Abstract
Biosynthesis of the hybrid polyketide-nonribosomal peptide antibiotic streptolydigin, 3-methylaspartate, is utilized as precursor of the tetramic acid moiety. The three genes from the Streptomyces lydicus streptolydigin gene cluster slgE1-slgE2-slgE3 are involved in 3-methylaspartate supply. SlgE3, a ferredoxin-dependent glutamate synthase, is responsible for the biosynthesis of glutamate from glutamine and 2-oxoglutarate. In addition to slgE3, housekeeping NADPH- and ferredoxin-dependent glutamate synthase genes have been identified in S. lydicus. The expression of slgE3 is increased up to 9-fold at the onset of streptolydigin biosynthesis and later decreases to ∼2-fold over the basal level. In contrast, the expression of housekeeping glutamate synthases decreases when streptolydigin begins to be synthesized. SlgE1 and SlgE2 are the two subunits of a glutamate mutase that would convert glutamate into 3-methylaspartate. Deletion of slgE1-slgE2 led to the production of two compounds containing a lateral side chain derived from glutamate instead of 3-methylaspartate. Expression of this glutamate mutase also reaches a peak increase of up to 5.5-fold coinciding with the onset of antibiotic production. Overexpression of either slgE3 or slgE1-slgE2 in S. lydicus led to an increase in the yield of streptolydigin.
INTRODUCTION
The vast majority of antibiotic and antitumor drugs belong either to the polyketide or the nonribosomal families of natural products. A related family comprises hybrid compounds containing polyketide and nonribosomal peptide moieties. Their biosynthesis involves the participation of a modular polyketide synthase (PKS) for the condensation of acyl coenzyme A (acyl-CoA) precursors and a nonribosomal peptide synthetase (NRPS) that condenses amino acids after their activation to an aminoacyl-AMP precursor. Both type I PKSs and NRPSs are multifunctional enzymes that are organized into modules and use a similar strategy for the assembly of these short carboxylic and amino acid building blocks. The minimal set of domains in a type I PKS includes ketosynthase (KS), acyltransferase, and acyl-carrier protein activities responsible for the catalysis of one cycle of polyketide chain elongation. These PKS modules can contain further domains such as ketoreductase (KR), dehydratase (DH), or enoylreductase to reduce the keto groups generated during the condensation process (9). In a similar way, a typical minimal NRPS module consists of condensation, adenylation, and peptidyl carrier protein (PCP) domains (9).
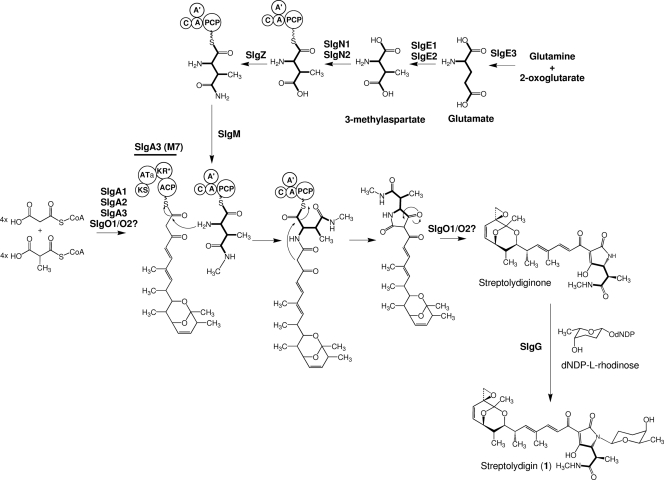
Streptolydigin (compound 1) (Fig. 1) is an inhibitor of bacterial RNA polymerase β-subunit produced by Streptomyces lydicus (27, 29) and a potent inhibitor of eukaryotic DNA polymerase terminal deoxynucleotidyltransferase (6, 7). The streptolydigin biosynthetic gene cluster has been isolated and characterized from the producer organism (21). Streptolydigin belongs to the hybrid polyketide-nonribosomal peptide family of natural products. The streptolydigin type I PKS, composed of one loading domain and seven extension modules distributed over three polypeptides, would condense four units of malonyl-CoA and four units of methyl-malonyl-CoA and is proposed to generate the polyketide core. The formation of the tetramic acid moiety of the molecule involves the participation of an NRPS system composed of at least two polypeptides. Early biosynthetic studies using labeled precursors have shown the incorporation of propionate, acetate, methionine, and glutamic acid (suggested to be in form of β-methylaspartate) into the main structure of streptolydigin (3, 4, 22, 23). In addition, recent work in our laboratory has demonstrated the involvement of SlgZ, an asparaginyl-tRNA synthetase-like enzyme, in the 3-methylaspartate tailoring process by amidation of a 3-methylaspartatyl-NRPS bound to generate 3-methylasparaginyl-NRPS (11). This activity might be followed by the methylation of NRPS-bound 3-methylasparagine by SlgM to obtain N-methyl-3-methylasparaginyl-NRPS that then would be condensed with the polyketide chain synthesized by the streptolydigin PKS (11) (Fig. 1). Three other genes in the cluster are predicted to encode two subunits of a glutamate mutase (GM) (slgE1 and slgE2) and a ferredoxin-dependent glutamate synthase (GS) (slgE3). We report here the characterization of these three genes and their role in supplying 3-methylaspartate as the precursor for streptolydigin biosynthesis. We have also identified two housekeeping GSs, dependent, respectively, on NADPH and ferredoxin. Deletion of the GM genes slgE1 and slgE2 led to the production of a novel streptolydigin derivative.
Fig. 1.
Proposed pathway for the biosynthesis of streptolydigin (compound 1). The incorporation of glutamate, in the form of 3-methylaspartate, to generate the tetramic acid lateral side chain is shown in thick lines. M7, PKS module 7; KS, ketosynthase; ATa, acyl transferase specific for malonyl-CoA; ACP, acyl carrier protein; KR*, inactive ketoreductase; C, condensation domain; A', SlgN1 adenylation domain; A, SlgN2 adenylation domain; PCP, peptidyl carrier protein.
MATERIALS AND METHODS
Strains and culture conditions.
