Abstract
HAMP domains are sensory transduction modules that connect input and output domains in diverse signaling proteins from archaea, bacteria, and lower eukaryotes. Here, we employed in vivo disulfide cross-linking to explore the structure of the HAMP domain in the Escherichia coli aerotaxis receptor Aer. Using an Aer HAMP model based on the structure of Archaeoglobus fulgidus Af1503-HAMP, the closest residue pairs at the interface of the HAMP AS-1 and AS-2′ helices were determined and then replaced with cysteines and cross-linked in vivo. Except for a unique discontinuity in AS-2, the data suggest that the Aer HAMP domain forms a parallel four-helix bundle that is similar to the structure of Af1503. The HAMP discontinuity was associated with a segment of AS-2 that was recently shown to interact with the Aer-PAS sensing domain. The four-helix HAMP bundle and its discontinuity were maintained in both the kinase-on and kinase-off states of Aer, although differences in the rates of disulfide formation also indicated the existence of different HAMP conformations in the kinase-on and kinase-off states. In particular, the kinase-on state was accompanied by significantly increased disulfide formation rates at the distal end of the HAMP four-helix bundle. This indicates that HAMP signaling may be associated with a tilting of the AS-1 and AS-2′ helices, which may be the signal that is transmitted to the kinase control region of Aer.
INTRODUCTION
HAMP domains (which are found in histidine kinases, adenylyl cyclases, methyl-accepting chemotaxis proteins, phosphatases [4], and some diguanylate cyclases and phosphodiesterases [12]) are signal transduction modules that transduce diverse input signals into output signals in proteins from archaea, bacteria, and lower eukaryotes (12, 25). Currently, more than 12,000 proteins are predicted to contain HAMP domains, and many of these proteins regulate two-component signaling pathways (SMART [http://smart.embl-heidelberg.de/]). The abundance of these domains, and their conservation across a wide range of species, argues for a strategic role for HAMP domains in signaling. This universal role and their mechanism of action are the focus of extensive current research.
In Escherichia coli, methyl-accepting chemoreceptors like Tsr and Tar each contain two subunits with two transmembrane helices, the second of which links a periplasmic sensing module with cytosolic HAMP and signal output (kinase control) domains. When an attractant molecule binds to the periplasmic sensing domain of these receptors, transmembrane helix 2 (TM2) is displaced ∼2 Å toward the cytoplasm, shifting the conformation of the HAMP domain to the signal-off conformation (reviewed in reference 13). In contrast with Tsr and Tar, the E. coli aerotaxis receptor Aer lacks a periplasmic domain. Aer is anchored in the membrane, but its sensing and HAMP domains are cytosolic (31) (Fig. 1). The cytoplasmic sensor is an N-terminal PAS (Per-ARNT-Sim) (24) domain with a bound flavin adenine dinucleotide (FAD) cofactor that monitors intracellular redox potential (6, 27, 31, 33) (Fig. 1). The cytoplasmic location of the PAS sensing domain permits direct contact with the HAMP domain (Fig. 1), removing the need to signal through the transmembrane segments (2, 9, 35) (Fig. 1). Precise PAS-HAMP signaling mechanisms are not understood, but previous studies have indicated that the Aer HAMP domain is essential for PAS folding, stability, and FAD binding (7, 16, 22). From these data, and the finding that HAMP-C253R is an allele-specific suppressor of PAS-N34D (37), we inferred that the Aer PAS and HAMP domains interact directly. Direct evidence for PAS-HAMP interactions was recently provided by the results of disulfide cross-linking experiments in which it was shown that residues on the PAS β-scaffold can specifically cross-link with residues in the Aer HAMP domain (9).
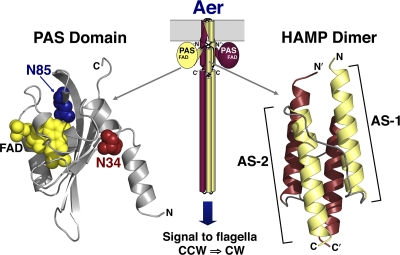
Fig. 1.
Cartoon showing the proposed arrangement of an Aer dimer (center), with helices represented as cylinders, loops represented as lines, and PAS domains represented as circles. The expanded ribbon diagrams show homology models of the Aer PAS domain (left) (modeled based on the coordinates of the Azotobacter vinelandii NifL PAS domain [19]) and the Aer HAMP domain (right) (modeled on the coordinates of the Archaeoglobus fulgidus Af1503 HAMP domain [18]). On the PAS ribbon diagram, spheres are used to show the FAD cofactor (yellow) and the residues where PAS kinase-on lesions were generated for this study (N34 [red] and N85 [blue]). Abbreviations: FAD, flavin adenine dinucleotide; AS, amphipathic sequence; CCW, counterclockwise; CW, clockwise.
HAMP monomers consist of two α-helices (amphipathic sequence 1 [AS-1] and AS-2) separated by a loop. This secondary structure has been demonstrated by disulfide cross-linking studies of the Tar (8) and Aer (36) receptors. The first three-dimensional structure of a HAMP domain was determined by nuclear magnetic resonance (NMR) for the atypical Archaeoglobus fulgidus protein Af1503 (18). This structure is homodimeric and consists of an unusual parallel four-helix coiled-coil arrangement for AS-1 and AS-2, with each connector packed into a groove between helices of the same subunit (18) (Fig. 1). A similar four-helix bundle arrangement was subsequently verified for the Tar HAMP domain by disulfide cross-linking (29) and for the Tsr HAMP domain by genetic analysis (40). More recently, the four-helix bundle structure was also confirmed by crystallography for three sequential HAMP domains from the soluble Pseudomonas aeruginosa receptor Aer-2 (1).
