary sequence of IR2.
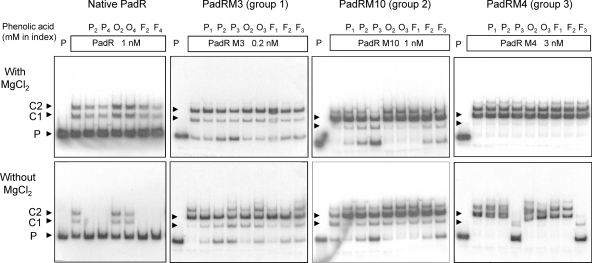
Effects of phenolic acids and MgCl2 on PadRM binding to the padC promoter.
It was previously shown that phenolic acids capable of inducing the PASR were able to abolish PadR binding to the padC promoter (32). In order to test whether single-residue substitutions in PadRMs could modify the response to phenolic acids, EMSAs with native PadR, PadRMs, and the padC promoter were performed in the presence or absence of MgCl2 and with or without preincubation with phenolic acids (Fig. 6), as previously described (32). For an effective comparison of our findings, we used the results obtained with our PadR-padC interaction site studies (Fig. 5) to choose for each PadRM the concentration yielding the specific complexes C1 and C2. Specific emphasis was placed on PadRMs yielding a high concentration of C2, which corresponds to the binding of two PadR dimers with the probe. For native PadR, binding was not altered by phenolic acids in the presence of MgCl2 but was abolished in the absence of MgCl2 with 2 or 4 mM p-coumaric or ferulic acid, two inducers of the PASR (Fig. 6), but not with o-coumaric acid, a noninducer of the PASR. Therefore, the absence of the C1 complex from the relevant lane indicates that the inactivation of PadR by phenolic acid in vitro results in suppression of binding with the two sites formed by the dIR1-2/IR1-2 pattern. Furthermore, o-coumaric acid, an isomer of p-coumaric acid, was not able to abolish the binding. For PadRM3 (group 1) at a concentration of 0.2 nM, we detected only a partial release of the probe with p-coumaric (P2 and P3) and ferulic (F2 and F3) acids in the absence of MgCl2. In addition, we detected no probe release under the same conditions for incubation with o-coumaric acid. The absence of a complete release of the probe in the absence of MgCl2 corroborates our finding that PadRM3 is inactivated by phenolic acids in vivo to a lesser extent than native PadR (Fig. 4B). A similar yet more robust liberation of the probe was noted for PadRM10 (group 2) at 1 nM, which was unexpected because this PadRM is inactivated by phenolic acids to a greater extent in vivo than native PadR. Lastly, PadRM4 (group 3) at 3 nM (the concentration giving a C2 major complex) demonstrated no significant changes in EMSA with addition of phenolic acid in the presence of MgCl2. However, we observed a release of the probe when PadRM4 was incubated with PASR acid inducers (P3 and F3) in the absence of MgCl2. This release was comparable to that observed with native PadR, except for the apparition of a low-intensity band corresponding to the size of the C1 complex. It is noteworthy that the in vivo findings do not correlate strictly with our EMSA results. Nevertheless, our data suggest that the residues mutated in PadRM3 and PadRM10 might be involved in the inactivation mechanism induced by phenolic acids.
Fig. 6.
EMSAs of native and modified His6-tagged PadRs M3, M10, and M4, representative of the three groups based on PAD activity with the native padC promoter at 0.2 nM (see Fig. 3B), in the presence or absence of MgCl2 or phenolic acids. Pn, p-coumaric acid; On, o-coumaric acid (isomer of p-coumaric acid unable to induce the PASR); Fn, ferulic acid; P, padC promoter probe. The concentration of each PadR protein was chosen to obtain the two specific complexes, C1 and C2, with the padC promoter (see Fig. 5).
Comparison of PadR and AphA secondary structures.