The bacterial strains used in the present study were S. lydicus NRRL2433, a streptolydigin producer; Escherichia coli DH10B (Invitrogen); and ET12567(pUB307) (12). The growth medium for S. lydicus and mutants was tryptone soy broth. For sporulation, MA medium was used and R5A was the streptolydigin production medium (8). Growth of S. lydicus in R5A liquid medium was monitored by measuring absorbance at 600 nm using the diphenylamine assay method for determining DNA content (2, 16). Culture conditions were those previously described (21). Intergeneric conjugation of Streptomyces-E. coli was performed according to standard procedures (12). The E. coli media were as described previously (25). When plasmid-containing clones were grown, the medium was supplemented with the appropriate antibiotics: 100 μg of ampicillin/ml, 20 μg of tobramycin/ml, 25 μg of apramycin/ml, 50 μg of thiostrepton/ml, 50 μg of hygromycin/ml, 10 μg of tetracycline/ml, 25 μg of chloramphenicol/ml, or 50 μg of nalidixic acid/ml.
DNA manipulation and plasmids.
DNA manipulations were performed according to standard procedures for E. coli (25) and Streptomyces (12). Platinum Pfx DNA polymerase (Invitrogen) and 2.5% dimethyl sulfoxide (DMSO) were used for all PCR amplifications. The PCR conditions used were as follows: 97°C for 5 min, followed by 30 cycles of 95°C for 30 s, 55°C for 45 s, and 68°C for 1 min, with a final extension cycle at 68°C for 10 min. All of the PCR products were cloned into pCR-BLUNT (Invitrogen) and then sequenced. Other plasmids used in the present study included pSL1180 (Amersham Pharmacia) for routine cloning, pEFBAoriT (11) for gene replacement, and pEM4T (17) for gene expression. pLHyg (20) was the source of the hygromycin resistance gene hyg.
Construction of plasmids for gene replacement and complementation of mutants.
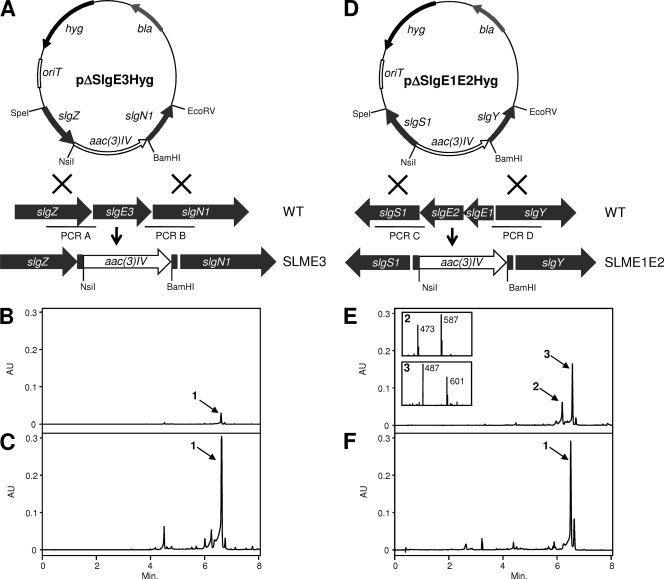
For deletion of slgE3 two DNA fragments of 1.0 and 1.1 kb, respectively, were amplified by PCR from cosmid Slg4A8 (21) using oligoprimers HEI9/HEI10 (PCR A) and HEI11/HEI12 (PCR B) (Table 1). The PCR A fragment (Fig. 2A) containing 34 bp from the slgE3 5′ end was cloned as a SpeI-NsiI fragment into SpeI-NsiI-digested pEFBAoriT. The resultant plasmid was digested with BamHI-EcoRVI, and the PCR B fragment (Fig. 2A), containing 22 bp from the slgE3 3′ end and digested with the same restriction enzymes, was cloned, yielding plasmid pΔslgE3. In this construct 1,267 bp of the slgE3 coding region have been substituted by the apramycin resistance gene aac3(IV). Deletion of genes slgE1 and slgE2 was accomplished by amplification of two DNA fragments (1.0 and 1.1 kb, respectively) from cosmid Slg4A8 by using the oligoprimers HEI15/HEI16 (PCR C) and HEI17/HEI18 (PCR D) (Table 1). The PCR C fragment (Fig. 2D), containing 18 bp from the slgE2 3′ end, was cloned as a SpeI-NsiI fragment into SpeI-NsiI-digested pEFBAoriT. The resultant plasmid was digested with BamHI-EcoRVI, and the PCR D fragment (Fig. 2D), containing 45 bp from the slgE1 5′ end and digested with the same restriction enzymes, was cloned, yielding plasmid pΔslgE1E2. In this construct, 1,744 bp of the slgE1-slgE2 coding region were substituted by the apramycin resistance gene aac3(IV). Finally, pΔslgE3 and pΔslgE1E2 were digested with XbaI, and the gene hyg from pLHyg was subcloned as a SpeI-NheI fragment to obtain plasmids pΔslgE3Hyg and pΔslgE1E2Hyg used for the generation of S. lydicus strains SLME3 and SLME1E2, respectively.
Table 1.
