Abstract
The RimM protein in Escherichia coli is important for the in vivo maturation of 30S ribosomal subunits and a ΔrimM mutant grows poorly due to assembly and translational defects. These deficiencies are suppressed partially by mutations that increase the synthesis of another assembly protein, RbfA, encoded by the metY-nusA-infB operon. Among these suppressors are mutations in nusA that impair the NusA-mediated negative-feedback regulation at internal intrinsic transcriptional terminators of the metY-nusA-infB operon. We describe here the isolation of two new mutations, one in rpoB and one in rpoC (encoding the β and β′ subunits of the RNA polymerase, respectively), that increase the synthesis of RbfA by preventing NusA from stimulating termination at the internal intrinsic transcriptional terminators of the metY-nusA-infB operon. The rpoB2063 mutation changed the isoleucine in position 905 of the β flap-tip helix to a serine, while the rpoC2064 mutation duplicated positions 415 to 416 (valine-isoleucine) at the base of the β′ dock domain. These findings support previously published in vitro results, which have suggested that the β flap-tip helix and β′ dock domain at either side of the RNA exit tunnel mediate the binding to NusA during transcriptional pausing and termination.
INTRODUCTION
The synthesis of ribosomes in bacteria such as Escherichia coli is highly efficient probably because of the action of ribosome maturation proteins that are not part of the mature ribosomal subunits. One of these, the RimM protein, binds to r-protein S19 in the 30S subunits (24) and also to S19 free in solution (46) and facilitates the incorporation of S19 during in vitro assembly of the 30S subunits (4). Mutants lacking RimM show a 7-fold-decreased growth rate and a reduced translational efficiency, resulting from a deficiency in the maturation of the 30S subunits (6, 7). Specific alterations in r-protein S13 or an increased expression of another ribosome maturation protein, RbfA, partially suppress the slow growth and translational deficiency of a ΔrimM102 mutant (6, 7).
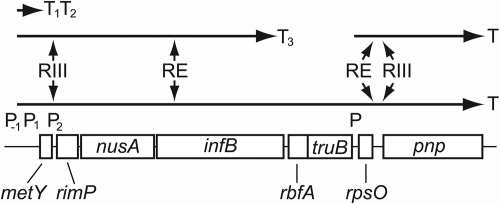
RbfA is encoded by the metY-nusA-infB operon (Fig. 1), which contains, in the direction of transcription, the metY gene encoding a minor form of the initiator tRNA (19), the rimP gene (formerly p15a or yhbC) for the ribosome maturation protein RimP (19, 31), the nusA gene for the transcriptional elongation factor NusA (11, 18, 41), the infB gene encoding the translation initiation factor IF2 (35, 39), the rbfA gene (12, 43), and the truB gene for the tRNA Ψ55 synthase (32, 43). The metY-nusA-infB operon contains two promoters upstream from metY, P−1 (15) and P1 (19, 26), and a minor promoter, P2, located between metY and rimP (37). The cleavage by RNase III at sites between metY and rimP on the polycistronic mRNA initiates the rapid degradation of the downstream RNA (37). There are two rho-independent transcriptional terminators, T1 (CCCCGATTTATCGGGGTTTTTT) and T2 (GGGCTTTAGGCCCTTTTTTT), between metY and rimP (18, 37) and one, T3 (GGGGCTAACAGCCCCTTTTT), between infB and rbfA (43).
Fig. 1.
Genetic organization of the metY-nusA-infB operon region of the chromosome. The gene metY encodes a minor form of the initiator tRNA, rimP encodes the ribosome maturation protein RimP, nusA encodes the transcriptional elongation factor NusA, infB encodes the translation initiation factor IF2; rbfA encodes the ribosome binding factor RbfA, truB encodes the tRNA Ψ55 synthase, rpsO encodes ribosomal protein S15, and pnp encodes polynucleotide phosphorylase. P−1, P1, P2, and P indicate the locations of promoters, and T1, T2, T3, and T indicate intrinsic transcriptional terminators, while RE and RIII show the cleavage sites for RNase E and RNase III, respectively. Horizontal arrows represent transcriptional products.
Of 29 previously isolated suppressor mutations that increased the growth rate of the ΔrimM102 mutant MW37, three were in rpsM, encoding r-protein S13 (6), and 23 were genetically linked to the metY-infB-nusA operon (7). At least 13 of the latter mutations increased the synthesis of RbfA: one was a duplication in which one copy of the rbfA gene was downstream from a promoter for the yhbM gene; another was an insertion of IS2 in infB that created a new promoter for rbfA; three deleted the transcriptional terminator between infB and rbfA; and eight were in the nusA gene, of which at least seven resulted in a deficiency in NusA-mediated feedback regulation at the two rho-independent terminators between metY and rimP (5). We describe here the identification of the three remaining suppressor mutations and the characterization of strains harboring these mutations. The mutations were localized to rpoB and rpoC, encoding the β and β′ subunit of the RNA polymerase, respectively, and shown to increase the expression of rbfA by abolishing NusA-mediated negative-feedback regulation at internal transcriptional terminators of the metY-nusA-infB operon.
MATERIALS AND METHODS
Strains and plasmids.
Strains and plasmids used are listed in Table 1. A library of mini-Tn10Cm insertions was constructed on the sdr-47 (suppressor to deletion of rimM) containing strain PW113 by using λNK1324 as described previously (22). Several clones with a mini-Tn10Cm linked to sdr-47 were identified by transducing strain MW37 (ΔrimM102) with phage P1 grown on the library, selecting for chloramphenicol resistance (Cmr) and screening for faster growth than that of strain MW37. One clone had a mini-Tn10Cm (xxx-2423::miniTn10Cm) ca. 50% linked to sdr-47 as demonstrated by backcrosses to strain MW37 (ΔrimM102). From one of the backcrosses, strain GOB125 (ΔrimM102 sdr-47 xxx-2423::miniTn10Cm) was isolated and used to transfer sdr-47 to a rimM+ background by P1 transduction of the wild-type strain MW100, resulting in strain GOB250 (sdr-47 xxx-2423::miniTn10Cm) that showed a temperature-sensitive growth phenotype. To examine whether also the suppressor mutations sdr-36 and sdr-46 were linked to xxx-2423::miniTn10Cm, phage P1 grown on strain GOB249 (rimM+ xxx-2423::miniTn10Cm), congenic to strain GOB250, was used to transduce the suppressor strains PW102 (ΔrimM102 sdr-36) and PW112 (ΔrimM102 sdr-46) selecting for Cmr. The growth of the obtained transductants was examined by scoring the colony sizes after single-cell streaks on rich medium plates and sdr-36 and sdr-46, as well as sdr+, derivatives were obtained showing that xxx-2423::miniTn10Cm was genetically linked also to sdr-36 and sdr-46. Accordingly, strains MW171 (ΔrimM102 sdr+ xxx-2423::miniTn10Cm), MW172 (ΔrimM102 sdr-46 xxx-2423::miniTn10Cm), and GOB307 (ΔrimM102 sdr-36 xxx-2423::miniTn10Cm) were generated. The suppressor mutations sdr-36 and sdr-46 were transferred from strains GOB307 and MW172, respectively, to the wild-type strain MW100 by P1 transduction and the resulting strains, MW177 (sdr-36 xxx-2423::miniTn10Cm) and MW181 (sdr-46 xxx-2423::miniTn10Cm) showed temperature-sensitive growth like the sdr-47 containing strain GOB250 (data not shown). Strain MW176 (sdr+ xxx-2423::miniTn10Cm) was isolated as a temperature-resistant clone from the former transduction. Both sdr-36 and sdr-46 were shown to be ca. 50% linked to xxx-2423::miniTn10Cm. Strains MW234 and MW237 were constructed by replacing xxx-2423::miniTn10Cm of strains MW181 and MW177, respectively, with thi-39::Tn10 from CAG18500 by P1 transduction. Strains GOB896 and GOB898 were constructed by transducing strain GOB434, containing a metY-metYt1t2-lacZ fusion in the lacI-lacZ region of the chromosome, with P1 grown on strain MW234 selecting for thi-39::Tn10 linked to sdr-46 and screening for the presence of sdr-46 or sdr+. Strains GOB900 and GOB902 were constructed in the same way using strain MW237 as donor of sdr-36. Strains GOB904, GOB906, GOB908, and GOB910 were constructed in similar ways using strain GOB435, which contains a metY-lacZ fusion, as the recipient. Similarly, strains MW286 (sdr-46) and MW288 (sdr+) are transductants of GOB838, which contains a metY-TrplS-lacZ fusion, using strain MW234 as the donor, while strain MW289 (sdr-36) was obtained using strain MW237 as the donor.