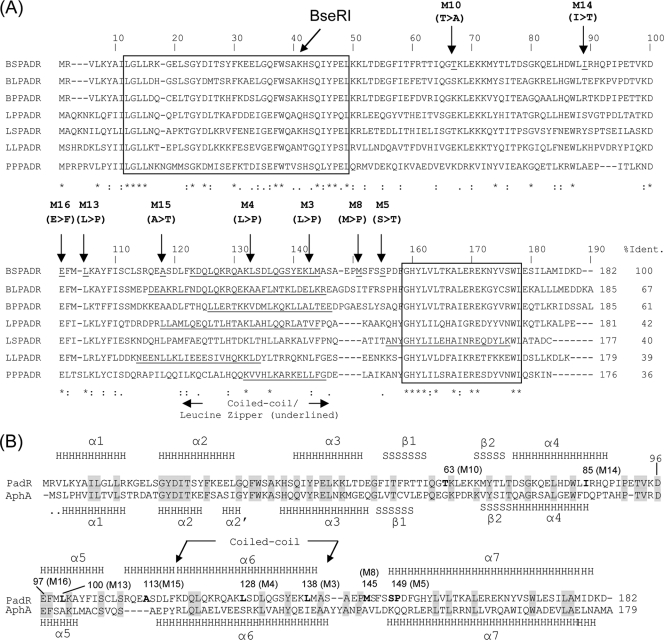
Multiple alignments of seven PadRs involved in the PASR were performed to identify conserved nucleotides, amino acid residues in the putative coiled-coil leucine zipper motif (predicted with COILS software [24]), and the conserved N- and C-terminal sequences (Fig. 7A). The positions of the mutations within this sequence were analyzed. Since we used the native BseRI restriction site at residue 39 for cloning of all mutated padR genes, there was no mutation produced upstream of residue 39. Therefore, except for three mutants (one M10 and two M14 mutants), all other mutants contained mutations from residue 97 (M16) to residue 149 (M5), which encompass the putative coiled-coil leucine zipper motif that is considered to be involved in the dimerization of regulator proteins, notably those belonging to the GntR superfamily, of which PadR is a member. This is in accordance with cross-linking (see the supplemental data) and EMSA results indicating that PadR dimerization is required for its interaction with the padC promoter.
Fig. 7.
(A) Multiple alignment of PadRs displaying the highest identity (% Ident.) to PadR from B. subtilis 168. BS, Bacillus subtilis 168; BL, Bacillus licheniformis; BP, Bacillus pumilus; LP, Lactobacillus plantarum; LS, Lactobacillus sakei; LL, Lactococcus lactis; PP, Pediococcus pentosaceus. Underlined sequences correspond to the putative coiled-coil leucine zipper motif predicted with COILS software (23). Conserved N- and C-terminal sequences are boxed. Vertical arrows indicate the positions of modified amino acid residues. The mutation in M16, which is a representative of the group 3 PadRs that are incapable of repressing padC expression, is at the site 97 (E) conserved residue. *, conserved residue. (B) Alignment and prediction of secondary structures (PSIPRED software) for B. subtilis 168 PadR and AphA from Vibrio cholerae (18). Gray boxed letters are conserved residues.
The hypothetical secondary structure of PadR predicted by the PSIPRED program (26) shows high similarities in structural elements with the secondary structure of AphA determined from its crystal structure (Fig. 7B) (11). The PadR hypothetical coiled-coil domain, determined by the use of COILS software (24), spans amino acids 118 to 140 (Fig. 7A).
DISCUSSION
In this work, we characterized the padC promoter region to which PadR binds in B. subtilis 168 when it is not exposed to phenolic acids, and thereby when expression of padC is completely repressed. We showed that the binding region of the padC gene contains a consensus IR1-2 inverted repeat, ATGT-8N-ACAT, and an overlapping upstream degenerate repetitive sequence, dIR1-2bis (gTGT-8N-ACAT). This pattern, named dIR1-2bis/IR1-2, which extends over 42 nt, corresponds to the sole region protected by 1 nM PadR. Footprinting and EMSA analyses revealed an extension of the protected region covering the upstream −35 box and a section from positions +22 to +32 (where +1 refers to the transcription initiation site) that could be responsible for additional low-affinity or less specific binding of PadR dimers with increasing concentrations of PadR from 2 to 10 nM (Fig. 5B and C). These additional binding sites could reinforce repression of the promoter under NI conditions. The length of the protected region with 1 nM PadR allows, depending on the concentration of PadR in EMSAs, the binding of one or two PadR dimers, which produces two complexes in EMSA, i.e., C1 and C2. As mentioned previously, IR1-2 is found in the padC (or padA) promoter in all bacteria displaying the PASR. While the dIR1-2bis sequence is also found in the padA promoter in L. plantarum (14), which does not form an operon with padR, dIR1-2bis is not found in the promoter of the P. pentosaceus padA gene, which does form a padAR operon with padR (7). We speculate that to repress padAR expression in P. pentosaceus, the binding of PadR to the padAR promoter may involve only one PadR dimer and may be reduced compared to that observed in B. subtilis 168 or L. plantarum. For B. subtilis 168 and L. plantarum, it is reasonable to think that under NI conditions in vivo, the binding of two PadR dimers, one with the first dyad, dIR1-2bis, and a second with the second dyad, IR1-2, as well as additional binding in the region covering the −35 box, is responsible for the complete repression of the padA (or padC) promoter. In P. pentosaceus, basal expression of padR is likely necessary under NI conditions to produce PadR and to autoregulate the promoter.