Primers used in this study
| Primer | Sequence (5′-3′)a | Description |
|---|---|---|
| HEI9 | AACTAGTCCCCGTCAGCACCCCACG | SpeI |
| HEI10 | AAAATGCATACCCCGGGGCAGACAGCC | NsiI |
| HEI11 | AAGGATCCGACGACGAGGGAGCCCTC | BamHI |
| HEI12 | AAGATATCTGAAGCCCATGCTCTCGG | EcoRV |
| HEI15 | AACTAGTTGTGGTCGCCGAGGACGA | SpeI |
| HEI16 | AAAATGCATGGCGGTCGAAGACCGATG | NsiI |
| HEI17 | AAGGATCCGCTGTGCCGTGCGTCGTC | BamHI |
| HEI18 | AAGATATCTGCAGACCGACCGCGGTC | EcoRV |
| HEI13 | AAGGATCCGAAGATCCCCGGCGTGGT | BamHI |
| HEI14 | AGAATTCGAACGGATTGCTCACGAG | EcoRI |
| CCL22B | AAGGATCCGATGGGCAAGGCAATGGA | BamHI |
| CCL21MF | AACAATTGCAGCACGAGTGCTCTCAT | MfeI |
| HEI-I | GTCGCCTCCGGCCGCTTC | Forward for gltS-α |
| HEI-J | SCCGACCGGGCAGGTGTC | Reverse for gltS-β |
| HEI-F | TTCTGCGTCCGCAACTCC | Forward for gltS-α and -β |
| HEI-C | CAGTCSGCRCCSGTGTCR | Reverse for gltS-α and -β |
| HEI-FR1 | CCSTACGACATGGCSYTGCT | Forward for gltS-FD |
| HEI-FR2 | CTTGCACACSGCGAGGAACT | Reverse for gltS-FD |
| NDGA | AGCGAGTACCTCGTCAACTCCGA | Forward for gltS-αb |
| NDGB | CATCATGACGCAGCCGGAGA | Reverse for gltS-α |
| NDPA | AGCGCGAGATCGCCAAGACC | Forward for gltS-β |
| NDPB | CGCTTGCGCAGCTTCTTGGC | Reverse for gltS-β |
| FDA | GACATGGCCCTGCTGAACGTCTC | Forward for gltS-FD |
| FDB | GGAGCCGACGCAGAGCTTGAAC | Reverse for gltS-FD |
| FDSLGE3A | TGGTGCCGCCGGTGTTCAT | Forward for slgE3 |
| FDSLGE3B | GGATCTCCTCGGTGACGGTGC | Reverse for sglE3 |
| CRIS3 | ATCGCGCTCAACGGCTAC | Forward for sglE2 |
| CRIS4 | TAGGGGATGTCCGACGAC | Reverse for sglE2 |
| RTNADG1 | ACCACGACATCTACTCCATCGAG | Forward for gltS-αc |
| RTNADG2 | CAGCTTCACGTGGATGCG | Reverse for gltS-α |
| RTNADP1 | TCAAGATGGAGAAGCGCCA | Forward for gltS-β |
| RTNADP2 | CGTACGGAACTTGGTGCCC | Reverse for gltS-β |
| RTGS1 | TCCGAGTTCGCCGACAA | Forward for gltS-FD |
| RTGS2 | GGTTTCAGGGAGACGCACTTGAT | Reverse for gltS-FD |
| RTGSSLG1 | CCAGCAGATCCGCTTCAT | Forward for slgE3 |
| RTGSSLG2 | AAGAGCTTCACCCACACCC | Reverse for sglE3 |
| RTGMA | GATTCACCACCTCGAAACCGT | Forward for sglE2 |
| RTGMB | TGATCGTCAGCAGTCTCGCTT | Reverse for sglE2 |
| HRDBqRT1 | CAACCCAGTGGAAGAACGTT | Forward for hrdB |
| HRDBqRT2 | TGCGGCACTGACCATCAG | Reverse for hrdB |
Restriction sites are underlined in the primer sequences.
This primer and the following one were used for RT-PCR studies.
This primer and the following one were used for qRT-PCR studies.
Fig. 2.
(A) Scheme representing the replacement in the chromosome of the wild-type slgE3 gene by a version mutated through the insertion of an apramycin resistance cassette. (B) UPLC analysis of mutant SLME3. (C) UPLC analysis of mutant SLME3 complemented by pEM4TslgE3. (D) Scheme representing the replacement of the wild-type slgE1-E2 genes by one mutated through the insertion of an apramycin resistance cassette. (E) UPLC analysis of mutant SLME1E2 and MS analysis of compounds 2 and 3. (F) UPLC analysis of mutant SLME1E2 complemented by pEM4TslgE1E2. aac(3)IV, apramycin resistance gene; hyg, hygromycin resistance gene; bla, β-lactamase gene; 1, streptolydigin; 2, streptolydigin B; 3, streptolydigin C; AU, arbitrary units.
Plasmids pEM4TslgE3 and pEM4TslgE1E2 were constructed for complementation of S. lydicus strains SLME3 and SLME1E2, respectively. The slgE3 gene was amplified by PCR from cosmid Slg4A8 by using the oligoprimers HEI13 and HEI14. The resultant 1.3-kp product was cloned as a BamHI-EcoRI fragment into BamHI-EcoRI-digested pEM4T, yielding plasmid pEM4TslgE3. The slgE1-slgE2 genes were amplified by PCR from cosmid Slg4A8 by using the oligoprimers CCL22B and CCL21MF. The resultant 1.8-kp product was cloned as a BamHI-MfeI fragment into BamHI-EcoRI-digested pEM4T, yielding plasmid pEM4TslgE1E2. In both constructs, the genes slgE3 and slgE1-slgE2 are under the control of the ermE* promoter.
Generation of S. lydicus mutant strains.
Constructs pΔslgE3Hyg and pΔslgE1E2Hyg were introduced into S. lydicus by intergeneric conjugation from E. coli ET12567(pUB307). For the generation of the S. lydicus strains SLME3 and SLME1E2, a single-crossover strain, which was apramycin and hygromycin resistant, was cultured in the absence of selection and then screened for the loss of hygromycin resistance and the retention of apramycin resistance because of a double recombination event. The deletion of slgE3 in SLME3 and slgE1-slgE2 in SLME1E2 were verified by Southern hybridization and PCR amplification by using the oligoprimers HEI9/HEI12 or HEI15/HEI18, respectively. Plasmids pEM4T, pEM4TslgE3, and pEM4TslgE1E2 were introduced into S. lydicus wild-type strain or mutants SLME3 and SLME1E2 by intergeneric conjugation and transconjugants were selected for resistance to thiostrepton.
UPLC, LC-MS, and NMR spectroscopy methods.
Streptolydigin production in S. lydicus wild-type or mutant strains grown on R5A medium was analyzed by ultrahigh-performance liquid chromatography (UPLC) and liquid chromatography-mass spectrometry (LC-MS) using previously described procedures (21). For structural elucidation streptolydigin C was purified and characterized by 1H and 13C nuclear magnetic resonance (NMR) experiments. The information regarding structural characterization of streptolydigin C is given in Table S1 in the supplemental material.
Bioactivity testing.
The antibiotic activity of streptolydigin C (compound 3, Fig. 3A) was assayed via antibiotic disc diffusion assay against Streptomyces albus. Streptolydigin was used as reference compound. In both cases, 2 μg of each compound was utilized according to a previously described procedure (21).