Table 1.
Bacterial strains, plasmids, and oligonucleotides
| Strain, plasmid, or oligonucleotide | Genotype or sequence | Source or referencea |
|---|---|---|
| Strains | ||
| CAG18500 | MG1655 thi-39::Tn10 | 45 |
| GOB125 | Hfr P4X ΔrimM102 yfiB::nptI rpoB2063 (sdr-47) rsd2423::miniTn10Cm | |
| GOB162 | Hfr P4X rbfA::Kmr | 7 |
| GOB249 | Hfr P4X rsd2423::miniTn10Cm | |
| GOB250 | Hfr P4X rpoB2063 (sdr-47) rsd2423::miniTn10Cm | |
| GOB307 | Hfr P4X ΔrimM102 yfiB::nptI rpoC2064 (sdr-36) rsd2423::miniTn10Cm | |
| GOB434 | Hfr P4X lacI′-TrplS-PmetY-metY-metY-T1T2-lacZ | 5 |
| GOB435 | Hfr P4X lacI′-TrplS-PmetY-metY-lacZ | 5 |
| GOB750 | GOB434 nusA94 argG2424::miniTn10Cm | 5 |
| GOB752 | GOB435 nusA94 argG2424::miniTn10Cm | 5 |
| GOB838 | Hfr P4X lacI′-rplSt-PmetY-metY-TrplS-lacZ | 5 |
| GOB868 | GOB838 nusA94 arg2424::miniTn10Cm | 5 |
| GOB896 | GOB434 rpoB2063 (sdr-46) thi-39::Tn10 | |
| GOB898 | GOB434 rpoB+thi-39::Tn10 | |
| GOB900 | GOB434 rpoC2064 (sdr-36) thi-39::Tn10 | |
| GOB902 | GOB434 rpoC+thi-39::Tn10 | |
| GOB904 | GOB435 rpoB2063 (sdr-46) thi-39::Tn10 | |
| GOB906 | GOB435 rpoB+thi-39::Tn10 | |
| GOB906 | GOB435 rpoB+thi-39::Tn10 | |
| GOB908 | GOB435 rpoC2064 (sdr-36) thi-39::Tn10 | |
| GOB910 | GOB435 rpoC+thi-39::Tn10 | |
| MW37 | Hfr P4X ΔrimM102 yfiB::nptI | 33 |
| MW100 | Hfr P4X | 51 |
| MW234 | Hfr P4X rpoB2063 (sdr-46) thi-39::Tn10 | |
| MW237 | Hfr P4X rpoC2064 (sdr-36) thi-39::Tn10 | |
| MW171 | Hfr P4X ΔrimM102 yfiB::nptI rsd2423::miniTn10Cm | |
| MW172 | Hfr P4X ΔrimM102 yfiB::nptI rpoB2063 (sdr-46) rsd2423::miniTn10Cm | |
| MW176 | Hfr P4X rsd2423::miniTn10Cm | |
| MW177 | Hfr P4X rpoC2064 (sdr-36) rsd2423::miniTn10Cm | |
| MW181 | Hfr P4X rpoB2063 (sdr-46) rsd2423::miniTn10Cm | |
| MW286 | GOB838 rpoB2063 (sdr-46) thi-39::Tn10 | |
| MW288 | GOB838 rpoB+thi-39::Tn10 | |
| MW289 | GOB838 rpoC2064 (sdr-36) thi-39::Tn10 | |
| PW102 | Hfr P4X ΔrimM102 yfiB::nptI sdr-36 = rpoC2064 (Dupl. of codons 115 to 116; AGTTAT) | 7 |
| PW112 | Hfr P4X ΔrimM102 yfiB::nptI sdr-46 = rpoB2063 (ATC to AGC in codon 905) | 7 |
| PW113 | Hfr P4X ΔrimM102 yfiB::nptI sdr-47 = rpoB2063 (ATC to AGC in codon 905) | 7 |
| PW157 | Hfr P4X ΔrimM102 yfiB::nptI zhc-2421::Tn10 | G. O. Bylund et al.b |
| Plasmids | ||
| pCW2 | Strr Spcr′rpoC yjaZ thiCEFSGH rsd2423::miniTn10Cm nudC hemE nfi yjaG hupA yjaH zraP zraS′ | |
| pJML007 | bla cat′ araC PBAD-nusA | 5 |
| pMW498 | bla cat′ araC PBAD-His6-nusA | |
| Oligonucleotides | ||
| nusA-BamHI-F | 5′-TTTTGGATCCAACAAAGAAATTTTGGCTGTAG-3′ | |
| nusA-HindIII-R | 5′-TTTTAAGCTTTATTACGCTTCGTCACCG-3′ |
Unless otherwise noted, the origin was this study.
G. O. Bylund, O. P. Persson, J. M. Lövgren, and P. W. Wikström, unpublished data.
Plasmid pMW498 expressing his6-nusA from the PBAD promoter was constructed in two steps. First, a DNA fragment containing the nusA gene was amplified by PCR using the oligonucleotides nusA-BamHI-F and nusA-HindIII-R as primers, digested with BamHI and HindIII, and ligated into BamHI/HindIII-digested His tag vector pSTN016 (31). From the resulting plasmid, pMW493, an EcoRI/HindIII fragment containing the ribosome binding site and his6-nusA was cloned into EcoRI/HindIII-digested pBAD30 (16).
Media and growth conditions.
The minimal medium used was morpholinepropanesulfonic acid (MOPS) (30) supplemented with 0.4% glucose. Rich medium was either rich MOPS (29) or LB (2) supplemented with E+B1 (49). Cultures were grown at 37°C, and the growth was monitored either on a Zeiss PMQ3 or a Shimadzu UV-1601 spectrophotometer at 420 or 600 nm or on a Klett-Summerson colorimeter equipped with a red filter.
Localization of xxx-2423::miniTn10Cm.
To localize xxx-2423::miniTn10Cm, chromosomal DNA from GOB125 (ΔrimM102 sdr-47 xxx-2423::miniTn10Cm) was digested with PstI and cloned into the low-copy-number vector pCL1921 (23), selecting for Cmr conferred by xxx-2423::miniTn10Cm. One resulting plasmid clone, pCW2, was demonstrated by DNA sequencing with vector specific primers to contain the 90 min region of the chromosome (data not shown). The xxx-2423::miniTn10Cm had been inserted into the start codon for the rsd (regulator of sigma D) gene, as demonstrated by DNA sequencing out of the xxx-2423::miniTn10Cm with a primer specific for its chloramphenicol acetyltransferase gene (data not shown). Therefore, the xxx-2423::miniTn10Cm was renamed rsd2423::miniTn10Cm.
Mapping of sdr-36, sdr-46, and sdr-47 by marker rescue.