To study whether PadR could be involved in regulating other genes, the consensus IR1-2 sequence was screened in the B. subtilis 168 strain by using PredictRegulon software (34). Bioinformatic analysis revealed an IR1-2 (ATGTaaatagttACAT) sequence present in the promoter regions of several genes from B. subtilis 168, among which the best score was obtained for a sequence in the promoters of the rbsRKDACB ribose operon (ATGTaaatagctACAT) (33), the bioA gene, and two genes (yesO and ydaI) of unknown function (http://genolist.pasteur.fr/SubtiList/) (data not shown). In these promoters, the 8-nt spacer between IR1 and IR2 is fairly conserved. For example, only one substitution is observed in the rbsRKDACB promoter. An intriguing future area of study will be the possible interplay between PadR and the expression of the rbsRKDACB operon, since this operon is involved in the metabolism of ribose, an essential component of RNA and key compounds of other metabolic pathways.
By random mutagenesis of the padR gene, we generated nine different padR mutants of interest and identified crucial sites for PadR function. These sites included residue 104 (L) in M13 from group 2 and residue 97 (E) in M16 from group 3. Both of these residues correspond to highly conserved residues in the PadR protein family (Fig. 7). Our findings suggest that these residues are essential for PadR function. In addition, residue 97 (E), which is also present in the α5 helix of AphA, has an identical location in PadR (Fig. 7B). Surprisingly, substitutions in the last 39 residues, including the most conserved C-terminal box, did not result in an interesting phenotype for our study. Similar findings have been shown for random mutagenesis of AphA from V. cholerae (18). We therefore hypothesize that either this C-terminal conserved region of PadR-like proteins is assimilated into a family signature not involved in the specific function of the PadR-like subfamily proteins or, conversely, mutations in this region cause severe conformational defects, resulting in product instability. Due to the strategy used for cloning padR mutant genes into the native BseRI site, we did not generate a mutant for the first 110 nucleotides of padR. As shown for AphA (11), this section might contain residues responsible for binding to the padC promoter. Future studies will require directed single codon substitutions in the conserved N-terminal domain without altering PadR structure to determine if this section is involved in binding to the padC promoter and then to determine which residues are essential for binding.
Although the overall primary amino acid sequence identity between PadR and AphA is low (28%) compared to that for other PadRs involved in the PASR, the predicted structure of PadR from B. subtilis 168 shares a high degree of resemblance with that of AphA, although at present there is no PASR described for V. cholerae. The DNA-binding domain of AphA, a region from residue 1 to residue 86, forms a WHTH DNA-binding domain of the GntR superfamily of transcriptional regulators, containing over 6,000 members distributed among almost all bacterial species, both eukaryotes and archaea. The GntR-like proteins bind to promoters as dimers, where each monomer recognizes a half-site of 2-fold symmetric DNA sequence. This is in accordance with our finding by EMSA of different sizes of complexes formed with increasing concentrations of PadR. PadR contains the structural elements α1, α2, α3, α4, β1, and β2. The amino acids which have been identified as important for DNA-binding activity of AphA, for example, G18, Y19, G30, H37, Q39, Y41, K63, and K37 (18), are conserved in PadR. The dimerization domain, including amino acids 98 to 179, forms a coiled-coil motif and corresponds to the structural elements α5, α6, and α7. This AphA motif of the dimerization domain is not similar to that of any other members of the PadR protein family (11).
In conclusion, the IR1-8N-IR2 DNA sequence, which corresponds to one of the two dyads formed by the dIR1-2bis/IR1-2 sequence, is present in all padC (or padA) promoters to which PadR binds. It is found in the promoter regions of several other genes in B. subtilis 168 and other bacteria, in particular Lactococcus lactis, Bacillus anthracis, and V. cholerae (aphA gene). Our findings suggest that PadR might modulate the expression of these genes, whose expression might be required directly or indirectly to reduce injury during the PASR. To decipher if PadR is a pleiotropic regulator, further interests of our group include (i) screening the entire genomes of B. subtilis 168 and other species, with or without PASR activity, for native or degenerate versions of dIR1-2bis/IR1-2; (ii) testing the interaction of PadR with modified padC promoters containing single nucleotide modifications in the dIR1-2bis/IR1-2 sequence but also in the whole protected sequence with 10 nM PadR to determine mutations that cause binding participating in repression under NI conditions; and (iii) proteomic studies with wild-type and mutant padR variants of B. subtilis 168 to compare phenolic acid-induced and noninduced proteomes.
