Abstract
Iron utilization by bacteria in aerobic environments involves uptake as a ferric chelate from the environment, followed by reduction to the ferrous form. Ferric iron reduction is poorly understood in most bacterial species. Here, we identified Bradyrhizobium japonicum frcB (bll3557) as a gene adjacent to, and coregulated with, the pyoR gene (blr3555) encoding the outer membrane receptor for transport of a ferric pyoverdine. FrcB is a membrane-bound, diheme protein, characteristic of eukaryotic ferric reductases. Heme was essential for FrcB stability, as were conserved histidine residues in the protein that likely coordinate the heme moieties. Expression of the frcB gene in Escherichia coli conferred ferric reductase activity on those cells. Furthermore, reduced heme in purified FrcB was oxidized by ferric iron in vitro. B. japonicum cells showed inducible ferric reductase activity in iron-limited cells that was diminished in an frcB mutant. Steady-state levels of frcB mRNA were strongly induced under iron-limiting conditions, but transcript levels were low and unresponsive to iron in an irr mutant lacking the global iron response transcriptional regulator Irr. Thus, Irr positively controls the frcB gene. FrcB belongs to a family of previously uncharacterized proteins found in many proteobacteria and some cyanobacteria. This suggests that membrane-bound, heme-containing ferric reductase proteins are not confined to eukaryotes but may be common in bacteria.
INTRODUCTION
Iron is essential for the growth and survival of most organisms. The metal is required for many cellular processes or incorporated into heme and iron sulfur clusters that function as cofactors of proteins. Iron exists in the insoluble ferric form in neutral or alkaline pH environments (6), and therefore, organisms have evolved highly specific systems for iron acquisition from their natural environments. However, too much iron can lead to the production of reactive oxygen species, which damage proteins and DNA (6). Thus, iron acquisition, storage, and usage are tightly regulated (1).
Siderophores are high-affinity compounds produced and secreted by microorganisms to chelate ferric iron from the environment (7). Many microorganisms produce siderophores, and the siderophores from Escherichia coli, Pseudomonas putida, Pseudomonas aeruginosa (7), and the smut fungus Ustilago maydis (39) have been particularly well characterized. In Gram-negative bacteria, Fe3+-siderophores are bound by TonB-dependent receptors on the outer membrane (17). TonB, ExbB, and ExbD transfer energy from a proton motive force of the inner membrane to the receptor on the outer membrane. The siderophore-specific receptor transports the ferric iron-bound siderophore into the periplasm. The siderophore is then bound by a periplasmic binding protein and delivered to an ABC transporter on the cytoplasmic membrane (5) for transport into the cytoplasm (10). Incorporation of the bound ferric iron into cellular components requires its release from the siderophore and reduction to the ferrous (Fe2+) form. These latter steps are less well understood than the systems that transport iron into the cell.
Ferric reductase activity has been demonstrated in whole cells or cell fractions in many bacterial species (31), but the proteins responsible for this activity have been identified only in a few cases (21, 32, 40). Most bacterial assimilatory ferric reductases are soluble and require flavin for activity. An exception is FhuF, an iron-sulfur cytoplasmic protein that reduces ferric ferrioxamine B (21).
Unlike bacterial ferric reductases, the eukaryotic enzymes are membrane-bound diheme proteins, some of which also contain flavin (9, 22, 28). Dancis et al. (9) first linked ferric reductase activity to ferric uptake in the yeast Saccharomyces cerevisiae. This activity is catalyzed by the heme- and flavin-containing proteins Fre1 and Fre2. The mammalian heme protein Dcytb is expressed under iron-limiting conditions in duodenal cells (22). Homologs of Dcytb are found in other mammalian cell types as well as other eukaryotic species (3, 4, 13, 36, 42). Although Fre1 and Dcytb have been well described and are necessary for ferric iron reduction in whole cells, ferric reductase activity has not been demonstrated in the purified proteins.
Bradyrhizobium japonicum exists as a free-living organism in the soil or as a symbiont of soybean. It is a member of the Alphaproteobacteria, a taxonomic group that includes symbionts, pathogens, and photosynthetic organisms. Regulation of iron metabolism differs substantially in B. japonicum and in the proteobacteria in general compared with other well-studied model organisms as described below. Moreover, B. japonicum can utilize ferric siderophores produced by other organisms as an iron source, but it does not synthesize them. This may make B. japonicum more competitive in the rhizosphere. Ferrichrome and rhodotorulic acid are siderophores secreted by fungi in the soil and are utilized by B. japonicum (27, 34). Five putative siderophore receptor genes have been identified in B. japonicum based on homology and on induction of their transcript levels under iron-limiting conditions (34, 41). The cognate siderophore has been identified for one receptor, ferrichrome, but the others are unknown (19, 34). The other components involved in ferric iron transport into cells and its subsequent reduction to the ferrous form have not been described in B. japonicum.
Induction of B. japonicum ferric siderophore receptors is dependent on the iron response regulator (Irr) protein, which is the major regulator of iron homeostasis in B. japonicum and in numerous other alphaproteobacteria (16, 33). This is markedly different from Fur-dependent iron regulation in E. coli and many other well-studied bacteria. The Fur protein is functional as a transcriptional repressor when bound to iron. Irr accumulates and functions under iron-limiting conditions and degrades in the presence of iron in response to heme. Whole-genome microarray analysis shows that Irr functions as a global regulator of iron homeostasis by upregulating genes involved in iron acquisition and downregulating genes encoding iron-containing proteins (29, 41). Irr binds to the iron control element (ICE), a cis-acting element in the promoters of target genes (25, 29, 30, 34).