Three large overlapping subfragments of each of the rpoB and rpoC genes from the wild-type strain MW100 were generated by PCR (endpoints are listed with respect to the translational start sites)—rpoB (−154 to +1889, +1202 to +2845, and +2046 to +4275) and rpoC (−324 to +1514, +1229 to +3008, and +2653 to +4454), mainly as described by Bartlett et al. (1)—and cloned into the plasmid vector pBR322 (3). The obtained plasmids were transferred by transformation into strains MW177 (sdr-36 rsd2423::miniTn10Cm), MW181 (sdr-46 rsd2423::miniTn10Cm), and GOB250 (sdr-47 rsd2423::miniTn10Cm) selecting at 44°C to examine whether they restored growth as a result of homologous recombination between the cloned wild-type fragments and the chromosome of the temperature-sensitive sdr mutants.
PCR amplification of chromosomal DNA and DNA sequencing.
Regions of the E. coli chromosome was amplified, from colonies resuspended in H2O, by PCR (25, 40). Pfu DNA polymerase from Stratagene cloning systems (La Jolla, CA) was used if the obtained fragments were to be cloned into plasmids, and Taq DNA polymerase from Roche Diagnostics Scandinavia AB (Bromma, Sweden) was used in all other cases. Obtained fragments were separated on agarose gels, cut out, and purified by using Gene-Clean from Bio 101, Inc. (La Jolla, CA). DNA sequencing of PCR fragments was done using Thermo Sequenase, whereas plasmid DNA sequencing was performed with a T7 sequencing kit, both of which were purchased from Amersham Pharmacia Biotech (Buckinghamshire, England).
Northern blot analysis.
Total RNA was prepared according to von Gabain et al. (50) and subjected to Northern blot analysis, mainly as described by Sambrook et al. (42). Equal amounts of the RNA from the different strains grown in LB medium were loaded onto the gels as determined both spectrophotometrically at 260 nm and by ethidium bromide staining of aliquots of the RNA electrophoresed in agarose gels. Probes were made with the Megaprime DNA labeling system from Amersham Pharmacia Biotech (Buckinghamshire, England) by using [α-32P]dATP and DNA fragments, purified as described above, as templates.
Western blot analysis.
Total cell extracts were subjected to sodium dodecyl sulfate-polyacrylamide (15%) gel electrophoresis, proteins in the gels were transferred to Hybond-ECL membranes from GE Healthcare (Uppsala, Sweden) and probed with the mouse penta-His antibody from Qiagen AB (Solna, Sweden) as recommended by the manufacturer or with an antiserum against RbfA raised in rabbit by AgriSera AB (Vännäs, Sweden). For the latter, 5% milk powder was used instead of 3% bovine serum albumin. The penta-His antibodies on the membranes were detected with a horseradish peroxidase (HRP)-conjugated rabbit anti-mouse antibody from Dako A/S, Denmark, by using an Amersham ECL Plus kit from GE Healthcare. Similarly, RbfA-antibodies were detected with an HRP-conjugated goat anti-rabbit antibody from Jackson Immunoresearch Laboratories, Inc. (West Grove, PA).
RESULTS
Three suppressor mutations to ΔrimM102 are in rpoB and rpoC.
In order to localize the three mutations sdr-36, sdr-46, and sdr-47, which suppress partially the slow growth of the ΔrimM102 mutant MW37, strain GOB125 containing a mini-Tn10Cm linked (ca. 50%) to sdr-47 was isolated (see Materials and Methods). The mini-Tn10Cm in GOB125 (ΔrimM102 sdr-47) was localized to the start codon of the rsd (regulator of sigma D) gene in the 90-min region of the chromosome (see Materials and Methods). This region of the chromosome from the sdr+ strain GOB249 (rimM+ rsd2423::miniTn10Cm) was able to cross out the suppressor mutations of strains PW102 (ΔrimM102 sdr-36) and PW112 (ΔrimM102 sdr-46) in P1 transduction experiments. Both sdr-36 and sdr-46 were ca. 50% linked to rsd2423::miniTn10Cm, indicating that they might be in the same gene as sdr-47. All three mutations were transferred to the wild-type strain MW100 by P1 transduction and the resulting strains, MW177 (sdr-36 rsd2423::miniTn10Cm), MW181 (sdr-46 rsd2423::miniTn10Cm), and GOB250 (sdr-47 rsd2423::miniTn10Cm) showed temperature-sensitive growth (data not shown). Since most of the previously isolated suppressor mutations to the ΔrimM102 mutation increase the expression of the rbfA gene, we speculated that also the mutations sdr-36, sdr-46, and sdr-47 might do so. Two candidate genes in this region of the chromosome for harboring mutations that could affect the expression of the rbfA gene were rpoB and rpoC, encoding the β and β′ subunits, respectively, of the RNA polymerase. Therefore, we tested in a marker rescue experiment whether different parts of the wild-type rpoB or rpoC genes carried on plasmids could restore the growth at 44°C of rimM+ strains containing sdr-36, sdr-46, or sdr-47 mutations as a result of recombination between the wild-type fragment in the plasmid and the mutant copy on the chromosome. The rpoB fragments +1202 to +2845 and fragments +2046 to +4275 (numbers are given with respect to the translation start site) both rescued the growth of strains MW181 (sdr-46) and GOB250 (sdr-47) at 44°C, whereas the rpoC fragment −324 to +1514 rescued strain MW177 (sdr-36) (data not shown). In the region that overlapped between the two rpoB fragments that rescued strains MW181 (sdr-46) and GOB250 (sdr-47), there was a single mutation on the chromosome of both strains, which changed codon 905 from ATC (Ile) to AGC (Ser). The chromosomal region of strain MW177 (sdr-36) that corresponded to the rpoC fragment that rescued this strain contained a duplication of codons 415 to 416 (AGTTAT) encoding Val and Ile. These mutations will hereafter be referred to as rpoB2063 and rpoC2064, respectively. In addition, the temperature sensitivity of strains containing the rpoB2063 and rpoC2064 mutations were complemented by plasmids pACTB1 and pACTC1, respectively, expressing wild-type copies of rpoB and rpoC (14), demonstrating that the two mutations were recessive (data not shown).
The rpoB2063 and rpoC2064 mutations increase the expression of the metY-infB-nusA operon.
The growth rate in LB medium at 37°C of the original rpoB2063 ΔrimM102 mutants PW112 and PW113 was 2-fold higher than that of the ΔrimM102 mutant MW37 (specific growth rates, k = ln2/g [where g is the generation time in hours], of 0.78 and 0.39, respectively), while that of the rpoC2064 mutant PW102 was 1.6-fold higher (k = 0.62). To examine whether the mechanism behind the suppression of the slow growth of the ΔrimM102 mutant by the rpoB2063 and rpoC2064 mutations was an increased expression of rbfA, as has been seen for most of the other suppressor mutations isolated to ΔrimM102, the amount of RbfA in total protein extracts from ΔrimM102, as well as rimM+, strains containing the rpoB2063 or rpoC2064 mutations was determined by Western blot analyses. Evidently, the amount of RbfA was >3-fold higher in the ΔrimM102 mutant MW172 containing the rpoB2063 mutation than in the congenic rpoB+ strain MW171 (Fig. 2). The overexpression of RbfA was less in the rpoC2064 ΔrimM102 strain GOB307, which correlates with the slower growth of rpoC2064 ΔrimM102 strains compared to rpoB2063 ΔrimM102 strains. In a rimM+ background, the rpoB2063 and rpoC2064 mutations increased the amount of RbfA approximately 6- and 5-fold, respectively (Fig. 2).
Fig. 2.
Determination of the amount of RbfA in different strains. (A) Western blot analysis of cellular extracts of the indicated strains with an antiserum against RbfA. (B) Quantification of the amount of RbfA based on the results from three independent Western blot analyses. The amounts have been normalized to that obtained for the rimM+ rpoB+ rpoC+ strain MW176. The standard deviation is indicated by error bars.