Supplementary Material
ACKNOWLEDGMENTS
T. K. C. Nguyen and N. P. Tran were supported by Ph.D. grants from the French Embassy in Vietnam. This work was also partially supported by the Conseil Régional de Bourgogne.
We are grateful to Marta Perego (The Scripps Research Institute, La Jolla, CA) and Didier Lereclus (Institut Pasteur and INRA, France) for generously providing vectors. We thank Christine Rojas for her technical assistance.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 17 June 2011.
REFERENCES
- 1. Agustiandari H., Lubelski J., van den Berg van Saparoea H. B., Kuipers O. P., Driessen A. J. M. 2008. LmrR is a transcriptional repressor of expression of the multidrug ABC transporter LmrCD in Lactococcus lactis. J. Bacteriol. 190:759–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alekshun M. N., Levy S. B., Mealy T. R., Seaton B. A., Head J. F. 2001. The crystal structure of MarR, a regulator of multiple antibiotic resistance, at 2.3 A resolution. Nat. Struct. Biol. 8:710–714 [DOI] [PubMed] [Google Scholar]
- 3. Arantes O., Lereclus D. 1991. Construction of cloning vectors for Bacillus thuringiensis. Gene 108:115–119 [DOI] [PubMed] [Google Scholar]
- 4. Aravind L., Anantharaman V., Balaji S., Babu M. M., Iyer L. M. 2005. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol. Rev. 29:231–262 [DOI] [PubMed] [Google Scholar]
- 5. Arita K., et al. 2007. Structural and biochemical characterization of a Cyanobacterium circadian clock-modifier protein. J. Biol. Chem. 282:1128–1135 [DOI] [PubMed] [Google Scholar]
- 6. Barthelmebs L., Diviès C., Cavin J.-F. 2000. Knockout of the p-coumarate decarboxylase gene from Lactobacillus plantarum reveals the existence of two other inducible enzymatic activities involved in phenolic acid metabolism. Appl. Environ. Microbiol. 66:3368–3375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Barthelmebs L., Lecomte B., Diviès C., Cavin J.-F. 2000. Inducible metabolism of phenolic acids in Pediococcus pentosaceus is encoded by an autoregulated operon which involves a new class of negative transcriptional regulator. J. Bacteriol. 182:6724–6731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cavin J.-F., Barthelmebs L., Diviès C. 1997. Molecular characterization of an inducible p-coumaric acid decarboxylase from Lactobacillus plantarum: gene cloning, transcriptional analysis, overexpression in Escherichia coli, purification, and characterization. Appl. Environ. Microbiol. 63:1939–1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cavin J.-F., Dartois V., Diviès C. 1998. Gene cloning, transcriptional analysis, purification, and characterization of phenolic acid decarboxylase from Bacillus subtilis. Appl. Environ. Microbiol. 64:1466–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Christov L. P., Prior B. A. 1993. Esterases of xylan-degrading microorganisms—production, properties, and significance. Enzyme Microb. Technol. 15:460–475 [DOI] [PubMed] [Google Scholar]
- 11. De Silva R. S., et al. 2005. Crystal structure of the virulence gene activator AphA from Vibrio cholerae reveals it is a novel member of the winged helix transcription factor superfamily. J. Biol. Chem. 280:13779–13783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. de Vries R. P., Faulds C. B., Visser J. 1999. The faeA gene from Aspergillus niger encoding a feruloyl esterase with activity on xylan and pectin is subject to a complex system of regulation. J. Sci. Food Agric. 79:443–446 [Google Scholar]
- 13. Dower W. J., Miller J. F., Ragsdale C. W. 1988. High efficiency transformation of Escherichia coli by high voltage electroporation. Nucleic Acids Res. 16:6127–6145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gury J., Barthelmebs L., Tran N. P., Diviès C., Cavin J.-F. 2004. Cloning, deletion, and characterization of PadR, the transcriptional repressor of the phenolic acid decarboxylase-encoding padA gene of Lactobacillus plantarum. Appl. Environ. Microbiol. 70:2146–2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gury J., et al. 2009. Inactivation of PadR, the repressor of the phenolic acid stress response, by molecular interaction with Usp1, a universal stress protein from Lactobacillus plantarum, in Escherichia coli. Appl. Environ. Microbiol. 75:5273–5283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hirooka K., et al. 2007. Dual regulation of the Bacillus subtilis regulon comprising the lmrAB and yxaGH operons and yxaF gene by two transcriptional repressors, LmrA and YxaF, in response to flavonoids. J. Bacteriol. 189:5170–5182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huillet E., Velge P., Vallaeys T., Pardon P. 2006. LadR, a new PadR-related transcriptional regulator from Listeria monocytogenes, negatively regulates the expression of the multidrug efflux pump MdrL. FEMS Microbiol. Lett. 254:87–94 [DOI] [PubMed] [Google Scholar]