In the present study, we identify a gene necessary for ferric iron reduction in B. japonicum that encodes a diheme membrane protein, features characteristic of eukaryotic ferric reductases. In addition, homologs of this protein are widely distributed among the proteobacteria.
MATERIALS AND METHODS
Strains and media.
Bradyrhizobium japonicum strain LO is a spontaneous nalidixic acid-resistant derivative of strain USDA122. LO was the parent strain used in the present work. All mutants used in this study are derivatives of the parent strain LO (14, 34). A mutant disrupted in the bll3557 (frcB) gene was made by replacing the open reading frame (ORF) with the Ω cassette, which confers resistance to streptomycin and spectinomycin, as described previously (34). B. japonicum strains were routinely grown at 29°C in GSY medium (11). Strain LODTM5 was grown in medium supplemented with 50 μg/ml kanamycin and 50 μg/ml streptomycin. The mutants constructed in this study were grown in medium supplemented with 50 μg/ml streptomycin and 100 μg/ml spectinomycin. For the iron experiments, modified GSY medium was used, which contains 0.5 g/liter yeast extract instead of 1 g/liter, and either no exogenous iron was added for low-iron medium or 12 μM FeCl3·6H2O was added for high-iron medium. The actual iron concentration of the unsupplemented medium was 0.3 μM, as determined with a Perkin-Elmer model 1100B atomic absorption spectrometer.
E. coli strains were grown in standard LB medium (10 g/liter tryptone, 5 g/liter yeast extract, and 5 g/liter NaCl) or YT medium (8 g/liter tryptone, 5 g/liter yeast extract, and 5 g/liter NaCl). Strain S905 is a hemA deficient derivative of strain DK905 (18) and requires media supplemented with 40 μg/ml δ-aminolevulinic acid (ALA). Strains harboring pBluescript SK+ or pET14b plasmid derivatives were grown in media supplemented with 100 μg/ml ampicillin.
Construction of plasmids.
All primers used are listed in Table S1 in the supplemental material. The open reading frame of bll3557 (frcB) was obtained by PCR using genomic DNA as the template. The forward primer included the NdeI restriction site, and the reverse primer included the XhoI site. The product was ligated into the NdeI and XhoI sites of the pET14b expression vector, yielding pET14b-3557 (pET14b with the open reading frame of bll3557). The plasmid was transformed into E. coli strain C43(DE3) for overexpression. E. coli C43(DE3) is more tolerant to expression of toxic or membrane proteins and was purchased from Lucigen Corporation (Middleton, WI).
pET14b-3557 was digested with XbaI and XhoI to release the ORF plus the plasmid-borne His tag and ribosome binding site. The fragment was cloned into the XbaI and XhoI sites of pBluescript SK+ for expression of the gene under the control of the plasmid-borne lac promoter in E. coli strain DH5α. The plasmid and empty vector were transformed into E. coli DH5α.
Alignment of annotated cytochromes b561 from diverse organisms.
Homologs from the following species were compared (NCBI accession numbers are shown in parentheses): Bradyrhizobium japonicum (NP_770197.1) Burkholderia glumae (YP_002907809), Shewanella oneidensis (NP_718925), E. coli (AP_002041.1), Brucella abortus (YP_222970.1), Neisseria gonorrhoeae (YP_002002877.1), Synechocystis sp. strain PCC 6803 (NP_440882.1), Helicobacter pylori (NP_207427.1), and Desulfuromonas acetoxidans (ZP_01313790.1).
Disk assay.
Utilization of various siderophores as iron sources by wild-type and mutant B. japonicum bacteria was assessed as growth on solid, modified GSY medium containing 25 μM EDDHA (ethylenediamine-di-o-hydroxyphenylacetic acid). Six-millimeter Whatman paper disks that had been presoaked with siderophores, 10 mM FeCl3, or double-distilled water were placed in the center of the plates. Cells (1 × 105) cells of strains LO, LODTM5, and mutants defective in the ferric siderophore receptor-encoding genes were spotted 3.3 cm away from the disk and visually assessed for growth. The siderophores were a generous gift from J.-M. Meyer (Université Louis-Pasteur, Strasbourg, France) and are listed in Table S2 in the supplemental material.
Overexpression and purification of FrcB.
Five milliliters of a culture of E. coli C43(DE3) harboring pET14b-3557 grown overnight was used to inoculate 1 liter of YT medium. One-liter cultures were incubated with shaking (225 rpm) at 37°C until mid-log phase, at which time 0.5 mM IPTG (isopropyl-1-thio-β-d-galactopyranoside) was added to induce protein expression. Cultures were shaken at 37°C for 4 additional hours. Cells were harvested by centrifugation at 10,000 × g, washed with 100 mM Tris (pH 7.5), and resuspended in 10 ml Tris, 1 mM phenylmethylsulfonyl fluoride, and 25.5 μg aprotinin per 5 g of cells. Cells were disrupted by passage twice through a French pressure cell at 1,200 lb/in2. The lysate was clarified by centrifugation at 4,000 × g, solubilized by gentle rocking at 4°C for 1 h in 5% glycerol, 150 mM NaCl, and 1.2% dodecyl maltoside (DM) (Thermo Fisher Scientific), and further clarified by centrifugation for 90 min at 45,000 × g. The cleared lysate was added to 1 ml of a 50% nickel-nitrilotriacetic acid (Ni-NTA) slurry (Qiagen Inc., Valencia, CA) in a sealed column, and rocked for 75 min at 4°C. The Ni-nitrilotriacetic acid slurry-protein mixture was washed four times with 5 ml of phosphate wash buffer (50 mM NaH2PO4 [pH 8.0], 300 mM NaCl, 20 mM imidazole, 0.2% DM) and once with 5 ml of phosphate wash buffer containing 10% glycerol. Purified His-tagged proteins were eluted with phosphate elution buffer (50 mM NaH2PO4 [pH 8.0], 300 mM NaCl, 250 mM imidazole, 10% glycerol, 0.2% DM). Eluted protein fractions were dialyzed twice, for 10 h each time against 400 ml buffer containing 100 mM Tris (pH 7.5), 10% glycerol, and 0.2% DM. The purified proteins were stored at −80°C.