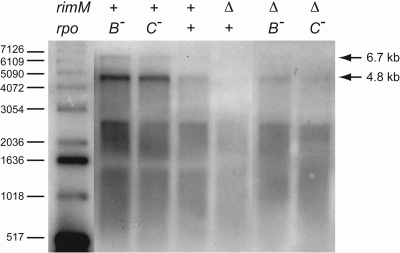
Conceivably, this increased amount of RbfA in strains that harbor the rpoB2063 or rpoC2064 mutations would result from mechanisms operating at the transcriptional level. Therefore, the effect of the two mutations on the transcriptional pattern of the metY-nusA-infB operon in both rimM+ and ΔrimM102 strains was examined by Northern blotting. The most abundant mRNA transcribed from this operon is 4.8 kb in size and results from initiation at the P−1 and P1 promoters, readthrough of the two rho-independent terminators between metY and rimP, termination at the rho-independent T3 terminator just upstream from rbfA, and processing by RNase III between metY and rimP (Fig. 1) (15, 19, 26, 37, 43). The production of the less-abundant 6.7-kb mRNA requires readthrough of the T3 terminator and termination at the terminator downstream from the pnp gene followed by processing at the RNase III site between rpsO and pnp (36, 38, 43). Visibly, the rpoB2063 and rpoC2064 mutations increased the amount of both the 4.8- and the 6.7-kb mRNAs in a rimM+ background (Fig. 3). In the ΔrimM102 mutant, the amount of the two mRNAs was lower than in the rimM+ background, as also has been seen previously (7). The amount of the 4.8-kb mRNA was higher in the rpoB2063 ΔrimM102 and rpoC2064 ΔrimM102 strains than in the suppressor-free ΔrimM102 mutant. The amount of the 6.7-kb mRNA covering the rbfA gene was just above detection level; however, it appeared to be more abundant in the two suppressor-containing strains. This was supported by the results from a real-time reverse transcriptase PCR experiment, in which the amounts of mRNA corresponding to the rbfA gene were 8.6- and 2.3-fold higher in the rpoB2063 ΔrimM102 strain MW172 and the rpoC2064 ΔrimM102 strain GOB307, respectively, than in the ΔrimM102 mutant PW157 (data not shown).
Fig. 3.
Northern blot analysis of metY-nusA-infB operon mRNA. Portions (5 μg) of total RNA were subjected to electrophoresis in an agarose gel containing formaldehyde, transferred to a Hybond N filter, and probed with a radiolabeled DNA probe corresponding to the rimP gene. The sizes of the [γ-32P]ATP kinase fragments of the 1-kb DNA ladder from Gibco-BRL Life Technologies, Inc. (Gaithersburg, MD), are indicated. The strains used (with relevant genetic markers in parentheses) were MW181 (rimM+ rpoB2063), MW177 (rimM+ rpoC2064), GOB249 (rimM+ rpoB+ rpoC+), PW157 (ΔrimM102 rpoB+ rpoC+), MW172 (ΔrimM102 rpoB2063), and GOB307 (ΔrimM102 rpoC2064). For complete genotypes, see Table 1.
The rpoB2063 and rpoC2064 mutations abolish NusA-mediated feedback regulation at internal rho-independent transcriptional terminators of the metY-nusA-infB operon.
The T1 and T2 rho-independent terminators between metY and rimP are a target for NusA-mediated negative-feedback regulation of the metY-nusA-infB operon (5, 27, 34), and different nusA mutations isolated as suppressors to the ΔrimM102 mutation increase the readthrough of these terminators (5). Therefore, we wanted to examine whether the rpoB2063 and rpoC2064 mutations also increased the readthrough of these terminators. The two mutations were introduced into two strains that harbor either of two transcriptional fusions between metY and lacZ, integrated into the lacI-lacZ region of the chromosome. One of the fusions contains the T1 and T2 terminators, while the other lacks them (5). The different strains were grown at 37°C in liquid LB, supplemented with medium E+B1, and their expression of the metY-lacZ fusions was determined. The ratio of the β-galactosidase activity in a strain containing the fusion with the terminators to that of a strain containing the fusion that lacks the terminators, in an otherwise identical genetic background, was used as a measure of the readthrough. Interestingly, the rpoB2063 and rpoC2064 mutations increased readthrough approximately 1.6- and 1.2-fold, respectively (Table 2). Further, the effect of these two mutations was also tested after a shift from 37°C to the nonpermissive temperature 44°C by diluting the cultures 10-fold into prewarmed medium and assaying after two generations of growth. The temperature shift did not affect the readthrough of the terminators in any of the strains (data not shown). To examine whether the increased readthrough resulted from an inability of NusA to stimulate termination at these terminators in the two mutants, the effect of increased synthesis of NusA on the readthrough was examined. Plasmid pJML007 containing the nusA gene under the control of the arabinose-inducible PBAD promoter was introduced into the different strains containing the metY-lacZ fusions. A nusA94 mutant that shows increased readthrough of these terminators was used as a control for that the synthesis of NusA from the plasmid could restore feedback regulation (5). The nusA94 mutant shows a temperature-sensitive growth phenotype and are unable to grow at 44°C. Therefore, the amount of arabinose to be used for achieving relevant expression of nusA from the plasmid was determined by testing different concentration of arabinose for the ability to support growth of the nusA94 mutant on LA plates at 44°C. A 0.02% concentration of arabinose restored growth at 44°C of the nusA94 strains that contained the metY-lacZ fusions (data not shown). Concentrations of arabinose of 0.05% and higher reduced the growth of the nusA94 strains, as well as that of the nusA+ strains. The growth of the rpoB2063 and rpoC2064 mutants, which also are unable to grow at 44°C, was not promoted by synthesis of NusA from the plasmid. The expression of the metY-lacZ fusions was determined by growing the different strains at 37°C in liquid LB, supplemented with medium E+B1, without or with 0.02% of arabinose, and measuring the β-galactosidase activity in cellular extracts. In the absence of arabinose, the readthrough values of the two terminators were 1.9 and 1.7 times higher in the nusA94 and rpoB2063 mutants, respectively, than in the nusA+ rpoB+ rpoC+ strain (Table 3). In the presence of arabinose, readthrough in the nusA+ rpoB+ rpoC+ strain was 2.4 times lower than in the absence of arabinose, suggesting that increased synthesis of NusA from the plasmid repressed the readthrough of the two terminators. Similarly, the addition of arabinose to the nusA94 strains repressed the readthrough almost 3-fold. However, the presence of arabinose did not significantly reduce the readthrough of the T1 and T2 terminators in the rpoB2063 and rpoC2064 mutants. Thus, the rpoB2063 and rpoC2064 mutants seem deficient in NusA-mediated negative-feedback regulation at these terminators.
Table 2.
Effect of rpoB2063 and rpoC2064 mutations on the readthrough of transcriptional terminators between metY and rimP a
| Expt |
rpoB genes |
rpoC genes |
||||
|---|---|---|---|---|---|---|
| Readthrough |
Relative readthrough (rpoB2063/rpoB+) | Readthrough |
Relative readthrough (rpoC2064/rpoC+) | |||
| rpoB+ | rpoB2063 | rpoC+ | rpoC2064 | |||
| 1 | 0.27 | 0.43 | 1.59 | 0.25 | 0.32 | 1.28 |
| 2 | 0.31 | 0.55 | 1.77 | 0.30 | 0.35 | 1.17 |
| 3 | 0.25 | 0.38 | 1.52 | 0.24 | 0.27 | 1.12 |
| 4 | 0.28 | 0.38 | 1.36 | 0.25 | 0.29 | 1.16 |
| Avg | 1.56 | 1.18 | ||||
Readthrough is calculated as [(P + T)/P]. The readthrough of the metY-T1T2 terminators was determined by comparing the β-galactosidase activity of the PmetY-metY-T1T2-lacZ (P + T) and PmetY-metY-lacZ (P) fusions in four independent experiments. The strains used were GOB906 (rpoB+ [P]), GOB898 (rpoB+ [P + T]), GOB904 (rpoB2063 [P]), GOB896 (rpoB2063 [P + T]), GOB910 (rpoC+ [P]), GOB902 (rpoC+ [P + T]), GOB908 (rpoC2064 [P]), and GOB900 (rpoC2064 [P + T]). For complete genotypes, see Table 1.