- 18. Kovacikova G., Lin W., Skorupski K. 2004. Vibrio cholerae AphA uses a novel mechanism for virulence gene activation that involves interaction with the LysR-type regulator AphB at the tcpPH promoter. Mol. Microbiol. 53:129–142 [DOI] [PubMed] [Google Scholar]
- 19. Kovacikova G., Lin W., Skorupski K. 2003. The virulence activator AphA links quorum sensing to pathogenesis and physiology in Vibrio cholerae by repressing the expression of a penicillin amidase gene on the small chromosome. J. Bacteriol. 185:4825–4836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kovacikova G., Skorupski K. 2001. Overlapping binding sites for the virulence gene regulators AphA, AphB and cAMP-CRP at the Vibrio cholerae tcpPH promoter. Mol. Microbiol. 41:393–407 [DOI] [PubMed] [Google Scholar]
- 21. Lavrrar J. L., McIntosh M. A. 2003. Architecture of a Fur binding site: a comparative analysis. J. Bacteriol. 185:2194–2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lin K.-C., Shiuan D. 1995. A simple method for DNaseI footprinting analysis. J. Biochem. Biophys. Methods 30:85–89 [DOI] [PubMed] [Google Scholar]
- 23. Lin W., Kovacikova G., Skorupski K. 2007. The quorum sensing regulator HapR downregulates the expression of the virulence gene transcription factor AphA in Vibrio cholerae by antagonizing Lrp- and VpsR-mediated activation. Mol. Microbiol. 64:953–967 [DOI] [PubMed] [Google Scholar]
- 24. Lupas A., Vandyke M., Stock J. 1991. Predicting coiled coils from protein sequences. Science 252:1162–1164 [DOI] [PubMed] [Google Scholar]
- 25. Madoori P. K., Agustiandari H., Driessen A. J. M., Thunnissen A. 2009. Structure of the transcriptional regulator LmrR and its mechanism of multidrug recognition. EMBO J. 28:156–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McGuffin L. J., Bryson K., Jones D. T. 2000. The PSIPRED protein structure prediction server. Bioinformatics 16:404–405 [DOI] [PubMed] [Google Scholar]
- 27. McSweeney C. S., Dulieu A., Webb R. I., Del Dot T., Blackall L. L. 1999. Isolation and characterization of a Clostridium sp. with cinnamoyl esterase activity and unusual cell envelope ultrastructure. Arch. Microbiol. 172:139–149 [DOI] [PubMed] [Google Scholar]
- 28. Msadek T., et al. 1998. ClpP of Bacillus subtilis is required for competence development, motility, degradative enzyme synthesis, growth at high temperature and sporulation. Mol. Microbiol. 27:899–914 [DOI] [PubMed] [Google Scholar]
- 29. Nguyen V. D., et al. 2007. The proteome and transcriptome analysis of Bacillus subtilis in response to salicylic acid. Proteomics 7:698–710 [DOI] [PubMed] [Google Scholar]
- 30. Perego M. 1993. Integrational vectors for genetic manipulations in Bacillus subtilis, p. 615–624 In Sonensheim A. L., Hoch J. A., Losick R. (ed.), Bacillus subtilis and other Gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, DC [Google Scholar]
- 31. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed., vol. 1 Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 32. Tran N. P., et al. 2008. Phenolic acid-mediated regulation of the padC gene, encoding the phenolic acid decarboxylase of Bacillus subtilis. J. Bacteriol. 190:3213–3224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Woodson K., Devine K. M. 1994. Analysis of a ribose transport operon from Bacillus subtilis. Microbiology 140:1829–1838 [DOI] [PubMed] [Google Scholar]
- 34. Yellaboina S., Seshadri J., Kumar M. S., Ranjan A. 2004. PredictRegulon: a web server for the prediction of the regulatory protein binding sites and operons in prokaryote genomes. Nucleic Acids Res. 32:W318–W320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang C. M., Nietfeldt J., Zhang M., Benson A. K. 2005. Functional consequences of genome evolution in Listeria monocytogenes: the lmo0423 and lmo0422 genes encode sigma(C) and LstR, a lineage II-specific heat shock system. J. Bacteriol. 187:7243–7253 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.