Absorption spectroscopy and heme quantification.
Ferrous heme bound to protein was determined by measuring the absorption of 8 μM purified, recombinant protein in 100 mM Tris (pH 7.5) after the addition of several crystals of sodium dithionite, which reduced ferric heme to ferrous heme. All spectra were recorded between 360 and 600 nm on a SpectraMax MS spectrophotometer (Molecular Devices).
Quantification of heme was performed using the pyridine hemochromagen method (35). A 500-μl sample contained 8 μM purified, recombinant protein, 100 mM NaOH, and 8.5 μl pyridine. Dithionite crystals were added to reduce the heme. A cuvette containing no protein was used as a reference. The heme-to-protein ratio was calculated using a published millimolar molecular extinction coefficient of 192 at 419 nm.
Mutagenesis of FrcB.
Mutations were made by single-round PCR in pSKBluescript-3557 (pBluescript SK+ with the open reading frame of bll3557) using complementary primers containing the nucleotide changes. The H20, H55, H148, and H169 codons were changed from CAC to the alanine (Ala) codon GCC. The H162 codon was changed from CAC to GCG. PCR products were digested with DpnI to remove the parent plasmids and transformed into E. coli DH5α. Nucleotide mutations were confirmed by sequencing.
Measurement of the accumulation of FrcB and mutant derivatives in E. coli.
Two hundred fifty microliters of stationary-phase, overnight culture of E. coli DH5α harboring pSKBluescript-3557 (wild-type pSKBluescript-3557 or mutant plasmids with histidine mutations) was used to inoculate 50 ml of YT medium. Cultures were grown to mid-log phase with shaking at 37°C, followed by the addition of 0.5 mM IPTG and continued incubation with shaking for 4 h. To investigate heme-dependent protein stability, pSKBluescript-3557 was transformed into the heme-deficient E. coli strain S905. E. coli S905 harboring pBluescript-3557 was grown overnight in 5 ml LB medium plus different concentrations of 5-aminolevulinic acid as stated in the text. We determined empirically that supplementation with 5 μg/ml of ALA was the minimum concentration that allowed growth of the heme auxotroph S905. Growth of the mutant with 5 μg/ml ALA lagged by about two doubling times compared to cells grown with 25 μg/ml ALA. In both sets of experiments, cells were harvested by centrifugation. Samples were mixed with 2× sodium dodecyl sulfate (SDS) sample buffer (20 mM sodium phosphate, 20% glycerol, 4% SDS, 0.2 M dithiothreitol, and 0.001% bromophenol blue) and boiled for 10 min. Aliquots were loaded onto a 15% SDS-polyacrylamide gel, the gel was run for 1 h at 160 V, and the contents of the gel were transferred to a polyvinylidene difluoride (PVDF) membrane and probed with a horseradish peroxidase (HRP)-conjugated, nickel-charged probe which has high affinity to the histidine (His) tag, followed by chemiluminescence and visualized by exposure to X-ray film. Probing with antibodies against GroEL (StressGen, Vancouver, Canada) was performed by standard immunoblot procedures (12).
Whole-cell ferric reductase assay.
E. coli strain DH5α cells harboring pBluescript-3557 or empty vector were grown in 500 ml LB medium overnight to stationary phase. Two liters of B. japonicum strain LO and mutants were grown to mid-log phase in either iron-limiting or 12 μM FeCl3·6H2O conditions in modified GSY medium as indicated in the text. Cells were harvested by centrifugation at either 8,000 × g (E. coli) or 9,500 × g (B. japonicum), washed twice with double-distilled H2O, and resuspended in 100 mM Tris (pH 7.5) to a concentration of 1 × 1010 cells/ml. Ferric reductase activity was measured in cells using a modified version of the protocol of Dailey and Lascelles (8). Ferrozine (125 mM), FeCl3 (100 mM), succinate (100 mM), and iron-nitrilotriacetic acid (Fe-NTA) (100 mM) were made fresh in 100 mM Tris buffer (pH 7.5). Fe-NTA stock was made by first dissolving 1.64 g NaHCO3 in 80 ml double-distilled H2O, followed by the addition of 1.91 g NTA and 2.7 g FeCl3·6H2O. Double-distilled H2O was added to bring the final volume to 100 ml. One-milliliter E. coli reaction mixtures contained 7.5 × 109 cells, 200 μM FeCl3, 100 μM succinate, and 1 mM ferrozine. One milliliter of the B. japonicum reaction mixture contained 7.5 × 109 cells, 200 μM Fe-nitrilotriacetic acid, and 1 mM ferrozine. Reactions were started by the addition of whole cells, blanked against cells harboring empty vector (E. coli) or parent strain LO (B. japonicum) at time zero. The reaction mixtures were then incubated in a 37°C water bath, and the absorbance at 562 nm was measured over time. Ferrous ferrozine produced was calculated using the ferrozine molar extinction coefficient 27.9 mM−1 cm−1 at 562 nm.