In Af1503, HAMP residues comprising the core of the helical bundle are packed in an unusual x-da (knob-to-knob) arrangement that can be converted in silico into a canonical a-d (knob-into-hole) conformation by a 26° counterrotation of each helix. Hulko et al (18) therefore proposed helix rotation as a signaling mechanism for HAMP domains, the so-called “gearbox” model. Although recent structural studies on Af1503 HAMP mutants lends some support to the gearbox rotation model (14), work on other receptors has led to suggestions of alternative HAMP signaling models with discrete kinase-on and kinase-off conformations, such as the scissors (29) and the combined helical rotation with tilting (1) models. These models share some features of the gearbox model. In contrast, the dynamic HAMP signaling model proposes that the HAMP kinase-off conformation involves the stable packing of the four-helix bundle, while the kinase-on conformation is associated with a more dynamic HAMP bundle (39, 40). Compact and dynamic HAMP states have similarly been shown by electron paramagnetic resonance (EPR) for HtrII from Natronomonas pharaonis (11). In this study, we employed an in vivo disulfide cross-linking approach to address the question of HAMP signaling in Aer. We confirm that the Aer HAMP domain forms a parallel four-helix bundle but with features that may be unique to PAS-HAMP-containing receptors and then demonstrate that the Aer HAMP domain has different conformations in the “on” and “off” signaling states of the receptor.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Wild-type Aer and cysteineless (C-less) Aer (Aer-C193S/C203A/C253A) were expressed from pGH1 (27) and pMB1 (23, 38), respectively. Both plasmids are derived from pTrc99A and express Aer under the control of an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible ptrc promoter. Plasmids in this study were expressed in E. coli strain BT3312, which lacks the two aerotaxis receptors Aer and Tsr (Δaer-1 Δtsr-7021) (28).
In silico modeling.
An Aer HAMP homology model, previously created from the coordinates of the A. fulgidus Af1503 HAMP domain and the sequence of the E. coli Aer HAMP domain, was used for this study (18, 36). The Af1503 and Aer HAMP domains both contain conserved sequence features that are common to canonical HAMP domains, as found in membrane-bound receptors (12). PyMOL (http://pymol.sourceforge.net) was used to view the Aer HAMP model, substitute side chains, and measure β-carbon distances.
Site-directed mutagenesis.
Site-directed mutagenesis was performed on pMB1 and pMB1-derived plasmids according to the instructions of the QuikChange II site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA). Plasmids were introduced into BT3312, and Aer expression was confirmed by Western blotting using anti-Aer2-166 antisera (28). Mutations were confirmed by the sequencing of the entire aer gene.
Aerotaxis phenotypes were determined for each of the Aer mutants by inoculating cells into succinate-minimal soft agar containing 50 μg ml−1 ampicillin, incubating the plates at 30°C for 15 to 20 h, and then observing the colony morphologies (32). To determine clockwise (CW) biases and oxygen responses, cells were grown in tryptone broth and induced with either 200 μM or 1 mM IPTG. Cells were placed into a gas perfusion chamber and monitored when the gas was switched between air and nitrogen, as described previously (27, 32).
In vivo cross-linking.
BT3312 cells expressing each of the Aer-cysteine mutants were grown to mid-log phase in H1 minimal salts medium supplemented with 30 mM succinate, 0.1% (wt/vol) Casamino Acids, and 100 μg ml−1 ampicillin. Cultures were then induced for 3 h with 50 μM IPTG, and steady-state Aer accumulation levels were compared with that of C-less Aer (pMB1). After Western blotting, band intensities were quantified and compared on a UVP BioSpectrum digital imaging system (UVP, Upland, CA). For disulfide-cross-linking experiments, all of the mutants were cultured and induced as described above, with the exception of Aer-V209C/E238C and Aer-E213C/E238C, which had lower steady-state levels and were induced with 100 μM IPTG. Cross-linking was performed at 25°C by exposing whole cells to 600 μM Cu(II)(1,10-phenanthroline)3 (CuPhe) for 10 min, a procedure similar to that described previously (3, 17, 36) but with modifications as described previously (21, 32). For all proximal di-Cys pairs, disulfide bond formation was also determined in time courses over 20 min. Cross-linked products were separated by SDS-PAGE and quantified on the UVP digital imaging system after Western blotting. The percent cross-linking was calculated by dividing the intensity of the cross-linked dimer band by the sum of the intensities of the monomer and dimer bands, multiplied by 100. Aer-V260C (36) and C-less Aer (23) were used as positive and negative cross-linking controls, respectively. Dimer bands were absent for C-less Aer, whereas Aer-V260C routinely formed ∼50% dimers. To compare cross-linking with and without N34D or N85S, statistical analyses were carried out by using a two-tailed Student t test. A P value of <0.05 was considered to be statistically significant.
RESULTS
Study design.
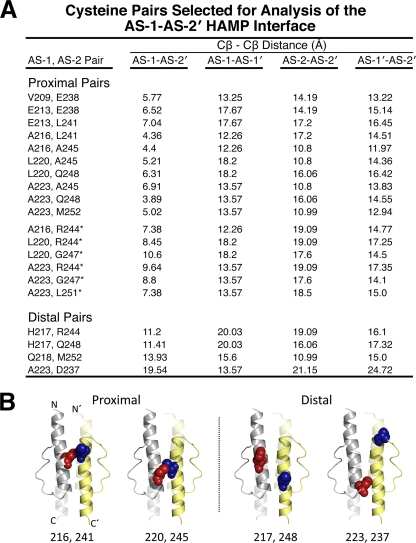
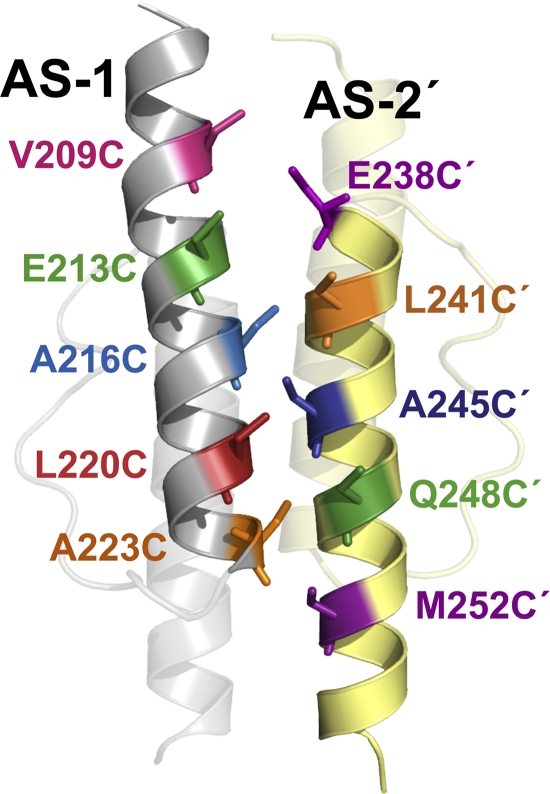
We previously probed the subunit interface of an Aer HAMP dimer by disulfide cross-linking and found that the symmetric (AS-1-AS-1′ and AS-2-AS-2′) dimer interfaces mirrored those of the Af1503 HAMP domain (18, 36). In the current study, we extended that analysis to examine structural relationships between the asymmetric AS-1 and AS-2′ helices and tested the hypothesis that, similar to Af1503, the Aer HAMP domain is a parallel four-helix bundle. For these studies, an Aer HAMP homology model was constructed from the coordinates of the Af1503 HAMP structure (18, 36) and was used to predict the closest contact distances between all AS-1 and AS-2′ residues at the asymmetric packing face (Fig. 2) (29). The goal was to identify AS-1 residues that were not at the symmetric dimer interface of Aer but were instead closer to an AS-2′ residue than to their cognate AS-1′ residue. Proximal AS-1-AS-2′ residue pairs were defined as having AS-1-AS-2′ β-carbon-β-carbon distances of ≤7 Å and AS-1-AS-1′, AS-2-AS-2′, and AS-1′-AS-2′ distances of >10 Å. All 10 residue pairs that satisfied these constraints are listed in Fig. 2A and are mapped onto the Aer HAMP model in Fig. 3. A subset of distal residue pairs (Fig. 2A), defined as having all four possible β-carbon-β-carbon distances of >10 Å, were selected as controls (compare examples of proximal and distal di-Cys pairs in Fig. 2B).