Table 3.
Effect of rpoB2063 and rpoC2064 on NusA-mediated transcriptional feedback regulation at the terminators between metY and rimP
| Genotype | Readthrougha |
Mean fold repression ± SDb | |||
|---|---|---|---|---|---|
| Expt 1 |
Expt 2 |
||||
| −Ara | +Ara | −Ara | +Ara | ||
| nusA+rpoB+rpoC+/pJML007 (nusA+) | 0.20 | 0.085 | 0.21 | 0.085 | 2.41 ± 0.06 |
| nusA94/pJML007 (nusA+) | 0.36 | 0.13 | 0.40 | 0.13 | 2.92 ± 0.15 |
| rpoB2063/pJML007 (nusA+) | 0.32 | 0.31 | 0.36 | 0.29 | 1.14 ± 0.10 |
| rpoC2064/pJML007 (nusA+) | 0.17 | 0.22 | 0.24 | 0.21 | 0.95 ± 0.18 |
Readthrough was calculated as [(P + T)/P]. The effect of induced expression of nusA on the readthrough of the metY-T1T2 terminators was determined in two independent experiments by comparing the β-galactosidase activity of the PmetY-metY-T1T2-lacZ (P + T) and PmetY-metY-lacZ (P) fusions in strains harboring plasmid pJML007 that carries nusA under the control of the arabinose-inducible PBAD promoter. Readthrough was determined with (+Ara) or without (−Ara) the addition of 0.02% arabinose. Only relevant genotypes are indicated. For complete genotypes. see Table 1. The strains used were GOB906/pJML007, GOB898/pJML007, GOB752/pJML007, GOB750/pJML007, GOB904/pJML007, GOB896/pJML007, GOB908/pJML007, and GOB900/pJML007.
The repression was calculated as the ratio of the readthrough in the absence of arabinose to that in the presence of arabinose.
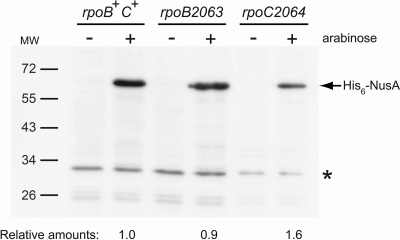
Formally, the lack of effect on the expression of β-galactosidase by the addition of arabinose to the rpoB2063 and rpoC2064 mutants could have resulted from the fact that NusA was not produced from plasmid pJML007 due to an inability of the mutant RNA polymerases to initiate transcription at the PBAD promoter. Therefore, the ability of arabinose to induce expression from this promoter in the rpoB2063 and rpoC2064 mutants MW181 and MW177, as well as the rpoB+ rpoC+ strain MW176, was tested in a Western blotting experiment. A His6-nusA construct was placed downstream from the PBAD promoter in plasmid pBAD30, i.e., the same vector that pJML007 is based on, and the amount of the His6-NusA protein in the three strains grown at 37°C in LB, without or with 0.02% arabinose, was examined using antibodies against the His tag. No detectable amount of the His6-NusA protein was observed in the absence of arabinose, whereas in the presence of arabinose a severalfold-higher expression was seen for all three strains (Fig. 4). Thus, the PBAD promoter is functional in the rpoB2063 and rpoC2064 mutants, suggesting that the lack of effect on the readthrough of the T1 and T2 terminators between metY and rimP upon addition of arabinose (Table 3) indeed resulted from an inability of NusA to support transcriptional termination at these terminators when the RNA polymerase contained either of the alterations brought about by the rpoB2063 and rpoC2064 mutations.
Fig. 4.
Expression of His6-nusA from the PBAD promoter in rpo+, rpoB2063, and rpoC2064 strains as determined by Western blot analysis. Total cellular extracts of strains grown in the presence (0.02%) or absence of arabinose were probed on filters with an antibody against the His tag. The strains used (with relevant genetic markers in parentheses) were MW176/pMW498 (rpoB+ rpoC+/his6-nusA), MW181/pMW498 (rpoB2063 rpoC+/his6-nusA), and MW177/pMW498 (rpoB+ rpoC2064/his6-nusA). For complete genotypes, see Table 1. MW, molecular mass marker with sizes in kilodaltons. The relative amounts of His6-NusA shown were obtained by using an unknown protein (*) for which the synthesis did not respond to addition of arabinose as an internal control and normalizing the obtained ratios to that for the rpoB+ rpoC+ strain.
The rpoB2063 mutation also affects transcriptional termination at the terminator of the trmD operon.
To investigate whether the effect of the rpoB2063 and rpoC2064 mutations on transcriptional termination was limited to terminators of the metY-nusA-infB operon or if the two mutations also affected termination at other terminators, the readthrough of the rho-independent major terminator, TrplS, of the trmD operon (which encodes ribosomal proteins S16 and L19, the ribosome maturation protein RimM, and the tRNAm1G37methyltransferase) (8, 9) was studied. Previously, the readthrough of the TrplS terminator was shown to be 2.2-fold higher in a nusA94 mutant than in a nusA+ strain when this terminator was present in the terminator-containing metY-lacZ fusion used above, in place of the metY T1 and T2 terminators (5). The rpoB2063 and rpoC2064 mutations were introduced into the metY-lacZ fusion strain containing the TrplS terminator, and their effect on the readthrough was determined. The rpoB2063 mutation increased the readthrough of this terminator 1.6-fold, while the rpoC2064 mutation seemed not to affect the readthrough (Table 4). Further, the effect of increased expression of nusA on the readthrough of the TrplS terminator was examined by introducing plasmid pJML007 (PBAD-nusA+) into the rpoB2063 and rpoC2064 mutants, as well as the nusA94 mutant, and inducing the expression of nusA+ by arabinose, as described above. The increased expression of nusA+ in the nusA94 mutant reduced the readthrough of the TrplS terminator 1.7-fold, partially restoring termination at this terminator (Table 4). However, increased expression of nusA+ in the rpoB2063 mutant did not reduce but on the contrary seemed to increase the readthrough. Induced expression of nusA+ in a nusA+ rpoB+ rpoC+ genetic background did not further reduce readthrough of the TrplS terminator, suggesting that termination at TrplS is close to maximal at normal levels of NusA. Increased expression of nusA+ in the rpoC2064 mutant seemed to slightly reduce readthrough. In conclusion, the rpoB2063 mutation not only affects transcriptional termination at terminators of the metY-nusA-infB operon but also that of the TrplS terminator of the trmD operon.
Table 4.
Effect of rpoB2063 and rpoC2064 mutations on the readthrough of the rho-independent transcriptional terminator, TrplS, of the trmD operona
| Genotype | Readthrough | Relative readthroughb | Genotype | Readthrough |
Mean fold repression ± SDc | |
|---|---|---|---|---|---|---|
| –Ara | +Ara | |||||
| rpoB+rpoC+ | 0.084 | 1.0 | nusA+rpoB+rpoC+/pJML007 (nusA+) | 0.090 | 0.099 | 1.00 ± 0.09 |
| 0.090 | 0.083 | |||||
| rpoB2063 | 0.13 | 1.55 | rpoB2063/pJML007 (nusA+) | 0.19 | 0.23 | 0.75 ± 0.08 |
| 0.16 | 0.24 | |||||
| rpoC2064 | 0.086 | 1.02 | rpoC2064/pJML007 (nusA+) | 0.098 | 0.083 | 1.19 ± 0.01 |
| 0.12 | 0.10 | |||||
| nusA94/pJML007 (nusA+) | 0.21 | 0.11 | 1.66 ± 0.24 | |||
| 0.17 | 0.12 | |||||
Readthrough was calculated as [(P + T)/P]. The readthrough of the TrplS terminator was determined by comparing the β-galactosidase activity of PmetY-metY-TrplS-lacZ (P + T) and PmetY-metY-lacZ (P) fusions. The effect of induced expression of nusA on the readthrough of this terminator was studied by measuring the β-galactosidase activity in strains harboring plasmid pJML007 that carries nusA under the control of the arabinose-inducible PBAD promoter. Readthrough was determined with (+Ara) or without (−Ara) the addition of 0.02% arabinose. Only relevant genotypes are indicated. For complete genotypes, see Table 1. The P + T strains used were MW288 (rpoB+ rpoC+), MW286 (rpoB2063), MW289 (rpoC2064), GOB838/pJML007 (rpoB+ rpoC+ nusA+/nusA+), MW286/pJML007 (rpoB2063/nusA+), MW289/pJML007 (rpoC2064/nusA+), and GOB868/pJML007 (nusA94/nusA+); the P strains used were GOB906 (rpoB+ rpoC+), GOB904 (rpoB2063), GOB908 (rpoC2064), GOB906/pJML007 (rpoB+ rpoC+ nusA+/nusA+), GOB904/pJML007 (rpoB2063/nusA+), GOB908/pJML007, (rpoC2064/nusA+), and GOB752/pJML007 (nusA94/nusA+).