Iron-dependent oxidation of FrcB.
Oxidation of FrcB was discerned spectrophotometrically by comparing the reduced sample to that oxidized by FeCl3. Dithionite (final concentration of 200 μM) was added to 20 μM protein. Fifty microliters of the reduced protein was then aliquoted into the reaction mixture containing 500 μM FeCl3 or air-saturated buffer in a final volume of 500 μl and a final FrcB concentration of 2 μM. The reduced spectrum between 390 and 600 nm was recorded immediately. The oxidizing reaction mixture was incubated at 29°C for 30 min before the spectrum was recorded and then subtracted from the reduced spectrum. A background spectrum of dithionite plus FeCl3 minus FeCl3 in the absence of protein was subtracted from the reduced minus FeCl3-oxidized spectrum. This was done to eliminate spectral features of oxidized FeCl3 below 500 nm.
Determination of steady-state mRNA levels.
Parent strain LO, irr, or bll3557 mutant cells were grown under iron-limiting and iron-replete conditions. RNA extraction, cDNA synthesis, and PCR were carried out as described previously (34). Primers were constructed to amplify a region within the open reading frame. gapA is a housekeeping gene which is not iron or Irr responsive. The data are expressed as the relative starting quantities (SQ) of the mRNA normalized to gapA and presented as the averages plus standard deviations for three samples.
5′ RACE.
The transcription start site for frcB was determined by rapid amplification of 5′ cDNA ends (5′ RACE) using a kit from Invitrogen (Carlsbad, CA) according to the manufacturer's instructions. RNA was prepared from cells grown in iron-limited media, where the genes of interest were expressed at high levels.
RESULTS
Identification of outer membrane receptors necessary for utilization of rhodotorulic acid and pyoverdine PL-8 as iron sources.
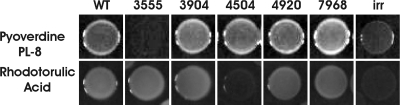
Previous work identified five putative outer membrane ferric siderophore receptor genes based on homology and on their induction under iron-limiting conditions (34, 41). However, these data cannot accurately predict which siderophores the receptors bind and transport into the periplasm. Moreover, B. japonicum does not contain genes that encode recognizable siderophore synthesis proteins, and thus, the genome does not yield clues to the identity of the receptors. Among the five putative proteins, only Bll4920 was identified as a ferrichrome receptor (19, 34). Rhodotorulic acid was shown previously to be an iron source for B. japonicum (27). Thus, we tested the ability of mutants defective in each of the receptor genes to grow on rhodotorulic acid as an iron source (Fig. 1). To do this, a paper filter disk containing rhodotorulic acid was placed at the center of an agar medium plate, and cells of the wild type or mutant strain were spotted onto the plate 3.3 cm from the disk and allowed to grow at 29°C. Whereas the parent strain grew with rhodotorulic acid, the blr4504 mutant strain did not. The wild type and all mutants grew with FeCl3 as an iron source (data not shown). Thus, Blr4504 is very likely the rhodotorulic acid receptor. We designated the blr4504 gene fhuE in keeping with the E. coli name for the gene encoding the rhodotorulic acid receptor and because Blr4504 is the B. japonicum protein with the highest homology with the E. coli FhuE protein.
Fig. 1.
Growth of wild-type (WT) and putative outer membrane receptor mutants on pyoverdine PL-8 or rhodotorulic acid as the sole iron source. The parent B. japonicum strain LO, irr mutant strain, and the ferric siderophore receptor mutants, defective in the blr3555 (3555), blr3904, blr4504, bll4920, or bll7968 genes were spotted on plates where the sole iron source was pyoverdine PL-8 or rhodotorulic acid, and the ability of the strains to grow was assessed visually.
We tested 28 additional siderophores from several Pseudomonas species (gift of J.-M. Meyer) (see Table S2 in the supplemental material) obtained on paper disks in the same manner as described above for rhodotorulic acid. Only one of them, pyoverdine PL-8, supported growth of the parent strain. Pyoverdine PL-8 is a siderophore produced by Pseudomonas fluorescens strain PL8; its structure has been determined, but the cognate outer membrane receptor has not been identified in that organism (23). Unlike the B. japonicum parent strain, the blr3555 mutant did not grow on pyoverdine PL-8 (Fig. 1), indicating that Blr3555 is the outer membrane receptor for that siderophore. The annotated genome shows blr3556 immediately downstream of blr3555 (http://genome.kazusa.or.jp/rhizobase/Bradyrhizobium). However, resequencing that region reveals that blr3555 and blr3556 comprise a single open reading frame (GenBank accession no. FJ430786.1), with a likely frameshift error in the annotated genome. This gene was designated pyoR.
The pyoR and fhuE genes are regulated by the Irr protein and thus are not activated by iron limitation in an irr mutant (34). Consistent with those observations, the irr strain grew very poorly on rhodotorulic acid or pyoverdine PL-8 as iron sources (Fig. 1).
The bll3557 gene found within the pyoR gene cluster encodes a heme protein.