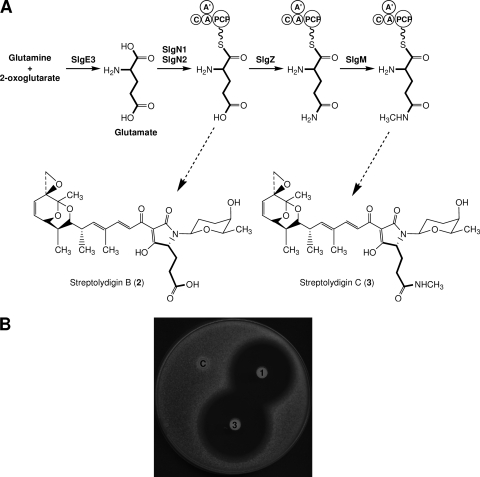
Fig. 3.
(A) Structures of streptolydigin B (compound 2) and novel derivative streptolydigin C (compound 3) showing the proposed origin of tetramic acid lateral side chain from glutamate (thick lines). C, condensation domain; A′, SlgN1 adenylation domain; A, SlgN2 adenylation domain; PCP, peptidyl carrier protein. (B) Antibiotic activity of streptolydigin (area 1) and streptolydigin C (area 3) against S. albus. Each paper disk was soaked with 2 μg of the corresponding compound. Control without antibiotic (area C).
Identification of S. lydicus housekeeping glutamate synthases.
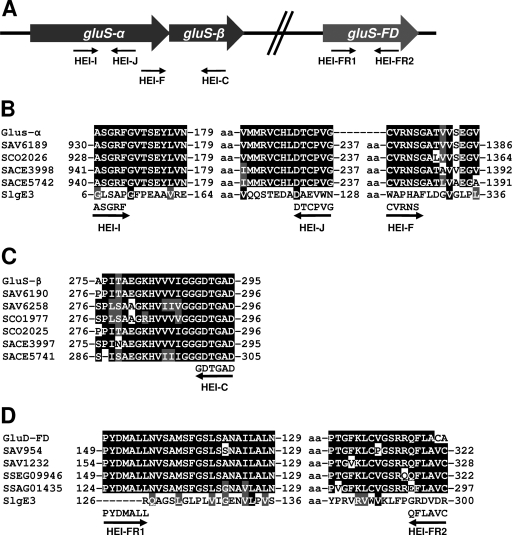
PCR amplification of glutamate synthase-encoding regions was performed using degenerate oligoprimers (Table 1) and total DNA from S. lydicus NRRL2433. The oligoprimers HEI-I and HEI-J were designed to locate the presence of an internal region of an NADPH-dependent glutamate synthase α subunit gene (gluS-α). They were derived from conserved amino acid sequences present in enzyme SAV6189 from S. avermitilis MA-4680, enzyme SCO2026 from S. coelicolor A3(2), and enzymes SACE3998 and SACE5742 from Saccharopolyspora erythraea NRRL2338 (see Fig. 5B). Oligoprimers HEI-F and HEI-C were designed to locate the presence of the 5′ end of an NADPH-dependent glutamate synthase α subunit (gluS-α) and the 3′ end of an NADPH-dependent glutamate synthase β subunit (gluS-β) genes. Their sequences were designed from previously mentioned enzymes (see Fig. 5B) and from NADPH-dependent glutamate synthase β subunits SAV6190 and SAV6258 from S. avermitilis MA-4680, SCO1977, and SCO2025 from S. coelicolor A3(2) and SACE3997 and SACE5741 from Saccharopolyspora erythraea NRRL2338 (see Fig. 5C). Oligoprimers HEI-FR1 and HEI-FR2 were designed to locate the presence of an internal region of a housekeeping ferredoxin-dependent glutamate synthase gene (gluS-FD). These primers were designed using the enzymes SAV954 and SAV1232 from S. avermitilis MA-4680, SSEG09946 from S. sviceus ATCC 29083, and SSAG01435 from Streptomyces sp. strain Mg1 (see Fig. 5D). DNA sequencing of PCR product obtained was performed on double-stranded DNA templates with the dideoxynucleotide chain termination method (26) and the Cy5 Autocycle sequencing kit (GE Healthcare). An Alf-express automatic DNA sequencer (GE Healthcare) was used. Computer-aided database searching and sequence analysis were carried out with the University of Wisconsin Genetics Computer Group software (5) and the BLAST program (1).
Fig. 5.
(A) Scheme representing PCR amplification of glutamate synthase gluS-α, gluS-β, and gluS-FD from the chromosome of S. lydicus NRRL2433. (B) Sequence alignments of NADPH-dependent glutamate synthase α subunits showing the conserved regions used for designing degenerated the oligoprimers HEI-I, HEI-J, and HEI-F. GluS-α, S. lydicus NRRL2433; SAV6189, S. avermitilis MA-4680; SCO2026, S. coelicolor A3(2); SACE3998 and SACE5742, Saccharopolyspora erythraea NRRL2338. (C) Sequence alignments of NADPH-dependent glutamate synthase β subunits showing the conserved regions used for designing degenerated oligoprimer HEI-C. GluS-β, S. lydicus NRRL2433; SAV6190 and SAV6258, S. avermitilis MA-4680; SCO1977 and SCO2025, S. coelicolor A3(2); SACE3997 and SACE5741, Saccharopolyspora erythraea NRRL2338. (D) Sequence alignments of ferredoxin-dependent glutamate synthase showing the conserved regions used for designing degenerated oligoprimers HEI-FR1 and HEI-FR2. SlgE3 and GluS-FD, S. lydicus NRRL2433; SAV954 and SAV1232, S. avermitilis MA-4680; SSEG09946; S. sviceus ATCC 29083; SSAG01435, Streptomyces sp. strain Mg1. aa, amino acids.
Isolation of total RNA.
Mycelium corresponding to cultures from S. lydicus NRRL2433 was obtained at 12, 24, 48, and 72 h during growth in R5A liquid medium. Mycelium corresponding to cultures from S. lydicus SLME3 and SLME1E2 was obtained at 72 h during growth in R5A liquid medium. Samples (15 ml) of each culture were mixed with 2 volumes of RNAprotect bacteria reagent (Qiagen) and, after vortexing, mycelia were harvested by centrifugation and immediately frozen at −70°C according to the manufacturer's instructions. The total RNA was extracted from frozen mycelium using the lysis method from the Kirby procedure (12) and purified with a RNeasy Midi kit (Qiagen) according to the manufacturer's instructions. RNA preparations were subjected to additional DNase I treatments (RNase-Free; Qiagen) to eliminate possible chromosomal DNA contamination. The RNA concentration was determined by measuring the absorbance at 260 nm.