Fig. 2.
Cysteine pairs selected for analysis of the AS-1-AS-2′ HAMP interface. (A) Summary of proximal and distal di-Cys pairs and relevant β-carbon distances. Four β-carbon distances were determined for each cysteine pair on an Aer HAMP model that was generated from the coordinates of the Af1503 HAMP domain (18). Proximal pairs were selected so that AS-1-AS-2′ distances were ≤7 Å, and the three other distances (AS-1-AS-1′, AS-2-AS-2′, and AS-1′-AS-2′) were all >10 Å. Distal di-Cys control pairs had all four β-carbon distances of >10 Å. An additional set of proximal cysteine pairs were selected based on experimental data rather than on distance constraints (indicated by an asterisk) (see the text for details). (B) Examples of proximal and distal AS-1-AS-2′ pairs mapped onto the HAMP model to demonstrate their predicted intersubunit separation. AS-1 substitutions are shown as red spheres, and AS-2′ substitutions are shown as blue spheres.
Fig. 3.
Aer HAMP model showing the nearest AS-1 and AS-2′ residues that were tested in proximal di-Cys pairs (as listed in Fig. 2A). HAMP subunits are yellow or gray, and part of the HAMP domain has been made transparent to highlight AS-1 and AS-2′. All side chains are shown as cysteine replacements.
To map the AS-1-AS-2′ interface using disulfide cross-linking, cysteine residues were serially substituted for each of the proximal and distal residue pairs by site-directed mutagenesis of a cysteineless (C-less) Aer expression vector (pMB1). C-less Aer (Aer-C193S/C203A/C253A) orchestrates aerotaxis in soft agar, although cells expressing C-less Aer have a slower rate of colony spreading than cells expressing wild-type (WT) Aer (Fig. 4A). This is likely caused by the C253A substitution in C-less Aer (15). Larger substitutions at C253 abolish Aer function (C253R) (37), possibly by disrupting HAMP packing or PAS-HAMP contacts. The changes that result from C253A decreased but did not abolish aerotaxis in soft agar (Fig. 4A) or in temporal gas perfusion assays (data not shown).
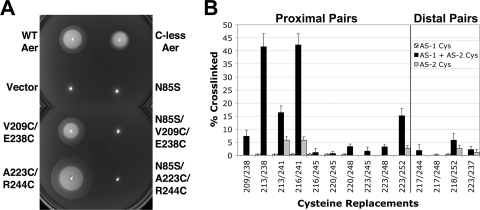
Fig. 4.
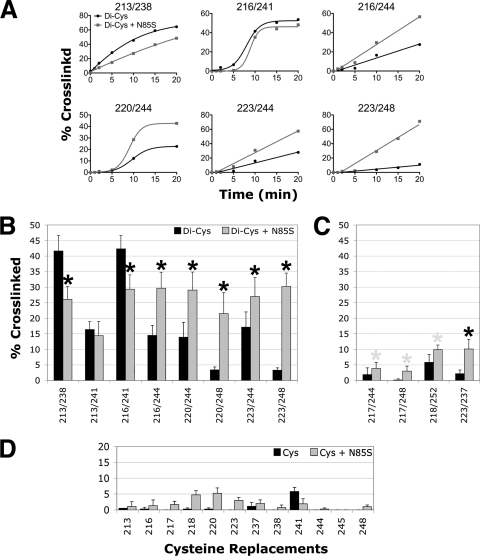
Behavior and in vivo disulfide cross-linking of di-Cys mutants. (A) Influence of HAMP-cysteine and PAS-N85S substitutions on Aer-mediated behavior in succinate-minimal soft agar containing 50 μg ml−1 ampicillin. E. coli BT3312 (aer tsr) mutants were inoculated into soft agar, incubated for 20 h, and compared with positive (WT Aer and C-less Aer [Aer-C193S/C203A/C253A]) and negative (pTrc99A vector) aerotaxis controls. Representative aerotactic (Aer-V209C/E238C) and “superswarming” (Aer-A223C/R244C) colonies are shown. All Aer-cysteine mutants containing N34D or N85S were nonaerotactic (Aer-N85S/V209C/E238C and Aer-N85S/A223C/R244C are shown). (B) Percent disulfide-linked dimer formation for Aer di-Cys receptors (black bars) and corresponding single-Cys controls (hatched and gray bars). Cross-linking was determined after the incubation of whole cells with CuPhe for 10 min at 25°C. Error bars indicate the standard deviations determined from two or more independent experiments.
In E. coli BT3312 (aer tsr), all of the di-Cys Aer mutants exhibited the classic dome shape of an aerotactic colony in succinate minimal soft agar, with an outermost ring of aerotactic bacteria at the lower edge (Fig. 4A) (32). Colony diameters ranged from 68% to 230% of those of wild-type Aer colonies (as expressed from pGH1 in BT3312), with seven of the mutants behaving as “superswarmers” (colony diameters >1.3 times the diameter of wild-type colonies [22]) (see example in Fig. 4A). There was no correlation between the behavior of the mutants in soft agar and the steady-state cellular accumulation levels of the modified Aer proteins, which varied from 54% to 125% of that of C-less Aer. The finding that Aer can signal in the presence of the introduced cysteine pairs suggests that the native structure of Aer is not seriously distorted by the introduced cysteines.