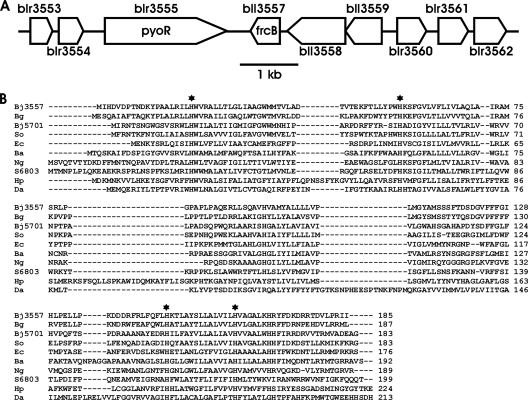
Microarray analysis shows that blr3555 is clustered with eight other genes from blr3553 to blr3562 that are all upregulated 3- to 23-fold at the RNA level under iron-limiting conditions (41) (Fig. 2 A) (see Table S3 in the supplemental material). bll3558 and bll3559 are predicted to encode regulatory proteins. blr3553, blr3554, blr3560, blr3561, and blr3562 encode proteins of unknown function. We focused on bll3557, because it is predicted to encode a protein belonging to a family of heme proteins of unknown function that are common within numerous subdivisions of the proteobacteria and that are also found in some cyanobacteria (Fig. 2B). This bacterial protein family, annotated as Cytb561, comprises small uncharacterized proteins that are part of the cytochrome_b_N superfamily in the conserved domain database (cl00859) (20) that includes the N-terminal portion of the eukaryotic cytochrome b subunit of the mitochondrial bc1 complex. bll3557 was also of interest, because most genes encoding iron-containing proteins are downregulated under iron-limiting conditions in B. japonicum (41), and thus, it was noteworthy that bll3557 was upregulated under those conditions. This gene was renamed frcB for ferric reductase cytochrome b on the basis of its characterization described below.
Fig. 2.
Context of frcB in the B. japonicum genome and homologs of FrcB in other organisms. (A) The blr3555 (pyoR) and bll3557 (frcB) genes are in a cluster of iron- and Irr-regulated genes. (B) ClustalW alignment of the B. japonicum (Bj) FrcB sequence with the sequences of annotated cytochromes b561 from the cyanobacterium Synechocystis sp. strain PCC 6803 (S6803), and other proteobacterial species, namely, Burkholderia glumae (Bg), Shewanella odeidensis (So), Escherichia coli (Ec), Brucella abortus (Ba), Neisseria gonorrhoeae (Ng), Helicobacter pylori (Hp), and Desulfuromonas acetoxidans (Da). Conserved histidine residues are indicated by black stars. Gaps introduced to maximize alignment are indicated by dashes.
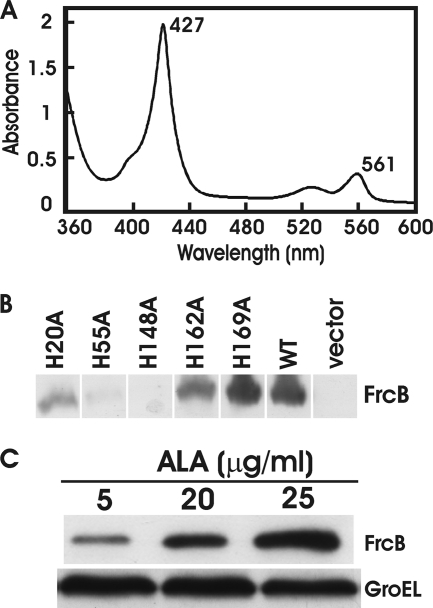
To determine whether frcB encodes a heme protein, the gene was overexpressed in E. coli and purified as described in Materials and Methods. Bioinformatic analysis predicts that FrcB is an integral membrane protein with four membrane-spanning domains (see Fig. S1 in the supplemental material) (15, 37). In support of this, the recombinant protein was found in the membrane fraction, and detergent was necessary for its solubilization throughout the purification process. The molecular mass of the purified protein was about 20 kDa, in good agreement with its predicted molecular mass. Recombinant FrcB reduced with dithionite had absorption spectra at 427 nm and 561 nm (Fig. 3 A) that were characteristic of a b-type cytochrome heme protein. The heme was readily extracted into an alkaline pyridine solution, showing that it was noncovalently bound to the protein. Moreover, the absorption peaks at 419 and 556 nm of the pyridine-coordinated heme group identifies it as protoheme (data not shown). Finally, quantification of the extracted heme revealed a stoichiometry of 2.1 heme molecules per monomer. Neither the addition of the heme precursor 5-aminolevulinic acid (ALA) to the overexpressing cells nor the addition of heme to the purified protein increased the heme-to-protein ratio (data not shown).
Fig. 3.
frcB encodes a diheme protein, and heme-bound FrcB was analyzed. (A) The absorption spectrum of purified, recombinant FrcB reduced with sodium dithionite by light between wavelengths of 360 and 600 nm. Peaks at 427 and 561 nm are noted. (B) Analysis of FrcB and histidine-to-alanine (His-to-Ala) mutants in E. coli cells by immunoblot analysis. An HRP-conjugated anti-His probe was used to detect His-tagged proteins. (C) Analysis of FrcB accumulation in E. coli hemA strain S905 whole cells, grown in media supplemented with 5, 20, or 25 μg/ml δ-aminolevulinic acid (ALA), by immunoblot analysis. An HRP-conjugated anti-His probe was used to detect His-tagged proteins. GroEL was used as a control for a protein not controlled by heme.
Evidence that heme is required for FrcB protein stability.