Gene expression analysis by reverse transcriptase PCR (RT-PCR).
Transcript detection analysis was carried out by using the SuperScript One-Step RT-PCR with Platinum Taq DNA polymerase (Invitrogen) with 100 ng of total RNA as a template. DMSO (5% [vol/vol], final) was added to all reactions, along with RNAguard RNase inhibitor (Amersham Biosciences; 32.2 U per reaction). The conditions were as follows: first-strand cDNA synthesis at 50°C for 30 min, followed by 94°C for 2 min, and then 30 amplification cycles of 95°C for 1 min, 61°C for 1 min, and 68°C for 2 min. Primers (18- to 23-mers; melting temperature [Tm], 65°C) (Table 1) were designed with the aid of the software Primer Express v.2.0 (Applied Biosystems) to generate PCR products of ∼600 bp. The exception was the slgE2-specific primers CRIS3/CRIS4, which were designed to give a product of 840 bp. Negative controls for each pair of primers were carried out with Platinum Taq DNA polymerase (Invitrogen) in the absence of reverse transcriptase to confirm that amplified products were not due to the presence of contaminating chromosomal DNA in RNA preparations. The oligonucleotide primers HRDB-GB1-F and HRDB-GB2-R for hrdB, encoding the constitutively expressed housekeeping sigma factor, were used as an internal control to assess the quality of RNA (24). The amount of RNA used for hrdB detection was 300 ng. The RT-PCR analysis was carried out at least three times for each pair of primers, and the RT-PCR products were separated in agarose gels and visualized by ethidium bromide staining. The identity of the PCR products was verified by direct sequencing with one of the amplification primers.
Gene expression analysis by qRT-PCR.
Transcript detection analysis by quantitative RT-PCR (qRT-PCR) was performed with 0.2 μg of RNA. cDNA was generated with the iScript cDNA synthesis kit (Bio-Rad). PCR amplification was performed in a 7500 Fast Real-Time PCR System (Applied Biosystems, Warrington, United Kingdom). The data were analyzed with the software provided by the supplier. Amplification was carried out in 25 μl containing 0.5 μg of cDNA, 1× Power SYBR green (Applied Biosystems), and each primer (Table 1) at a concentration of 0.4 μM. After incubation at 95°C for 10 min, amplification proceeded with 40 cycles of 95°C for 15 s and 60°C for 1 min. The efficiencies of the primer sets were measured using a dilution series of cDNA. The raw threshold cycle (CT) values were converted to relative expression levels by the 2−ΔΔCT method (15) to quantify the relative gene expression. Based on the sequence of hrdB amplified using primers HRDB-GB1-F and HRDB-GB2-R, two new oligonucleotides (HRDBqRT1 and HRDBqRT2) were designed and used for amplification of the hrdB transcript as an internal control to quantify the relative expression of target genes.
RESULTS
Insertional inactivation of glutamate synthase slgE3.
SlgE3 shows high similarity to ferredoxin-dependent GSs and to alpha subunits of NADPH-dependent GSs from different microorganisms with the highest similarity scores being shown by the GSs from the archaean bacteria Caldivirga maquilingensis IC-167 (ZP_01710164), Thermofilum pendens Hrk 5 (YP_920540), and Methanothermobacter thermautotrophicus strain Delta H (NP_275248). The involvement of SlgE3 in streptolydigin biosynthesis was demonstrated by deleting slgE3 from S. lydicus (Fig. 2A). Analysis of the products accumulated by mutant SLM3 (Fig. 2B) showed the production of very small amounts of streptolydigin with a UPLC retention time of 6.48 min. Analysis of this peak by MS showed two ions with masses of 601 and 487 m/z [M+H]+ corresponding to the unfragmented compound and the aglycone fragment, respectively. The mutant SLM3 produced only 2 to 5% of the level of streptolydigin produced by the wild type. Production of streptolydigin in mutant SLME3 was fully restored by the introduction of pEM4TslgE3 containing slgE3 under the control of the ermE* promoter (Fig. 2C). These experiments indicate that the active participation of SlgE3 is essential for the usual production yields of streptolydigin in the wild-type strain. In addition, the results also suggest that a second GS can partially sustain the supply of glutamate for the biosynthesis of the antibiotic.
Insertional inactivation of glutamate mutase slgE1 and slgE2.
SlgE1 and SlgE2 show significant similarities to VinH and VinI, respectively, S and E subunits of coenzyme B12 (adenosylcobalamin)-dependent mutase from S. halstedii (19). They are also similar to the pairs NikU-NikV, SanU-SanV, and GlmA-GlmB, S and E subunits of GMs from S. tendae, S. ansochromogenes, and Actinoplanes friuliensis involved in the biosynthesis of nikkomycin (13, 14) and friulimycin (10), respectively. The participation of slgE1 and slgE2 in streptolydigin biosynthesis was assessed by the simultaneous deletion of both genes from S. lydicus, thus generating mutant SLME1E2 (Fig. 2D). Analysis of the products accumulated by this mutant showed the production of two compounds (compounds 2 and 3) with UV spectra characteristic of streptolydigin (Fig. 2E). Compound 2 showed a UPLC retention time of 6.10 min and masses of 587 and 473 m/z [M+H]+, ions corresponding to the unfragmented compound and the aglycone fragment, respectively. This compound was identical to streptolydigin B (Fig. 3A), previously characterized form S. lydicus mutant SLMZ (11). Compound 3 presented UPLC retention time of 6.50 min and masses of 601 and 487 m/z [M+H]+. According to these masses, this compound could correspond to streptolydigin. However, after purification of compound 3 and coinjection with authentic samples of streptolydigin in UPLC, these compounds did not comigrate. Structural elucidation of compound 3 was carried out using one-dimensional 1H, two-dimensional 1H COSY, 1H, 13C HSQC-edited, and HMBC NMR experiments (see Table S1 in the supplemental material). This allowed the identification of this compound as a novel streptolydigin derivative carrying a tetramic acid lateral side chain directly derived from glutamate instead of 3-methyl-aspartate and decorated by amidation of the carboxyl group and subsequent methylation of the amino group. We designated this compound streptolydigin C (Fig. 3A). The production of streptolydigin by mutant SLME1E2 was restored by the introduction of pEM4TslgE1E2 containing slgE1 and slgE2 under the control of the ermE* promoter (Fig. 2F).