For determination of the relative readthrough, the readthrough was normalized to that obtained with nusA+ rpoB+ rpoC+ strains.
The mean fold repression was calculated as the ratio of the readthrough in the absence to that in the presence of arabinose (0.02%).
DISCUSSION
We have identified here a mutation in each of the rpoB and rpoC genes encoding the β and β′ subunits, respectively, of the RNA polymerase. The two mutations, rpoB2063 and rpoC2064, isolated as suppressors of the slow growth of a strain lacking the ribosome maturation protein RimM, were found to increase the synthesis of RbfA, another ribosome maturation protein. Most of the suppressor mutations isolated in the same screen showed increased expression of rbfA; however, the mechanism varied (5). Eight of the suppressor mutations lead to alterations in the transcriptional elongation factor NusA which resulted in a reduced ability of NusA to stimulate transcription termination at internal rho-independent terminators of the metY-nusA-infB operon, thereby increasing expression of the distally located rbfA (5). The rpoB2063 and rpoC2064 mutations characterized here seem to increase the readthrough of the terminators by the same mechanism as the nusA mutations.
The level of arabinose that induced sufficient expression of NusA from a plasmid to complement the temperature sensitivity of a nusA98 mutant also reduced the readthrough of the T1 and T2 terminators between metY and rimP in this mutant down to the level that was seen in a plasmid-free nusA+ background (5). Further, the reduction in the readthrough was 1.8-fold both in the nusA98 and the nusA+ background. These findings suggested that the amount of NusA produced from the plasmid corresponded to that found in a plasmid-free nusA+ background. Thus, an ∼2-fold increased amount of NusA reduced the readthrough of these terminators almost 2-fold. NusA has been shown to stimulate pausing of the elongating RNA polymerase in vitro at different pause sites at equimolar amounts of NusA and RNA polymerase, whereas a 10-fold excess of NusA relative to RNA polymerase was needed to terminate transcription at rho-independent terminators (44). Thus, the role of NusA in feedback regulation at the T1 and T2 terminators between metY and rimP might be primarily to enhance pausing of the elongating RNA polymerase thereby facilitating transcript release or providing time for the formation of the terminator structures. However, NusA might affect termination directly at these terminators. In a large-scale computational search for bacterial transcriptional terminators, the rimP leader, which contains the T1 and T2 terminators, was the most ubiquitous transcription attenuator found (28). The stem of the T2 terminator was found to contain the most conserved motif of all 5′ and 3′ terminators identified, suggesting that the sequence signature was specific for the rimP leader. Such high sequence conservation and the lack of any visible anti-terminator structure were suggested to indicate regulation by a termination or anti-termination protein such as NusA that specifically would recognize the conserved nucleic acid motif. The role of NusA at the T1 and T2 terminators at which it feedback regulates its own synthesis might be different from that at other terminators where the binding of NusA might be more unspecific. Thus, NusA might directly stimulate intrinsic termination at the T1 and T2 terminators even at lower concentrations.
The rpoB2063 mutation reduced significantly termination at the T1 and T2 terminators between metY and rimP and an increased synthesis of NusA could not restore termination, suggesting that the alteration in the β subunit abolished NusA-mediated negative-feedback regulation. The effect of the rpoC2064 mutation on the readthrough of these two terminators was small; however, increased expression of nusA in the rpoC2064 mutant did not increase termination, suggesting that NusA could not promote termination at these terminators as a result of the alteration in the β′ subunit. The previously characterized nusA mutations have been shown to increase the readthrough also of the rho-independent terminator between infB and rbfA (5). Similarly, the rpoB2063 and rpoC2064 mutations increased the readthrough of the same terminator 2- and 1.5-fold, respectively, as judged from the quantification of the readthrough (6.7 kb) and terminated transcripts (4.8 kb) in a Northern blotting experiment (data not shown).
Previously, the nusA94 mutation was found to increase readthrough not only of the terminators of the metY-nusA-infB operon but also of the rho-independent trmD operon terminator TrplS (5). We have shown here that the rpoB2063 mutation also increased the readthrough of TrplS, indicating that rpoB2063 might have a general effect on NusA-dependent transcriptional termination. The rpoC2064 mutation, on the other hand, did not seem to affect the readthrough of TrplS. In wild-type cells, termination at TrplS was not affected by increased synthesis of NusA, while at the T1 and T2 terminators higher levels of NusA reduced the readthrough, probably reflecting that at the latter NusA has a role to regulate its own synthesis up or down in response to different conditions and not only to maximize termination efficiency. In the rpoB2063 mutant, increased amounts of NusA increased the readthrough of TrplS, while in the rpoC2063 mutant it slightly decreased the readthrough. This contrasts with the lack of effect at the T1 and T2 terminators in the two mutants at increased levels of NusA. Thus, these dissimilarities might reflect slightly different roles of NusA at the different terminators or that the two mutant RNA polymerases interact differently with different terminators.
Three mutations in rpoB that increased the readthrough of rho-dependent and rho-independent terminators have been shown to be incompatible with either of two mutations in nusA (21), one of which is identical to nusA98 that increased the readthrough of the two terminators between metY and rimP (5). It was suggested that an enhanced readthrough of terminators accounted for the incompatibility of the nusA and rpoB mutations (21). In an attempt to introduce either of two other nusA alleles, nusA91 and nusA94, both of which also increase the readthrough of the two terminators between metY and rimP (5), into rpoB2063 and rpoC2064 mutants by P1 transduction and selecting for a tightly linked resistance marker, 10- to 20-fold-lower numbers of transductants were obtained compared to when rpoB+ rpoC+ strains were used as recipients (data not shown). Faster-growing clones were nusA+, while slower-growing ones were unstable and shown to contain both the nusA+ and the respective nusA mutant allele showing that they were the result of insertion of the respective nusA mutant allele into one of two copies of the nusA region in a preexisting duplication. Thus, these findings suggest that mutants containing either nusA91 or nusA94 combined with rpoB2063 or rpoC2064 are not viable, possibly because of enhanced terminator readthrough. The essentiality of NusA has been attributed to its ability to increase transcriptional pausing and hence secure a tight coupling of transcription and translation (53). Thus, ribosomes bound to mRNA would prevent the transcriptional termination factor Rho from accessing it and thereby reduce premature intragenic transcriptional termination. However, more recently NusA was found to have an essential role in supporting rho-dependent termination during silencing of horizontally transferred DNA elements such as cryptic prophages, which can be toxic for the host cell when expressed (10). The rpoB2063 and rpoC2064 mutants show an increased sensitivity to bicyclomycin (data not shown), an antibiotic that targets the Rho protein (54). Thus, the temperature sensitivity of different nusA mutants, as well as of the rpoB2063 and rpoC2064 mutants, might result from inefficient regulation of rho-dependent termination at the higher temperatures. Conceivably, the inability to combine the nusA91 or nusA94 mutations with the rpoB2063 or rpoC2064 mutations even at lower temperatures might be explained by additive effects on rho-dependent termination.