Protoheme in b-type cytochromes is usually coordinated by histidine residues. Alignment of FrcB with homologs from diverse proteobacteria and cyanobacteria identified 5 histidine residues that are conserved throughout (Fig. 2B). We made a series of frcB mutations that result in histidine-to-alanine variants at the conserved positions 20, 55, 148, 162, and 169. In attempts to overexpress the gene variants to produce pure protein, we were successful only with the H169A variant, and little or no protein of the other variants was obtained. FrcB(H169A) contained 1.9 heme molecules per monomer, as was found in the wild-type protein (data not shown).
To assess whether the inability to obtain pure protein for the four His-to-Ala variants was due to accumulation levels in E. coli, we examined the gene variants borne on pBluescript SK for expression in E. coli strain DH5α by Western blot analysis using antibodies directed against the His tag. Protein accumulation was much lower for proteins carrying variants H20A, H55A, H148A, and H162A compared to the wild type (Fig. 3B), but variant His169A protein was expressed at approximately wild-type levels (Fig. 3B).
The low level of expression of the four His-to-Ala variants suggested that those amino acid residues may be necessary for protein stability. Moreover, if they are heme-binding ligands, then heme may be required for protein stability. To test this idea, we expressed wild-type frcB in the E. coli heme-deficient mutant strain S905. This mutant is defective in the gtrA (hemA) gene encoding the heme synthesis enzyme glutamyl tRNA reductase, which synthesizes the heme precursor δ-aminolevulinic acid. Thus, the gtrA mutant requires exogenous ALA for growth. We expressed the frcB gene in the gtrA strain and grew cells in media with various concentrations of ALA (Fig. 3C). FrcB protein levels were low in cells grown with 5 μg/ml ALA but were substantially higher in cells grown with 25 μg/ml ALA. These findings are consistent with the hypothesis that heme is required for FrcB stability and that conserved histidine residues 20, 55, 148, and 162 are likely required for heme coordination.
The frcB gene is necessary for maximum ferric reductase activity in iron-limited cells.
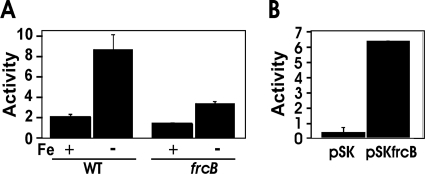
The arrangement and spacing of the nine genes in the blr3555 gene cluster indicate that they do not comprise an operon. Nevertheless, the arrangement and spacing of the genes suggest that the clustered and coregulated genes may be involved in a common process. Moreover, ferric reductase activity is required for iron assimilation into cells, and known eukaryotic ferric reductases contain heme as a prosthetic group (9, 22, 28). Although FrcB does not share sequence homology with the eukaryotic reductases, it is expressed under conditions where that activity would be needed. To address this further, we measured ferric reductase activity in whole cells of the parent strain and in a mutant strain containing a deletion in most of the frcB open reading frame and replaced the sequence with an antibiotic-encoding cassette (see Materials and Methods). Ferric reductase was measured using the ferrozine assay as described previously (8). Ferric reductase activity was detected in wild-type cells grown in iron-replete media and was induced by about 4-fold in cells from iron-limited cultures (Fig. 4 A). However, this activity was diminished in the frcB mutant, suggesting that FrcB is involved in inducible ferric reductase activity. Deletion of frcB did not abolish activity, which may explain the lack of a growth phenotype of the mutant under iron-limiting conditions (data not shown).
Fig. 4.
Ferric reductase activity in E. coli and B. japonicum cells. The production of ferrous ferrozine produced by whole cells was measured and is presented as nanomoles of ferrous iron produced per hour. Ferric reductase activity is expressed as nanomoles of Fe2+ produced/hour/109 cells, and the values are presented as the averages plus standard deviations (error bars) of three replicate samples. (A) B. japonicum wild-type (WT) or frcB mutant strains were grown in low-iron media (Fe −) or supplemented with 12 μM FeCl3 (Fe +). (B) E. coli DH5α harboring empty vector (pSK) or vector plus bll3557 (frcB) (pSKfrcB), grown in LB medium.
Expression of frcB in E. coli from a high-copy-number plasmid confers ferric reductase activity on cells.
The frcB gene was introduced into E. coli strain DH5α on pBluescript SK and under the control of the plasmid-borne lacZ promoter. Strain DH5α harboring the empty vector showed low ferric reductase activity (Fig. 4B), but cells that expressed frcB from the plasmid had much higher activity. Thus, frcB confers ferric reductase activity on a heterologous host.
Oxidation of purified recombinant FrcB by iron in vitro.
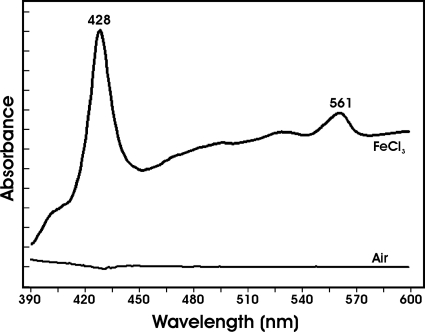
Ferric reductases mediate electron transfer from a reductant to ferric iron. In our hands, reductants that reduced the heme groups of purified FrcB, such as dithionite or ascorbate, were also able to reduce ferric iron nonenzymatically in the absence of protein. To circumvent this problem, we measured the oxidation of reduced FrcB by ferric iron rather than ferric iron reduction. To do this, FrcB was reduced with dithionite, and the spectrum was compared to the spectrum of dithionite-treated protein in the presence of 100 μM FeCl3 (Fig. 5). The reduced minus FeCl3-treated spectra gave absorption peaks at 561 and 424 nm, showing that iron oxidized the heme groups of the protein. The control with air only yielded no spectral features, thus FrcB was not auto-oxidized. These findings, along with the in vivo data, strongly support the conclusion that FrcB is a ferric reductase.