Streptolydigin C, when tested for its antibacterial activity against S. albus, showed a halo of inhibition slightly bigger than that of streptolydigin (4.0 versus 3.2 cm in diameter) (Fig. 3B) or streptolydigin LA (3.0 cm in diameter) (21) and showed considerably more activity than that of streptolydigin B (2.5 cm in diameter) reported previously (11).
Expression of slgE3 and slgE1-slgE2 in S. lydicus.
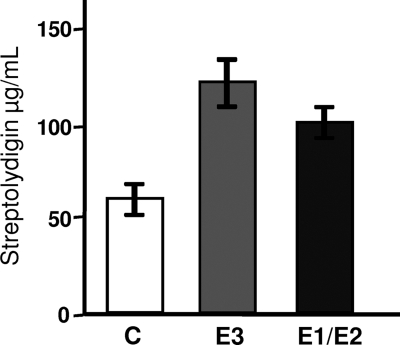
The effect of slgE3 and slgE1-slgE2 on the production of streptolydigin was also assessed by expressing these genes in S. lydicus wild type by using constructs pEM4TslgE3 and pEM4TslgE1E2. In both cases the genes were under the control of the ermE* promoter. Overexpression of slgE3 led to a 2-fold increase in streptolydigin production compared to the control S. lydicus/pEM4T (Fig. 4). Overexpression of slgE1-slgE2 led to a 1.6-fold increase in streptolydigin production compared to S. lydicus/pEM4T (Fig. 4).
Fig. 4.
Effect of slgE3 or slgE1-slgE2 overexpression in S. lydicus on streptolydigin production. Cultures were performed on R5A solid medium, and streptolydigin production was determined by high-pressure liquid chromatography analysis. Experiments were run in triplicate. C, S. lycidus/pEM4T; E3, S. lydicus/pEM4TslgE3; E1/E2, S. lydicus/pEM4TslgE1E2.
Identification of housekeeping glutamate synthases.
The reduced amount of streptolydigin produced by mutant SLME3 implies the possible participation of another GS, different from SlgE3, in supplying glutamate for streptolydigin biosynthesis. To identify additional GS-encoding genes in the chromosome of S. lydicus, we used the PCR and oligoprimers derived from consensus amino acid sequences obtained by comparison of NADPH- and ferredoxin-dependent GS from different actinomycetes (Table 1 and Fig. 5). The PCR product obtained using the oligoprimers HEI-I and HEI-J was a fragment of 620 bp. Sequencing of this fragment and comparison of its deduced amino acid sequence with proteins in databases showed that it would code for an internal region of an NADPH-dependent GS α subunit (gluS-α) (Fig. 5A and B). The highest similarity score obtained was with a putative NADPH-dependent GS large subunit from Streptomyces sp. strain C (ZP_05506017) (99% identity, 100% similarity). The sequence of the 1,316-bp PCR product obtained using oligoprimers HEI-F and HEI-C showed 437 bp coding for the C-terminal end of an NADPH-dependent GS α subunit (gluS-α) and 887 bp for the N-terminal end of an NADPH-dependent GS β subunit (gluS-β) (Fig. 5A to C). The highest scores were with an NADPH-dependent GS large subunit from Streptomyces flavogriseus ATCC 33331 (ZP_04995602) (84% identity, 89% similarity) and an NADPH-dependent GS small subunit from Streptomyces sp. strain ACTE (ZP_06271158) (80% identity, 85% similarity). A PCR product of 520 bp was obtained using the oligoprimers HEI-FR1 and HEI-FR2, which could code for an internal region of a ferredoxin-dependent GS (gluS-FD) (Fig. 5). The best match was a ferredoxin-dependent GS from Streptomyces sp. strain AA4 (ZP_05479083) (85% identity, 91% similarity). The nucleotide sequences of the three PCR products have been deposited in the EMBL database with accession codes FR819656, FR819657, and FR819658, respectively. These results suggest the existence of at least two additional GS in S. lydicus: an NADPH-dependent GS constituted by two subunits (GluS-α and GluS-β) and a ferredoxin-dependent GS formed by a single polypeptide (GluS-FD).
We also attempted the identification of additional GM encoding genes in the chromosome of S. lydicus using a similar approach. Oligoprimers were designed from consensus amino acid sequences obtained after comparison of GM from different actinomycetes (data not shown). Furthermore, slgE1 and slgE2 were also used as probes for Southern hybridization. None of these approaches was successful, indicating that probably only the GM encoded by slgE1 and slgE2 is present in the streptolydigin producer organism, which would account for the results obtained by deleting these genes in S. lydicus (see above).
Gene expression analysis of glutamate synthases and glutamate mutase.
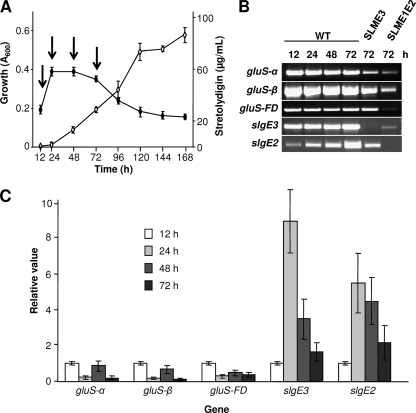
The expression of GS genes slgE3, gluS-α, gluS-β, and gluS-FD and GM gene slgE2 was analyzed in S. lydicus and in mutants SLME3 and SLME1E2 by RT-PCR in order to determine their temporal expression patterns in connection with the biosynthesis of streptolydigin. When monitored in R5A liquid medium, biosynthesis of streptolydigin occurred in a growth-phase-dependent manner and the antibiotic was first detected at 24 h when cultures entered the stationary phase (Fig. 6A). Total RNA was isolated at 12, 24, 48, and 72 h from cultures of the S. lydicus wild-type strain and at 72 h from cultures of the mutant strains. The expression of all four GSs was apparently constitutive, including the expression of slgE3. As expected, the three housekeeping GS genes were expressed in both streptolydigin-nonproducing mutants SLME3 and SLME1E2. In addition, expression of slgE3 was also confirmed in GM mutant SLME1E2, but it was absent from GS mutant SLME3. Expression of GM slgE2 increased after streptolydigin biosynthesis. This gene was also expressed in GS mutant SLME3 (Fig. 6B).