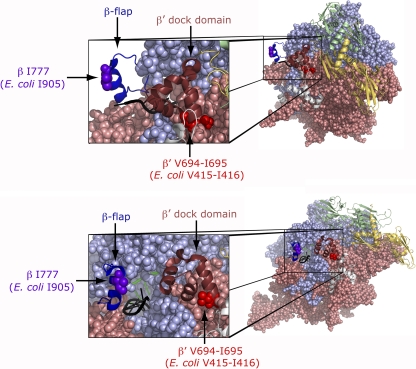
In its role to stimulate pausing and termination of transcription elongation, NusA interacts with the RNA polymerase at the RNA exit channel by contacting the flap tip of the β subunit (β flap-tip) and the dock domain of the β′ subunit (17, 52). The removal of the β flap-tip helix (residues 900 to 909) has been shown to abolish completely NusA enhancement of pausing in vitro (47). Interestingly, the rpoB2063 mutation alters the amino acid in position 905 (I905S), which is in the center of the β flap-tip helix (Fig. 5) and just next to F906 that has been shown to cross-link both to an RNA pause hairpin loop and to NusA (47). Thus, the location of the alteration in the β subunit of the rpoB2063 mutant suggests that the reduced termination at the internal terminators of the metY-nusA-infB operon stem directly from a reduced ability of the mutant RNA polymerase to interact with NusA and/or the internal terminators. The inability of increased levels of NusA to restore termination suggests that the I905S substitution has a more dramatic effect than just reducing the affinity of the β flap-tip helix for NusA. Possibly, the I905S substitution confers dramatic structural changes to the β flap-tip helix so that it can no longer bind NusA.
Fig. 5.
Location of the amino acid alterations, conferred by the rpoB2063 and rpoC2064 mutations in the structure of Thermus thermophilus elongating complex (PDB ID, 2O5I) (48). Two different views differing by ca. 90° are shown where the two α subunits (yellow and green, respectively) and ω (light-gray) are shown as secondary-structure cartoons, while β (light-blue) and β′ (salmon) are in space-fill. In the close-ups, the β-flap domain is shown in dark blue as a cartoon with the position altered in the rpoB2063 mutant in purple as space-fill, whereas the β′ dock domain is shown in maroon as a cartoon and at its base are the two amino acids duplicated in the rpoC2064 mutant shown in red as space-fill. The RNA emerging from the RNA exit tunnel is in black and the DNA is in green. The structures were prepared with PyMOL (13; http://www.pymol.org).
Recently, the amino acid in position 29 of the N-terminal domain of NusA was shown by hydroxyl radical foot-printing to be close to the β′ dock domain, residues 381 to 414 in E. coli (17). In addition, the most common suppressor to the G181D alteration in NusA, which was one of the alterations that showed an increased readthrough of internal terminators of the metY-nusA-infB operon (5), is the E402K substitution in the β′ dock domain (20). These findings suggest that NusA also interacts with the β′ dock domain. The rpoC2064 mutation characterized here resulted in a duplication of amino acids V415 and I416, which are at the base of the β′ dock domain (Fig. 5). Conceivably, the addition of these two extra amino acid residues might alter the orientation and/or structure of the β′ dock domain which would affect its interaction with NusA. The lack of effect of increased levels of NusA on the readthrough of internal transcriptional terminators of the metY-nusA-infB operon in the rpoC2064 mutant is in agreement with structural changes in the β′ dock domain preventing the binding of NusA. However, the increase in readthrough of these terminators was small in the rpoC2064 mutant compared to that in the rpoB2063 mutant, although termination in both mutants was equally insensitive to increased amounts of NusA. Thus, the assumed structural changes in the β′ dock domain seem to have affected termination at the internal terminators of the metY-nusA-infB operon to become in part NusA independent.
We have presented here the first in vivo results that directly support an interaction of NusA with the β flap-tip and the β′ dock domain of the RNA polymerase during transcriptional termination. It would be interesting to map genetically the interaction points between NusA and the β flap-tip by constructing mutants with conservative changes in the β flap-tip that do not destroy its structure but reduce the ability of NusA to stimulate termination at the terminators of the metY-nusA-infB operon and then isolate compensatory alterations in NusA expressed from a plasmid.
ACKNOWLEDGMENTS
Plasmids pACTB1 and pACTC1 were generous gifts from Derek Dykxhoorn. We thank Glenn Björk and Marcus Johansson for critical reading of the manuscript.
This research was supported by The Carl Trygger Foundation and The Magnus Bergvall Foundation.
Footnotes
Published ahead of print on 17 June 2011.
REFERENCES
- 1. Bartlett M. S., Gaal T., Ross W., Gourse R. L. 1998. RNA polymerase mutants that destabilize RNA polymerase-promoter complexes alter NTP-sensing by rrn P1 promoters. J. Mol. Biol. 279:331–345 [DOI] [PubMed] [Google Scholar]
- 2. Bertani G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62:293–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bolivar F., et al. 1977. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 2:95–113 [PubMed] [Google Scholar]
- 4. Bunner A. E., Nord S., Wikström P. M., Williamson J. R. 2010. The effect of ribosome assembly cofactors on in vitro 30S subunit reconstitution. J. Mol. Biol. 398:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bylund G. O., Lövgren J. M., Wikström P. M. 2001. Characterization of mutations in the metY-nusA-infB operon that suppress the slow-growth of a ΔrimM mutant. J. Bacteriol. 183:6095–6106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bylund G. O., Persson B. C., Lundberg L. A. C., Wikström P. M. 1997. A novel ribosome-associated protein is important for efficient translation in Escherichia coli. J. Bacteriol. 179:4567–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bylund G. O., Wipemo L. C., Lundberg L. A. C., Wikström P. M. 1998. RimM and RbfA are essential for efficient processing of 16S rRNA in Escherichia coli. J. Bacteriol. 180:73–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Byström A. S., Hjalmarsson K. J., Wikström P. M., Björk G. R. 1983. The nucleotide sequence of an Escherichia coli operon containing genes for the tRNA(m1G)methyltransferase, the ribosomal proteins S16 and L19 and a 21-K polypeptide. EMBO J. 2:899–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Byström A. S., von Gabain A., Björk G. R. 1989. Differentially expressed trmD ribosomal protein operon of Escherichia coli is transcribed as a single polycistronic mRNA species. J. Mol. Biol. 208:575–586 [DOI] [PubMed] [Google Scholar]
- 10. Cardinale C. J., et al. 2008. Termination factor Rho and its cofactors NusA and NusG silence foreign DNA in Escherichia coli. Science 320:935–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Craven M. G., Friedman D. I. 1991. Analysis of the Escherichia coli nusA10(Cs) allele: relating nucleotide changes to phenotypes. J. Bacteriol. 173:1485–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dammel C. S., Noller H. F. 1995. Suppression of a cold-sensitive mutation in 16S rRNA by overexpression of a novel ribosome-binding factor, RbfA. Genes Dev. 9:626–637 [DOI] [PubMed] [Google Scholar]
- 13. DeLano W. L. 2006. The PyMOL molecular graphics system, PyMOL 0.99rc6 ed. http://www.pymol.org
- 14. Dykxhoorn D. M., St. Pierre R., Linn T. 1996. Synthesis of the β and β′ subunits of Escherichia coli RNA polymerase is autogenously regulated in vivo by both transcriptional and translational mechanisms. Mol. Microbiol. 19:483–493 [DOI] [PubMed] [Google Scholar]