Fig. 5.
Dithionite-reduced FrcB is oxidized in the presence of FeCl3. FrcB (2 μM) reduced with dithionite as described in the text was treated with 500 μM FeCl3 or air-saturated buffer, and the absorption spectrum of each was subtracted from the reduced spectrum. Peaks at 428 and 561 nm are noted. The tick marks on the y axis represent a change in absorption of 0.05.
The frcB gene is regulated by Irr.
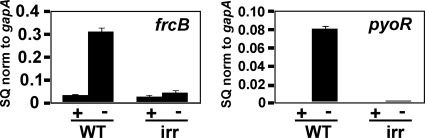
The nine-gene cluster from blr3553 to bll3562 is regulated by iron on the basis of the results of microarray analysis (41), and this was confirmed for frcB by quantitative real-time PCR (qPCR) (Fig. 6). Transcript levels were very low in cells grown in iron-replete media but were induced under iron-limiting conditions, consistent with the pattern of ferric reductase activity. Control of the pyoR (blr3555) gene by iron is mediated by Irr (34), and therefore, we wanted to determine whether frcB was regulated similarly. Thus, we measured steady-state levels of frcB in cells of the parent strain and an irr mutant, using pyoR as a control for an Irr-regulated gene (Fig. 6). The induction of frcB transcripts under iron-limiting conditions was not observed in the irr strain. Thus, Irr is normally a positive effector of that gene, and frcB is coregulated with pyoR.
Fig. 6.
Effects of an irr mutation on iron-dependent expression of frcB mRNA. mRNAs from cells of the parent strain or the irr mutant grown in medium supplemented with no added iron (−) or with 12 μM FeCl3 (+) were analyzed by quantitative real-time PCR (qPCR). The data are expressed as the relative starting quantity (SQ) of the mRNA normalized (norm) to the housekeeping gene gapA, and the values are presented as the averages plus standard deviations (error bars) of three replicate samples. pyoR was used for comparison.
DISCUSSION
Prokaryotic ferric reductases are mostly soluble, flavin-containing proteins based on the characterization of enzyme activity in cell fractions (31) and on the few proteins that have been purified (32, 40). In contrast, FrcB is bound to the membrane and contains two heme groups per monomer, as is observed for eukaryotic ferric reductases (9, 22, 28). Association of FrcB with the membrane raises the possibility that iron reduction occurs concomitantly with transport into the cytoplasm, and additional work is needed to clarify this. Ferric reductase activity has not been demonstrated for any of the purified eukaryotic heme proteins. It is plausible that ferric reductase activity requires the integrity of the membrane or cell or requires additional proteins. In the current study, we demonstrate that iron was able to oxidize reduced FrcB; therefore, it has reductase activity. Further work will be needed to determine the physiological reductant substrate and whether additional protein components are needed to reduce FrcB.
FrcB is part of the cytochrome b561 (cytb561) family of proteins, and homologs are found throughout the proteobacteria and in some cyanobacterial species. Thus, the current work suggests that ferric reductase heme proteins are not confined to eukaryotes but may be common in prokaryotes. Mammalian Dcytb is a member of a protein family also designated cytb561. However, the common designation of the prokaryotic and eukaryotic proteins is fortuitous and based on the absorption peak of the heme moieties, not on amino acid homology. Cytochromes with a protoheme prosthetic group will typically absorb in the 555- to 565-nm region of the spectrum and are functionally diverse proteins.
Accumulation of FrcB in E. coli cells was strictly dependent on heme, as determined by its dependence on exogenous δ-aminolevulinic acid (ALA) in a heme-dependent strain. This suggests that heme is required for FrcB stability. Moreover, mutants containing conserved histidine substitutions accumulated to only very low levels in cells. These amino acids are likely to be coordinating residues. Heme stabilizes cytochrome c-type cytochromes, which bind the protein covalently through cysteine ligation (12), and stabilizes some b-type cytochromes as well (24).
Iron metabolism is globally controlled by the Irr protein in B. japonicum (41), and genes involved in iron acquisition are positively regulated in an Irr-dependent manner under iron-limiting conditions (34). The current study shows that frcB is clustered with a ferric siderophore receptor gene and is coregulated with it. Collectively, these findings support the hypothesis that the ferric reductase activity of FrcB is involved in iron acquisition and metabolism in iron-limited cells. The frcB mutant showed diminished but measurable activity (Fig. 4A), suggesting the presence of an additional reductase in B. japonicum. This likely explains the lack of a growth phenotype of the mutant under the conditions tested.
Pyoverdines are a group of structurally related siderophores produced by fluorescent Pseudomonas species (38), and rhodotorulic acid is fungal in origin. We found that B. japonicum has a specific outer membrane receptor for each of these compounds. Unlike Sinorhizobium meliloti and Rhizobium leguminosarum, B. japonicum does not have a known siderophore biosynthesis pathway (26). Because pyoverdines and rhodotorulic acid are specific to limited microbial genera, it is extremely unlikely that the ancestor of B. japonicum had the capacity to synthesize them, followed by loss in the extant species. Rather, B. japonicum probably acquired the ability to transport pyoverdine and rhodotorulic acid as an adaptation to its availability in the rhizosphere.
Supplementary Material
ACKNOWLEDGMENTS
We thank J.-M. Meyer for the gift of siderophores.