Fig. 6.
(A) Growth and streptolydigin production by S. lydicus wild-type strain in R5A medium. Growth was monitored by measuring the absorbance at 600 nm for determining DNA content by the diphenylamine assay. Streptolydigin production was spectrophotometrically quantified at 360 nm by peak area integration (⋄). Experiments were run in triplicate. The arrows show time points when total RNA was isolated. (B) Transcriptional analysis of S. lydicus GSs (slgE3, gluS-α, gluS-β, and gluS-FD) and GM (slgE2) by RT-PCR. RNA samples were obtained, during growth in R5A liquid medium, at 12, 24, 48, and 72 h from S. lydicus wild type and at 72 h from S. lydicus SLME3 and SLME1E2. (C) Quantification of gene expression by qRT-PCR. The hrdB gene was used as an internal control to quantify the relative expression of target genes. The expression level of each gene at 12 h was taken as the calibrator. Error bars were calculated from three independent determinations of mRNA abundance in each sample.
Expression of GSs and GM was quantified by qRT-PCR using the same RNA samples. The value obtained for the 12 h sample was fixed arbitrarily at 1 for reference (Fig. 6C). The expression level of the housekeeping GSs (gluS-α, gluS-β, and gluS-FD) decreased to one-fourth, coinciding with the entry into the stationary phase (24-h sample). However, expression of the streptolydigin-related GS slgE3 and GM slgE2 considerably increased at that time (24 h) 9- and 5.5-fold, respectively (Fig. 6C), coinciding with the onset of streptolydigin production (Fig. 6A). Afterward, at 48 and 72 h, the expression of both genes decreased but kept at 72 h a 2-fold expression with respect to the 12-h sample.
DISCUSSION
Streptolydigin biosynthesis requires the incorporation of a 3-methylasparagine moiety. Several genes from the streptolydigin biosynthesis gene cluster have been proposed to encode enzymes that participate in 3-methylaspartate biosynthesis. SlgE3 has been proposed to be involved in the specific supply of glutamate derived from 2-oxoglutarate and glutamine. SlgE1 and SlgE2 have been proposed to catalyze the conversion of glutamate into 3-methylaspartate. This amino is thought to be activated and loaded onto the SlgN2 NRPS PCP domain by catalyzed by the SlgN1 and SlgN2 adenylation domains acting together. Once generated, the NRPS-bound substrate would be further processed by the activity of 3-methylasparagine synthetase SlgZ, followed by N methylation by SlgM to generate NRPS-bound N-methyl-3-methylasparagine (Fig. 1) (11). N-Methyl-3-methylasparagine would be condensed with the preformed streptolydigin polyketide chain, followed by the remaining biosynthetic steps: release and cyclization of the polyketide-nonribosomal peptide to generate the tetramic acid moiety, epoxidation to obtain streptolydiginone (streptolydigin aglycon), and finally attachment of l-rhodinose to obtain the final product (Fig. 1) (21).
The participation of streptolydigin GS and GM enzymes in the biosynthetic process has been confirmed by inactivation of the corresponding genes and their expression in S. lydicus wild type. Deletion of slgE3 reduced the production of streptolydigin to low levels, indicating the importance of ferredoxin-dependent GS SlgE3 in supporting antibiotic biosynthesis by supplying glutamic acid. On the other hand, expression of slgE3 in S. lydicus enhanced antibiotic production. Furthermore, the viability of mutant SLME3 is not compromised by the absence of SlgE3 since three other GS encoding genes have been identified in S. lydicus. Two of these genes (gluS-α and gluS-β) encode the α and β subunits of an NADPH-dependent GS, while the third (gluS-FD) encodes a ferredoxin-dependent GS. These housekeeping GSs, which support primary metabolism, have been shown to be expressed throughout S. lydicus growth, their expression decreasing when the strain reaches the stationary phase. In contrast, the expression of slgE3 sharply increases coinciding with the onset of streptolydigin biosynthesis.
Deletion of slgE1 and slgE2 abolished streptolydigin production leading to the accumulation of two streptolydigin derivatives streptolydigin B and streptolydigin C both containing glutamate but differing in an N-methyl group present in streptolydigin C and absent in streptolydigin B. The production of streptolydigin B has been observed before in an SLMZ mutant affected in ammonia-dependent 3-methylasparagine synthetase slgZ, which implies that the streptolydigin NRPS is flexible enough to accept glutamate instead of 3-methylaspartate as a substrate for the biosynthesis of streptolydigin or streptolydigin B (11). In addition, the production of streptolydigin C, with a tetramic lateral side chain modified by amidation and methylation, implies the flexibility of both SlgZ and SlgM for amidating and methylating glutamyl-NRPS and glutaminyl-NRPS intermediates, respectively. A similar effect has been shown by disruption of a GM gene involved in the biosynthesis of vicenistatin. Inactivation of vinI from S. halstedii led to abolition of vicenistatin production and to the accumulation of desmethylvicenistatins (18). On the other hand, the expression of slgE1 and slgE2 in S. lydicus improved streptolydigin production, a positive effect that has also been observed in the production of nikkomycin by S. ansochromogenes after the expression of sanU and sanV (14). In accordance with its proposed involvement in streptolydigin biosynthesis, an increase in slgE2 expression was observed coinciding with the production of the tetramic acid.
Comparison of the antibacterial activity of streptolydigin, streptolydigin B, and streptolydigin C gives some clues to a structure-function relationship. Substitution of 3-methylaspartate (as in streptolydigin) by glutamate (as in streptolydigin C) apparently increases the antibacterial activity. In addition, the presence of an acetamide moiety (as in streptolydigin C) clearly improves the biological activity in comparison with a compound lacking this group (as in streptolydigin B). This acetamide moiety has been shown to be important for the interaction of streptolydigin with the RNA polymerase (28).