Cross-linking of the AS-1-AS-2′ interface of the Aer HAMP domain.
The extent of disulfide cross-linking in response to the oxidant copper phenanthroline (CuPhe) depends on several parameters in addition to the distance between introduced cysteines. These parameters include the flexibility and orientation of the region being analyzed, the proclivity to form redox-active sulfanions, and the accessibility of the sulfanions to oxidizing reagents (8, 10, 17). To promote the formation of disulfide bonds, each di-Cys Aer receptor and all relevant single-Cys Aer controls (3, 36) were oxidized in vivo with 600 μM CuPhe for 10 min, unless specified otherwise. Disulfide-linked Aer dimers were separated from monomers by SDS-PAGE under nonreducing conditions, visualized by Western blotting, and quantitated by densitometry.
Figure 4B summarizes the extent of disulfide formation (after 10 min with CuPhe) for each of the predicted proximal and distal di-Cys receptors (black bars) and each of the relevant single-Cys controls (AS-1-AS-1′ and AS-2-AS-2′) (hatched bars and gray bars, respectively). For representative di-Cys pairs, time courses for cross-linking are shown in Fig. 6A. The cross-linking observed between AS-1 and AS-2 (Fig. 4B, black bars) occurred exclusively between cognate subunits (AS-1-AS-2′-linked dimers) rather than between AS-1 and AS-2 of the same subunit. Intrasubunit AS-1-AS-2 bonds would have resulted in compact monomers with different mobilities on SDS-PAGE gels (5), and these were not detected for any of the di-Cys pairs. The most extensive cross-linking between AS-1 and AS-2′ occurred with Aer-E213C/E238C and Aer-A216C/L241C (Fig. 4B), each of which has cysteine substitutions at the membrane-proximal end of the four-helix bundle (i.e., the top of the HAMP diagram in Fig. 3). Notably, the di-Cys pair A216C-L241C had one of the shortest predicted AS-1-AS-2′ distances in this study (Fig. 2A). Three additional proximal di-Cys pairs (V209C/E238C, E213C/L241C, and A223C/M252C) produced intermediate levels of disulfide-linked Aer dimers that were all greater than those of their relevant single-Cys controls (Fig. 4B). The extents of disulfide formation for these five proximal di-Cys pairs were therefore consistent with a parallel four-helix HAMP structure. Of note, A223C/M252C was the only proximal di-Cys pair that yielded substantial cross-linking in the lower, membrane-distal, portion of the four-helix bundle (Fig. 4B). The five remaining proximal di-Cys pairs, which contained AS-2 residue A245C or residue Q248C, exhibited less dimer formation, even though they had predicted AS-1-AS-2′ distances of between 3.9 and 6.9 Å, well within the range expected to generate disulfide-linked dimers (Fig. 2A and 4B).
Fig. 6.
Influence of signaling state on HAMP disulfide-linked dimer formation. (A) Percent cross-linking over 20 min for representative proximal di-Cys pairs in the presence of PAS-N85S (gray lines) or the wild-type PAS sequence (black lines). (B and C) Percent dimer formation for proximal di-Cys pairs (B) and distal di-Cys pairs (C) in receptors containing PAS-N85S (gray bars) or the wild-type PAS sequence (black bars) (data from Fig. 4B and 5B). Black asterisks indicate statistically significant differences in the extents of dimer formation in the presence of N85S (P < 0.05). Some differences could reflect changes in the intersubunit disulfide formation rates between the AS-1-AS-1′ and/or AS-2-AS-2′ controls (D), and gray asterisks are used to identify these. (D) Percent dimer formation for receptors containing a single cysteine substitution in AS-1 (residues 213 to 223) or AS-2 (residues 237 to 248) in the presence (gray bars) or absence (black bars) of N85S. Error bars indicate the standard deviations determined from two or more independent experiments.
Possible discontinuity in the HAMP AS-2 helix.
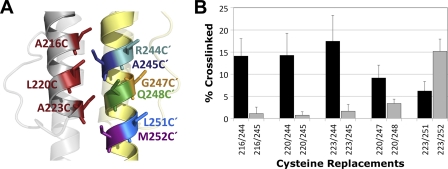
Of all the proximal di-Cys pairs, those containing the A245C substitution generated the lowest extents of disulfide-linked dimers (Fig. 4B). This was unexpected and differed from equivalent di-Cys substitutions in the Tar HAMP domain, which rapidly formed dimers in an in vitro cross-linking assay (29). We therefore considered the possibility that a discontinuity in AS-2 alters the orientation of the Aer-A245C side chain so that it is different from the orientation of the corresponding residue in Tar or Af1503. To investigate an alternative side-chain orientation in Aer, we examined cross-linking between AS-1 residues A216C, L220C, and A223C and the nearest neighbor of A245C, R244C (Fig. 5) (V246 was not tested because it is located at the HAMP dimer interface). The R244C-containing pairs formed more dimers than did the A245C-containing pairs (Fig. 5B), a finding that is incongruent with the predicted orientation of R244C (Fig. 5A) and the predicted AS-1-AS-2′ distances (Fig. 2A). Since the side chain of residue 244 was predicted to face away from the HAMP domain (Fig. 5A), we considered the possibility that R244C forms disulfide bonds with a neighboring dimer. To test this we constructed the tri-Cys receptor Aer-A184C/A223C/R244C. Because A184C forms intradimeric cross-links exclusively (2), any interdimeric cross-linking of residue 223C or 244C would produce oligomers larger than just dimers. Cells expressing the tri-Cys receptor were treated with CuPhe for 15 min at 30°C, during which time Aer-A184C/A223C/R244C produced only dimers (data not shown). This suggests that Aer A223C and R244C cross-link exclusively within dimers and that residue 244′ is either closer to AS-1 than residue 245′ and/or that the side chain of residue 244′ is oriented differently from how it is modeled in Fig. 5A. Since a reorientation of residues could not occur within the normal constraints of an α-helix, these data suggest that there is a discontinuity of the AS-2 helix before residue 244.
Fig. 5.
Incongruity between the HAMP model and cross-linking at residues 245C and 248C. (A) Aer HAMP model showing the location of residues 216C, 220C, and 223C in AS-1 and their predicted orientations relative to those of residues 244C′, 245C′, 247C′, 248C′, 251C′, and 252C′ in AS-2′. (B) Comparison between percent dimer formations for Aer di-Cys receptors containing R244C or A245C, G247C or Q248C, and L251C or M252C. The higher cross-linking values for R244C and G247C suggest that residues 244′ and 247′ are closer to AS-1 and/or that the orientation of these residues is different from how they are modeled in Fig. 5A. Error bars indicate the standard deviations determined from two or more independent experiments.