- 15. Granston A. E., Thompson D. L., Friedman D. I. 1990. Identification of a second promoter for the metY-nusA-infB operon of Escherichia coli. J. Bacteriol. 172:2336–2342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guzman L.-M., Belin D., Carson M. J., Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ha K. S., Toulokhonov I., Vassylyev D. G., Landick R. 2010. The NusA N-terminal domain is necessary and sufficient for enhancement of transcriptional pausing via interaction with the RNA exit channel of RNA polymerase. J. Mol. Biol. 401:708–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ishii S., et al. 1984. The nucleotide sequence of the cloned nusA gene and its flanking region of Escherichia coli. Nucleic Acids Res. 12:3333–3342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ishii S., Kuroki K., Imamoto F. 1984. tRNAMetf2 gene in the leader region of the nusA operon in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 81:409–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ito K., Nakamura Y. 1996. Localization of nusA-suppressing amino acid substitutions in the conserved regions of the β′ subunit of Escherichia coli RNA polymerase. Mol. Gen. Genet. 251:699–706 [DOI] [PubMed] [Google Scholar]
- 21. Jin D. J., et al. 1988. Effects of rifampicin resistant rpoB mutations on antitermination and interaction with nusA in Escherichia coli. J. Mol. Biol. 204:247–261 [DOI] [PubMed] [Google Scholar]
- 22. Kleckner N., Bender J., Gottesman S. 1991. Uses of transposons with emphasis on Tn10, p. 139–180, Escherichia coli and Salmonella typhimurium, vol. 204 Academic Press, Inc., New York, NY [Google Scholar]
- 23. Lerner C. G., Inouye M. 1990. Low copy number plasmids for regulated low-level expression of cloned genes in Escherichia coli with blue/white insert screening capability. Nucleic Acids Res. 18:4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lövgren J. M., et al. 2004. The PRC-barrel domain of the ribosome maturation protein RimM mediates binding to ribosomal protein S19 in the 30S ribosomal subunits. RNA. 10:1798–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mullis K. B., Faloona F. A. 1987. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 155:335–350 [DOI] [PubMed] [Google Scholar]
- 26. Nakamura Y., Mizusawa S. 1985. In vivo evidence that the nusA and infB genes of Escherichia coli are part of the same multi-gene operon which encodes at least four proteins. EMBO J. 4:527–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nakamura Y., Plumbridge J., Dondon J., Grunberg-Manago M. 1985. Evidence for autoregulation of the nusA-infB operon of Escherichia coli. Gene 36:189–193 [DOI] [PubMed] [Google Scholar]
- 28. Naville M., Gautheret D. 2010. Premature terminator analysis sheds light on a hidden world of bacterial transcriptional attenuation. Genome Biol. 11:R97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Neidhardt F. C., Bloch P. L., Pedersen S., Reeh S. 1977. Chemical measurement of steady-state levels of ten aminoacyl-tRNA synthetases in Escherichia coli. J. Bacteriol. 129:378–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Neidhardt F. C., Bloch P. L., Smith D. F. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nord S., Bylund G. O., Lövgren J. M., Wikström P. M. 2009. The RimP protein is important for maturation of the 30S ribosomal subunit. J. Mol. Biol. 386:742–753 [DOI] [PubMed] [Google Scholar]
- 32. Nurse K., Wrzesinski J., Bakin A., Lane B. G., Ofengand J. 1995. Purification, cloning, and properties of the tRNA Ψ55 synthase from Escherichia coli. RNA 1:101–112 [PMC free article] [PubMed] [Google Scholar]
- 33. Persson B. C., Bylund G. O., Berg D. E., Wikström P. M. 1995. Functional analysis of the ffh-trmD region of the Escherichia coli chromosome by using reverse genetics. J. Bacteriol. 177:5554–5560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Plumbridge J. A., Dondon J., Nakamura Y., Grunberg-Manago M. 1985. Effect of NusA protein on expression of the nusA infB operon in Escherichia coli. Nucleic Acids Res. 13:3371–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Plumbridge J. A., et al. 1982. Cloning and mapping of a gene for translational initiation factor IF2 in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 79:5033–5037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Portier C., Dondon L., Grunberg-Manago M., Régnier P. 1987. The first step in the functional inactivation of the Escherichia coli polynucleotide phosphorylase messenger is a RNase III processing at the 5′ end. EMBO J. 6:2165–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Régnier P., Grunberg-Manago M. 1989. Cleavage by RNase III in the transcripts of the metY-nusA-infB operon of Escherichia coli releases the tRNA and initiates the decay of the downstream mRNA. J. Mol. Biol. 210:293–302 [DOI] [PubMed] [Google Scholar]
- 38. Régnier P., Portier C. 1986. Initiation, attenuation and RNase III processing of transcripts from the Escherichia coli operon encoding ribosomal protein S15 and polynucleotide phosphorylase. J. Mol. Biol. 187:23–32 [DOI] [PubMed] [Google Scholar]
- 39. Sacerdot C., Dessen P., Hershey J. W. B., Plumbridge J. A., Grunberg-Manago M. 1984. Sequence of the initiation factor IF2 gene: unusual protein features and homologies with elongation factors. Proc. Natl. Acad. Sci. U. S. A. 81:7787–7791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Saiki R. K., et al. 1985. Enzymatic amplification of β-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science 230:1350–1354 [DOI] [PubMed] [Google Scholar]
- 41. Saito M., Tsugawa A., Egawa K., Nakamura Y. 1986. Revised sequence of the nusA gene of Escherichia coli and identification of nusA11(Ts) and nusA1 mutations which cause changes in a hydrophobic amino acid cluster. Mol. Gen. Genet. 205:380–382 [DOI] [PubMed] [Google Scholar]
- 42. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 43. Sands J. F., Régnier P., Cummings H. S., Grunberg-Manago M., Hershey J. W. 1988. The existence of two genes between infB and rpsO in the Escherichia coli genome: DNA sequencing and S1 nuclease mapping. Nucleic Acids Res. 16:10803–10816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sigmund C. D., Morgan E. A. 1988. Nus A protein affects transcriptional pausing and termination in vitro by binding to different sites on the transcription complex. Biochemistry 27:5622–5627 [DOI] [PubMed] [Google Scholar]
- 45. Singer M., et al. 1989. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol. Rev. 53:1–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Suzuki S., et al. 2007. Structural characterization of the ribosome maturation protein, RimM. J. Bacteriol. 189:6397–6406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Toulokhonov I., Artsimovitch I., Landick R. 2001. Allosteric control of RNA polymerase by a site that contacts nascent RNA hairpins. Science 292:730–733 [DOI] [PubMed] [Google Scholar]
- 48. Vassylyev D. G., Vassylyeva M. N., Perederina A., Tahirov T. H., Artsimovitch I. 2007. Structural basis for transcription elongation by bacterial RNA polymerase. Nature 448:157–162 [DOI] [PubMed] [Google Scholar]
- 49. Vogel H. J., Bonner D. M. 1956. Acetylornithinase of Escherichia coli: partial purification and some properties. J. Biol. Chem. 218:97–106 [PubMed] [Google Scholar]
- 50. von Gabain A., Belasco J. G., Schottel J. L., Chang A. C., Cohen S. N. 1983. Decay of mRNA in Escherichia coli: investigation of the fate of specific segments of transcripts. Proc. Natl. Acad. Sci. U. S. A. 80:653–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wikström P. M., Byström A. S., Björk G. R. 1988. Non-autogenous control of ribosomal protein synthesis from the trmD operon in Escherichia coli. J. Mol. Biol. 203:141–152 [DOI] [PubMed] [Google Scholar]
- 52. Yang X., et al. 2009. The structure of bacterial RNA polymerase in complex with the essential transcription elongation factor NusA. EMBO Rep. 10:997–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zheng C., Friedman D. I. 1994. Reduced Rho-dependent transcription termination permits NusA-independent growth of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 91:7543–7547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zwiefka A., Kohn H., Widger W. R. 1993. Transcription termination factor rho: the site of bicyclomycin inhibition in Escherichia coli. Biochemistry 32:3564–3570 [DOI] [PubMed] [Google Scholar]