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by grants of the Spanish Ministry of Science and Innovation (BFU2006-00404) and Red Temática de Investigación Cooperativa de Centros de Cáncer (Ministry of Health, ISCIII-RETIC RD06/0020/0026) to J.A.S. We also acknowledge Obra Social Cajastur for financial support to C.O., the Spanish Ministry of Science and Innovation for a Ph.D. student fellowship (FPI) to C.G., and the Spanish Ministry of Science and Innovation (SAF2008-01845) and the Centro de Investigación Príncipe Felipe for their financial support.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 10 June 2011.
REFERENCES
- 1. Altschul S. F., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Burton K. 1956. A study of the conditions and mechanism of the diphenylamine reaction for the colorimetric estimation of DNA. Biochem. J. 62:315–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen H., Harrison P. H. 2004. Investigation of the origin of C2 units in biosynthesis of streptolydigin. Org. Lett. 6:4033–4036 [DOI] [PubMed] [Google Scholar]
- 4. Chen H., Olesen S. G., Harrison P. H. 2006. Biosynthesis of streptolydigin: origin of the oxygen atoms. Org. Lett. 8:5329–5332 [DOI] [PubMed] [Google Scholar]
- 5. Devereux J., Haeberli P., Smithies O. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dicioccio R. A., Srivastava B. I. 1976. Selective inhibition of terminal deoxynucleotidyltransferase from leukemic cells by streptolydigin. Biochem. Biophys. Res. Commun. 72:1343–1349 [DOI] [PubMed] [Google Scholar]
- 7. Dicioccio R. A., et al. 1980. Structure-activity relationship, selectivity and mode of inhibition of terminal deoxyribonucleotidyltransferase by streptolydigin analogs. Biochem. Pharmacol. 29:2001–2008 [DOI] [PubMed] [Google Scholar]
- 8. Fernández E., et al. 1998. Identification of two genes from Streptomyces argillaceus encoding glycosyltransferases involved in transfer of a disaccharide during biosynthesis of the antitumor drug mithramycin. J. Bacteriol. 180:4929–4937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fischbach M. A., Walsh C. T. 2006. Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: logic, machinery, and mechanisms. Chem. Rev. 106:3468–3496 [DOI] [PubMed] [Google Scholar]
- 10. Heinzelmann E., et al. 2003. A glutamate mutase is involved in the biosynthesis of the lipopeptide antibiotic friulimicin in Actinoplanes friuliensis. Antimicrob. Agents Chemother. 47:447–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Horna D. H., et al. 11 March 2011. Biosynthesis of the RNA polymerase inhibitor streptolydigin in Streptomyces lydicus: tailoring modification of 3-methyl-aspartate. J. Bacteriol. 193:2647–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kieser T., Bibb M. J., Buttner M. J., Chater K. F., Hopwood D. A. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom [Google Scholar]
- 13. Lauer B., et al. 2001. Molecular characterization of co-transcribed genes from Streptomyces tendae Tü901 involved in the biosynthesis of the peptidyl moiety and assembly of the peptidyl nucleoside antibiotic nikkomycin. Mol. Gen. Genet. 264:662–673 [DOI] [PubMed] [Google Scholar]
- 14. Li Y., Ling H., Li W., Tan H. 2005. Improvement of nikkomycin production by enhanced copy of sanU and sanV in Streptomyces ansochromogenes and characterization of a novel glutamate mutase encoded by sanU and sanV. Metab. Eng. 7:165–173 [DOI] [PubMed] [Google Scholar]
- 15. Livak K. J., Schmittgen T. D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 16. Méndez C., Braña A. F., Manzanal M. B., Hardisson C. 1985. Role of substrate mycelium in colony development in Streptomyces. Can. J. Microbiol. 31:446–450 [DOI] [PubMed] [Google Scholar]
- 17. Menéndez N., Nur-e-Alam M., et al. 2006. Deoxysugar transfer during chromomycin A3 biosynthesis in Streptomyces griseus subsp. griseus: new derivatives with antitumor activity. Appl. Environ. Microbiol. 72:167–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ogasawara Y., Kakinuma K., Eguchi T. 2005. Involvement of glutamate mutase in the biosynthesis of the unique starter unit of the macrolactam polyketide antibiotic vicenistatin. J. Antibiot. 58:468–472 [DOI] [PubMed] [Google Scholar]
- 19. Ogasawara Y., et al. 2004. Cloning, sequencing, and functional analysis of the biosynthetic gene cluster of macrolactam antibiotic vicenistatin in Streptomyces halstedii. Chem. Biol. 11:79–86 [DOI] [PubMed] [Google Scholar]
- 20. Olano C., et al. 2004. Biosynthesis of the angiogenesis inhibitor borrelidin by Streptomyces parvulus Tü4055: cluster analysis and assignment of functions. Chem. Biol. 11:87–97 [DOI] [PubMed] [Google Scholar]
- 21. Olano C., et al. 2009. Deciphering biosynthesis of the RNA polymerase inhibitor streptolydigin and generation of glycosylated derivatives. Chem. Biol. 16:1031–1044 [DOI] [PubMed] [Google Scholar]
- 22. Pearce C. J., Rinehart K. L., Jr 1983. The use of doubly-labeled 13C-acetate in the study of streptolydigin biosynthesis. J. Antibiot. 36:1536–1538 [DOI] [PubMed] [Google Scholar]
- 23. Pearce C. J., Ulrich S. E., Rinehart K. L., Jr 1980. Biosynthetic incorporation of propionate and methionine into streptolydigin. J. Am. Chem. Soc. 102:2510–2512 [Google Scholar]
- 24. Rodríguez M., et al. 2008. Identification of transcriptional activators for thienamycin and cephamycin C biosynthetic genes within the thienamycin gene cluster from Streptomyces cattleya. Mol. Microbiol. 69:633–645 [DOI] [PubMed] [Google Scholar]
- 25. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 26. Sanger F., Nicklen S., Coulson A. R. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U. S. A. 74:5463–5467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Siddhikol C., Erbstoeszer J. W., Weisblum B. 1969. Mode of action of streptolydigin. J. Bacteriol. 99:151–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Temiakov D., et al. 2005. Structural basis of transcription inhibition by antibiotic streptolydigin. Mol. Cell 19:655–666 [DOI] [PubMed] [Google Scholar]
- 29. von Meyenburg K., et al. 1978. Reevaluation of the mode of action of streptolydigin in Escherichia coli: induction of transcription termination in vivo. Antimicrob. Agents Chemother. 13:234–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.