Although it is plausible that the formation of A245C-containing disulfide bonds was restricted by the local chemical environment and/or by side-chain constraints, this scenario does not explain the relatively rapid cross-linking observed for di-Cys pairs containing R244C. We considered the possibility that residue 244C was more accessible to solvent and therefore more readily oxidized, which would yield artifactually high levels of cross-linking. However, both R244C and A245C showed nearly identical solvent accessibilities, as determined by using the disulfide probe polyethylene glycol (PEG)-maleimide (K. J. Watts, unpublished data). Overall, the data are consistent with a deformation of the AS-2 helix in an Aer HAMP dimer that has not been observed for other HAMP domains.
If there is a discontinuity in AS-2, it may result in a distortion of the helix beyond residue A245C. Since di-Cys pairs containing Q248C exhibited only slightly higher reactivities than di-Cys pairs containing A245C (Fig. 4B), we examined cross-linking between AS-1 residues L220C and A223C and AS-2 residue G247C. G247C is located almost one turn C terminal to R244C and, like R244, is predicted to face away from the AS-1-AS-2′ interface (Fig. 5A). A223C dimerized to similar extents with G247C and with Q248C (data not shown), whereas L220C formed more dimers with G247C than with Q248C (Fig. 5B) (G247C alone does not dimerize). These data suggest that the discontinuity in AS-2 continues beyond residue 245 and are consistent with an altered side-chain orientation that is not present in the structure of Af1503. The deformation, which appears to be a rotation, includes multiple residues, but it is not clear whether it extends to the end of AS-2. L251C, which is located one heptad C terminal to R244, cross-linked with A223C, suggesting that the discontinuity in AS-2 rotates L251C with respect to the Af1503-based HAMP model (Fig. 5B) (L251C alone does not form disulfide bonds). However, there was also substantial cross-linking between M252C and A223C (Fig. 5B), suggesting that any helical rotation at M252C is less than the apparent rotation at A245C.
Cross-linking snapshots of HAMP signaling.
To explore conformational changes in the Aer HAMP domain that are associated with signaling, we compared both the rates and extents of disulfide formation in the kinase-off and kinase-on states of the receptor. In native Aer, the kinase-on state of the receptor is initiated by the reduction of PAS-FAD. This signal is relayed to the HAMP domain and kinase control module to generate the kinase-on conformation of Aer. This conformation in turn triggers the autophosphorylation of CheA, CW flagellar rotation, and cell tumbling (31). Since CuPhe creates an oxidizing environment, the cross-linking data presented above most likely represent the kinase-off conformation of the Aer HAMP domain. Moreover, oxidation by CuPhe is catalytically dependent on oxygen, and efficient disulfide formation with CuPhe is not possible in an anaerobic environment. To capture the kinase-on state of Aer in an aerobic environment, we introduced the CW-inducing substitution N85S into the PAS domain (Fig. 1). Aer-N85S was recently identified as a kinase-on mutant in a PAS mutant screen (9). By analogy with other PAS-FAD proteins (19, 34), the side chain of N85 in Aer is predicted to project inwards to contact and stabilize the isoalloxazine ring of FAD and not interact directly with the HAMP domain (9) (Fig. 1).
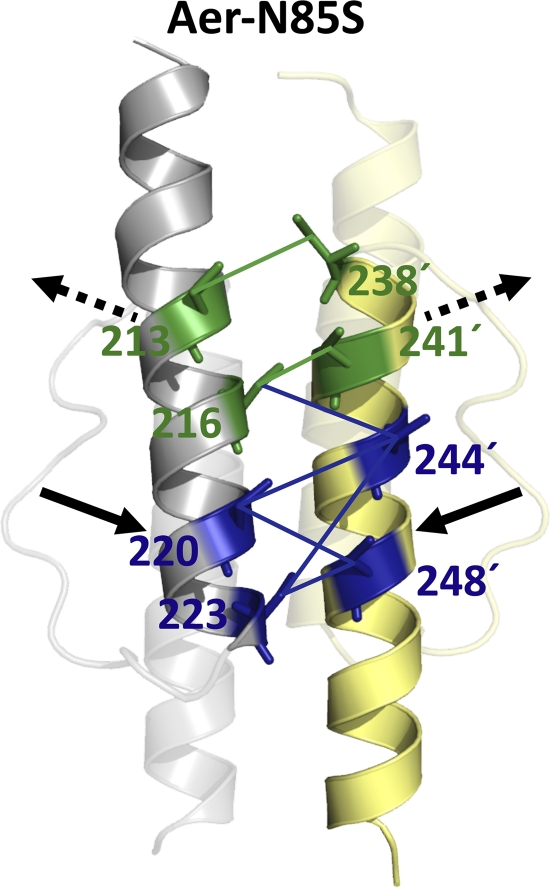
Aer-N85S-containing receptors were constructed for both proximal and distal di-Cys pairs, and BT3312 cells expressing each of these receptors were nonaerotactic in succinate semisoft agar (see examples in Fig. 4A). When viewed under the microscope in air, all of the mutants were more CW biased than their parents, although their biases varied from moderately CW biased to CW locked. These CW biases persisted after CuPhe was added, indicating that the kinase-on state of the receptor is maintained under the conditions used for disulfide cross-linking. This enabled us to use disulfide cross-linking to investigate the structure of the Aer HAMP domain in the kinase-on state and to conclude that changes caused by N85S most likely reflected differences in residue proximities and/or flexibilities during the kinase-on state. Figure 6 summarizes the rates (Fig. 6A) and extents (Fig. 6B to D) of disulfide formation determined for di-Cys receptors in the kinase-off state (black curves and bars) and their kinase-on, N85S-containing, counterparts (gray curves and bars). In the fixed-time assays, statistically significant changes in di-Cys cross-linking are indicated (Fig. 6B and C, asterisks). Where similar changes were also observed for the single-Cys controls (Fig. 6D), the di-Cys mutants are highlighted by gray asterisks in Fig. 6D and were not analyzed further. In the cross-linking time courses, the shapes of the reaction curves presumably reflect the different environments of the different cysteine pairs (Fig. 6A). However, there were also clear differences between the reaction rates of the kinase-off and kinase-on states, confirming conclusions based on the extents of cross-linking at 10 min. Five N85S-containing receptors showed significant increases in cross-linking at the bottom (the membrane-distal) half of the HAMP four-helix bundle compared with the kinase-off state (Fig. 6A and 7). In contrast, significant decreases in cross-linking were observed at the upper (membrane-proximal) portion of the four-helix bundle for two of the N85S-containing receptors (E213C/E238C and A216C/L241C) (Fig. 6 and 7). These findings indicate that the orientations of the helices are different in the kinase-on and kinase-off states, even though the HAMP domain retains the four-helix bundle architecture in both states (Fig. 7). The simplest interpretation of these data is that the HAMP helices move closer together at the bottom of the bundle in the kinase-on state but move further apart at the top of the bundle. However, results for the upper portion of the bundle were less conclusive, since there were fewer data for this region and the decrease in disulfide formation observed with Aer-E213C/L241C was not significant (Fig. 6).
Fig. 7.
Model showing differences between HAMP AS-1 and AS-2′ in the kinase-on versus the kinase-off state. Proximal AS-1-AS-2′ di-Cys pairs that showed significantly decreased (green) or increased (blue) collisional frequencies in the presence of the PAS kinase-on lesion N85S are shown. Collisions were inferred from the rates and extents of disulfide bond formation (Fig. 6). Only those di-Cys pairs that had significantly altered extents of disulfide bond formation in the presence of N85S are shown. Arrows designate helical movements that are consistent with the data (solid arrows indicate high confidence, and broken arrows indicate lower confidence).
In experiments similar to those with Aer-N85S, we also investigated changes in HAMP cross-linking effected by the CW-inducing PAS substitution N34D (Fig. 1) (37). However, the changes in the rates and extents of disulfide formation were less in the family of N34D-containing receptors than in the N85S receptors and were close to the precision limits of the disulfide cross-linking technique (results not shown). As a result of the noise associated with those measurements, we were unable to obtain statistically reliable data for N34D-containing receptors.
A significant cross-linking change was also observed for the distal di-Cys control Aer-N85S/A223C/D237C (Fig. 6C). This increase in dimer formation was unexpected, since residues 223 and 237 are located at opposite ends of the HAMP four-helix bundle (19.5 Å apart) (Fig. 2B). To determine whether disulfide bonds between residues 223C and 237C′ might have formed in nascent peptides before folding was complete, we inhibited protein synthesis by treating cells with 500 μg ml−1 chloramphenicol for 15 min prior to cross-linking. After chloramphenicol treatment, the extent of cross-linking observed for Aer-N85S/A223C/D237C was similar to the extent before treatment. In contrast, chloramphenicol treatment decreased the extents of cross-linking for other distal di-Cys controls, whereas for several proximal di-Cys mutants it either did not change or increased dimer formation (data not shown). The reason for the increased dimer formation between A223C and D237C in the presence of N85S is unclear.
DISCUSSION
In this study, we used a homology model based on the three-dimensional structure of Af1503-HAMP to predict that the Aer HAMP domain is a parallel four-helix bundle. Overall, the extents of dimer formation generated from engineered cysteine replacements at the symmetric (AS-1-AS-1′ and AS-2-AS-2′) (36) and asymmetric (AS-1-AS-2′) (Fig. 3 and 4B) HAMP dimer interfaces support a four-helix bundle structure for the Aer HAMP domain. This arrangement appears to be broadly similar to the resolved HAMP structures of Af1503 (14, 18) and Aer-2 (1) and similar to the structure of the Tar HAMP domain that was determined previously by disulfide cross-linking (29). The four-helix HAMP fold was maintained in the kinase-on and kinase-off signaling states of the Aer receptor, although the chemical reactivities of many of the engineered cysteine residues differed between the two states (Fig. 6 and 7). The simplest explanation for this is that the different HAMP signaling states have different conformations. The reactivities of cysteine residues at the bottom of the HAMP four-helix bundle increased in the kinase-on state, whereas the reactivities of cysteine residues at the top of the bundle tended to decrease (Fig. 6). These findings are consistent with a helical tilt motion between the kinase-off and kinase-on signaling states.
In addition to providing evidence for conformational changes associated with HAMP signaling states, this study is the first to demonstrate a conformational change in a HAMP domain associated with the signal-on state of a PAS domain. N85S, with its predicted location buried within the FAD-binding pocket of the PAS domain (9), was associated with greater cross-linking changes than was N34D (with its predicted location near the PAS N-terminal cap) (9). N85S may therefore cause more global conformational changes than does N34D. In the presence of N85S, the extent of dimer formation was significantly lower at the top of the four-helix bundle for E213C/E238C and A216C/L241C but was inconclusive for E213C/L241C (Fig. 6). In Fig. 7, broken arrows are used to indicate that the tops of the AS-1 and AS-2′ helices may move apart in the kinase-on state but that this finding was not conclusive. In contrast, solid arrows in Fig. 7 denote an increased confidence that conformational changes occurred at the bottom of the four-helix bundle. The cross-linking data do not provide evidence for, or against, the gearbox model of HAMP signaling. However, transitional changes from x-da to canonical knob-into-hole packing in Af1503 are associated with helical rotations and bundle shape changes that are most prominent at the bottom of the four-helix HAMP bundle (14, 18). In particular, the C-terminal ends of AS-2 were splayed outward in Af1503 signal-off mutants, which is qualitatively analogous to the differences observed for Aer HAMP in this study.
In comparison with models like the gearbox signaling model that propose static kinase-on and kinase-off conformations, an alternative HAMP signaling model was recently proposed, in which the kinase-on state was associated with a dynamic x-da bundle (39, 40). This raises the possibility that the cross-linking differences that we observed in the kinase-on state are the result of a more relaxed HAMP structure, in which case di-Cys pairs that were previously well placed to cross-link might show decreased dimer formation and vice versa. Interestingly, the HAMP region that appeared to move closer together (i.e., that showed increased extents of disulfide formation) in the kinase-on state overlaps with the site of HAMP lesions that promote the kinase-on state of Aer (36).
In the HAMP AS-2 helix, the reactivities of cysteine replacements at residues R244 and G247 were not consistent with either the structural predictions of the Af1503-based HAMP model (Fig. 5) or Aer HAMP models created from the recently solved Aer-2 HAMP structures (1; data not shown). All Aer HAMP models show AS-2 as a continuous α-helix, with residues 245 and 248 projecting into the AS-1-AS-2′ interface and residues 244 and 247 projecting away from it (Fig. 5A). However, residue 244C′ in particular readily formed disulfide bonds with AS-1 residues 216C, 220C, and 223C in both the kinase-on and kinase-off states, whereas residue 245C′ did not (Fig. 5B and 6). Moreover, residue 244C did not cross-link with an adjacent Aer dimer, and the solvent accessibility (and, thus, the CuPhe accessibility) of residue 244C was similar to that of residue 245C. Consequently, there is no obvious explanation for the reactivity of residue 244C, unless residue 244C′ is oriented toward residues 216, 220, and 223, which would require a 60° to 90° rotation of the helical backbone. The unexpected reactivity of residue 247C′ in forming disulfide bonds with residue 220C suggests that residue 247C is also rotated. According to the HAMP model, the orientation of residue 247C′ for bonding to residue 220C is less favorable than the orientation of either residue 244C′ or 248C′. Thus, the high reactivities of residues 244C′ and 247C′ were unexpected and are best accommodated by a rotation of the AS-2 helix.
The discontinuity identified in the Aer HAMP AS-2 helix is not present in the Tar or Af1503 HAMP domains (18, 29). There is no residue in the primary sequence of AS-2 that is known to distort an α-helix, but the free energy required to distort the helix could be supplied by an external peptide that interacts with AS-2 and is not present in Tar or Af1503. One possibility is that the discontinuity in AS-2 is dependent on an interaction with the PAS domain. In Aer, direct PAS-HAMP signaling interactions were previously proposed (7, 22, 35–37), and the region of AS-2 in Aer that includes HAMP residues R244 and G247 is an attractive candidate for PAS interactions because residues in this region are predicted to project outward from the surface of the HAMP domain (36), but they show poor solvent accessibility (Watts, unpublished). We recently showed that HAMP-Q248C can be specifically cross-linked to residues on the Aer-PAS β-scaffold (9), suggesting that the PAS domain does interact with the HAMP domain in this region. A more extensive analysis of this PAS-HAMP interaction surface is currently in progress.
The Af1503 HAMP domain, upon which the structure of the Aer HAMP domain was modeled, is at present an orphan domain with no signaling role. It is possible that HAMP domains that are constrained by lateral interactions with another signaling domain or protein will be shown in the future to have a discontinuity in the AS-2 helix, as demonstrated in the present study. Constraints on the AS-2 helix are apparently an integral component of the signaling mechanism. In chemoreceptors, such as Tsr or Tar, HAMP domains receive signals directly from a transmembrane segment, and there is no evidence of a discontinuity in AS-2 (8, 40). In methyl-accepting chemoreceptors, constraints on AS-2 are imposed by the motion of the transmembrane helix (TM2) and by the adaptive-methylation module (25), as opposed to the type of constraints imposed by PAS or other sensory input domains. This suggests that there may be fundamental differences in signaling in chemoreceptors compared to Aer-type proteins. In either system, the stimulus-induced change in the HAMP conformation produces a signal that communicates with the downstream kinase control region, the site of the receptor's signal output.
Within the functional unit of chemoreceptors, the trimer of dimers, HAMP dimers may alternate between an expanded trimer-of-dimer conformation in the kinase-off state and a compact conformation in the kinase-on state (20). Such conformational changes would require flexibility at the junction of the HAMP and signaling domains, which could be accommodated by the presence of a short unstructured sequence between the two domains (36). Recent studies suggested that the “on” conformation of the HAMP domain stabilizes the four-helix adaptation region in the proximal signaling domain, causing a concomitant destabilization of the distal kinase control region of the receptor (25, 30, 39). There is also evidence that minor alterations in local helical structures can be transmitted to the chemoreceptor signaling tip by destabilizing the coiled-coil structure of a receptor (26). In the current study, a deformation was evident in the Aer HAMP AS-2 region, and this is an attractive region for future studies. Although its role and functional significance are currently unknown, it may be a key to an understanding of the general mechanism used by Aer and other PAS-HAMP proteins to pass signals onto downstream components.
ACKNOWLEDGMENTS
We thank Lauren Abraham and Nathan Abraham for technical assistance, Andrei Lupas and Murray Coles for access to unpublished data, and Andrei Lupas, Murray Coles, John S. Parkinson, and Brian Crane for helpful discussions.
This work was supported by a grant from the National Institute of General Medical Sciences (grant GM029481) to B.L.T.
Footnotes
Published ahead of print on 10 June 2011.
REFERENCES
- 1. Airola M. V., Watts K. J., Bilwes A. M., Crane B. R. 2010. Structure of concatenated HAMP domains provides a mechanism for signal transduction. Structure 18:436–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Amin D. N., Taylor B. L., Johnson M. S. 2007. Organization of the aerotaxis receptor Aer in the membrane of Escherichia coli. J. Bacteriol. 189:7206–7212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Amin D. N., Taylor B. L., Johnson M. S. 2006. Topology and boundaries of the aerotaxis receptor Aer in the membrane of Escherichia coli. J. Bacteriol. 188:894–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aravind L., Ponting C. P. 1999. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol. Lett. 176:111–116 [DOI] [PubMed] [Google Scholar]
- 5. Bass R. B., Butler S. L., Chervitz S. A., Gloor S. L., Falke J. J. 2007. Use of site-directed cysteine and disulfide chemistry to probe protein structure and dynamics: applications to soluble and transmembrane receptors of bacterial chemotaxis. Methods Enzymol. 423:25–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bibikov S. I., Biran R., Rudd K. E., Parkinson J. S. 1997. A signal transducer for aerotaxis in Escherichia coli. J. Bacteriol. 179:4075–4079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Buron-Barral M. D. C., Gosink K. K., Parkinson J. S. 2006. Loss- and gain-of-function mutations in the F1-HAMP region of the Escherichia coli aerotaxis transducer Aer. J. Bacteriol. 188:3477–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Butler S. L., Falke J. J. 1998. Cysteine and disulfide scanning reveals two amphiphilic helices in the linker region of the aspartate chemoreceptor. Biochemistry 37:10746–10756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Campbell A. J., Watts K. J., Johnson M. S., Taylor B. L. 2010. Gain-of-function mutations cluster in distinct regions associated with the signalling pathway in the PAS domain of the aerotaxis receptor, Aer. Mol. Microbiol. 77:575–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Careaga C. L., Falke J. J. 1992. Structure and dynamics of Escherichia coli chemosensory receptors. Engineered sulfhydryl studies. Biophys. J. 62:209–216, discussion 217-219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Doebber M., et al. 2008. Salt-driven equilibrium between two conformations in the HAMP domain from Natronomonas pharaonis: the language of signal transfer? J. Biol. Chem. 283:28691–28701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dunin-Horkawicz S., Lupas A. N. 2010. Comprehensive analysis of HAMP domains: implications for transmembrane signal transduction. J. Mol. Biol. 397:1156–1174 [DOI] [PubMed] [Google Scholar]
- 13. Falke J. J., Hazelbauer G. L. 2001. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem. Sci. 26:257–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ferris H. U., et al. 2011. The mechanisms of HAMP-mediated signaling in transmembrane receptors. Structure 19:378–385 [DOI] [PubMed] [Google Scholar]
- 15. Gosink K. K., del Carmen Buron-Barral M., Parkinson J. S. 2006. Signaling interactions between the aerotaxis transducer Aer and heterologous chemoreceptors in Escherichia coli. J. Bacteriol. 188:3487–3493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Herrmann S., Ma Q., Johnson M. S., Repik A. V., Taylor B. L. 2004. PAS domain of the Aer redox sensor requires C-terminal residues for native-fold formation and flavin adenine dinucleotide binding. J. Bacteriol. 186:6782–6791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hughson A. G., Hazelbauer G. L. 1996. Detecting the conformational change of transmembrane signaling in a bacterial chemoreceptor by measuring effects on disulfide cross-linking in vivo. Proc. Natl. Acad. Sci. U. S. A. 93:11546–11551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hulko M., et al. 2006. The HAMP domain structure implies helix rotation in transmembrane signaling. Cell 126:929–940 [DOI] [PubMed] [Google Scholar]
- 19. Key J., Hefti M., Purcell E. B., Moffat K. 2007. Structure of the redox sensor domain of Azotobacter vinelandii NifL at atomic resolution: signaling, dimerization, and mechanism. Biochemistry 46:3614–3623 [DOI] [PubMed] [Google Scholar]
- 20. Khursigara C. M., Wu X., Zhang P., Lefman J., Subramaniam S. 2008. Role of HAMP domains in chemotaxis signaling by bacterial chemoreceptors. Proc. Natl. Acad. Sci. U. S. A. 105:16555–16560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lai W. C., Hazelbauer G. L. 2007. Analyzing transmembrane chemoreceptors using in vivo disulfide formation between introduced cysteines. Methods Enzymol. 423:299–316 [DOI] [PubMed] [Google Scholar]
- 22. Ma Q., Johnson M. S., Taylor B. L. 2005. Genetic analysis of the HAMP domain of the Aer aerotaxis sensor localizes flavin adenine dinucleotide-binding determinants to the AS-2 helix. J. Bacteriol. 187:193–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ma Q., Roy F., Herrmann S., Taylor B. L., Johnson M. S. 2004. The Aer protein of Escherichia coli forms a homodimer independent of the signaling domain and FAD binding. J. Bacteriol. 186:7456–7459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nambu J. R., Lewis J. O., Wharton K. A., Jr, Crews S. T. 1991. The Drosophila single-minded gene encodes a helix-loop-helix protein that acts as a master regulator of CNS midline development. Cell 67:1157–1167 [DOI] [PubMed] [Google Scholar]
- 25. Parkinson J. S. 2010. Signaling mechanisms of HAMP domains in chemoreceptors and sensor kinases. Annu. Rev. Microbiol. 64:101–122 [DOI] [PubMed] [Google Scholar]
- 26. Pollard A. M., Bilwes A. M., Crane B. R. 2009. The structure of a soluble chemoreceptor suggests a mechanism for propagating conformational signals. Biochemistry 48:1936–1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rebbapragada A., et al. 1997. The Aer protein and the serine chemoreceptor Tsr independently sense intracellular energy levels and transduce oxygen, redox, and energy signals for Escherichia coli behavior. Proc. Natl. Acad. Sci. U. S. A. 94:10541–10546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Repik A., et al. 2000. PAS domain residues involved in signal transduction by the Aer redox sensor of Escherichia coli. Mol. Microbiol. 36:806–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Swain K. E., Falke J. J. 2007. Structure of the conserved HAMP domain in an intact, membrane-bound chemoreceptor: a disulfide mapping study. Biochemistry 46:13684–13695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Swain K. E., Gonzalez M. A., Falke J. J. 2009. Engineered socket study of signaling through a four-helix bundle: evidence for a yin-yang mechanism in the kinase control module of the aspartate receptor. Biochemistry 48:9266–9277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Taylor B. L. 2007. Aer on the inside looking out: paradigm for a PAS-HAMP role in sensing oxygen, redox and energy. Mol. Microbiol. 65:1415–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Taylor B. L., Watts K. J., Johnson M. S. 2007. Oxygen and redox sensing by two-component systems that regulate behavioral responses: behavioral assays and structural studies of Aer using in vivo disulfide cross-linking. Methods Enzymol. 422:190–232 [DOI] [PubMed] [Google Scholar]
- 33. Taylor B. L., Zhulin I. B. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63:479–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ukaegbu U. E., Rosenzweig A. C. 2009. Structure of the redox sensor domain of Methylococcus capsulatus (Bath) MmoS. Biochemistry 48:2207–2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Watts K. J., Johnson M. S., Taylor B. L. 2006. Minimal requirements for oxygen sensing by the aerotaxis receptor Aer. Mol. Microbiol. 59:1317–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Watts K. J., Johnson M. S., Taylor B. L. 2008. Structure-function relationships in the HAMP and proximal signaling domains of the aerotaxis receptor Aer. J. Bacteriol. 190:2118–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Watts K. J., Ma Q., Johnson M. S., Taylor B. L. 2004. Interactions between the PAS and HAMP domains of the Escherichia coli aerotaxis receptor Aer. J. Bacteriol. 186:7440–7449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Watts K. J., Sommer K., Fry S. L., Johnson M. S., Taylor B. L. 2006. Function of the N-terminal cap of the PAS domain in signaling by the aerotaxis receptor Aer. J. Bacteriol. 188:2154–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhou Q., Ames P., Parkinson J. S. 2011. Biphasic control logic of HAMP domain signaling in the Escherichia coli serine chemoreceptor. Mol. Microbiol. 80:596–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhou Q., Ames P., Parkinson J. S. 2009. Mutational analyses of HAMP helices suggest a dynamic bundle model of input-output signalling in chemoreceptors. Mol. Microbiol. 73:801–814 [DOI] [PMC free article] [PubMed] [Google Scholar]