This work was supported by National Institutes of Health grant R01 GM067966 to M.R.O.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 24 June 2011.
REFERENCES
- 1. Andrews S. C., Robinson A. K., Rodriguez-Quinones F. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27:215–237 [DOI] [PubMed] [Google Scholar]
- 2. Reference deleted.
- 3. Berczi A., Su D., Asard H. 2007. An Arabidopsis cytochrome b561 with trans-membrane ferrireductase capability. FEBS Lett. 581:1505–1508 [DOI] [PubMed] [Google Scholar]
- 4. Berczi A., Su D., Lakshminarasimhan M., Vargas A., Asard H. 2005. Heterologous expression and site-directed mutagenesis of an ascorbate-reducible cytochrome b561. Arch. Biochem. Biophys. 443:82–92 [DOI] [PubMed] [Google Scholar]
- 5. Braun V. 2006. Energy transfer between biological membranes. ACS Chem. Biol. 1:352–354 [DOI] [PubMed] [Google Scholar]
- 6. Braun V., Killmann H. 1999. Bacterial solutions to the iron-supply problem. Trends Biochem. Sci. 24:104–109 [DOI] [PubMed] [Google Scholar]
- 7. Chu B. C., et al. 2010. Siderophore uptake in bacteria and the battle for iron with the host: a bird's eye view. Biometals 23:601–611 [DOI] [PubMed] [Google Scholar]
- 8. Dailey H. A., Jr., Lascelles J. 1977. Reduction of iron and synthesis of protoheme by Spirillum itersonii and other organisms. J. Bacteriol. 129:815–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dancis A., Klausner R. D., Hinnebusch A. G., Barriocanal J. G. 1990. Genetic evidence that ferric reductase is required for iron uptake in Saccharomyces cerevisiae. Mol. Cell. Biol. 10:2294–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ferguson A. D., Deisenhofer J. 2002. TonB-dependent receptors—structural perspectives. Biochim. Biophys. Acta 1565:318–332 [DOI] [PubMed] [Google Scholar]
- 11. Frustaci J. M., Sangwan I., O'Brian M. R. 1991. Aerobic growth and respiration of a δ-aminolevulinic acid synthase (hemA) mutant of Bradyrhizobium japonicum. J. Bacteriol. 173:1145–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gao T., O'Brian M. R. 2007. Control of DegP-dependent degradation of c-type cytochromes by heme and the cytochrome c maturation system in Escherichia coli. J. Bacteriol. 189:6253–6259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Glanfield A., et al. 2010. A cytochrome b561 with ferric reductase activity from the parasitic blood fluke, Schistosoma japonicum. PLoS Negl. Trop. Dis. 4:e884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hamza I., Chauhan S., Hassett R., O'Brian M. R. 1998. The bacterial Irr protein is required for coordination of heme biosynthesis with iron availability. J. Biol. Chem. 273:21669–21674 [DOI] [PubMed] [Google Scholar]
- 15. Hirokawa T., Boon-Chieng S., Mitaku S. 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14:378–379 [DOI] [PubMed] [Google Scholar]
- 16. Johnston A. W., et al. 2007. Living without Fur: the subtlety and complexity of iron-responsive gene regulation in the symbiotic bacterium Rhizobium and other alpha-proteobacteria. Biometals 20:501–511 [DOI] [PubMed] [Google Scholar]
- 17. Krewulak K. D., Vogel H. J. 2008. Structural biology of bacterial iron uptake. Biochim. Biophys. Acta 1778:1781–1804 [DOI] [PubMed] [Google Scholar]
- 18. Leong S. A., Ditta D. S., Helinski D. R. 1982. Heme synthesis in Rhizobium. Identification of a cloned gene coding for d-aminolevulinic acid synthetase from Rhizobium meliloti. J. Biol. Chem. 257:8724–8730 [PubMed] [Google Scholar]
- 19. LeVier K., Guerinot M. L. 1996. The Bradyrhizobium japonicum fegA gene encodes an iron-regulated outer membrane protein with similarity to hydroxamate-type siderophore receptors. J. Bacteriol. 178:7265–7275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Marchler-Bauer A., et al. 2011. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 39:D225–D229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Matzanke B. F., Anemuller S., Schunemann V., Trautwein A. X., Hantke K. 2004. FhuF, part of a siderophore-reductase system. Biochemistry 43:1386–1392 [DOI] [PubMed] [Google Scholar]
- 22. McKie A. T. 2008. The role of Dcytb in iron metabolism: an update. Biochem. Soc. Trans. 36:1239–1241 [DOI] [PubMed] [Google Scholar]
- 23. Meyer J. M., et al. 2002. Siderophore-mediated iron uptake in fluorescent Pseudomonas: characterization of the pyoverdine-receptor binding site of three cross-reacting pyoverdines. Arch. Biochem. Biophys. 397:179–183 [DOI] [PubMed] [Google Scholar]
- 24. Mukhopadhyay K., Lecomte J. T. 2004. A relationship between heme binding and protein stability in cytochrome b5. Biochemistry 43:12227–12236 [DOI] [PubMed] [Google Scholar]
- 25. Nienaber A., Hennecke H., Fischer H. M. 2001. Discovery of a haem uptake system in the soil bacterium Bradyrhizobium japonicum. Mol. Microbiol. 41:787–800 [DOI] [PubMed] [Google Scholar]
- 26. O'Brian M. R., Fabiano E. 2010. Mechanisms and regulation of iron homeostasis in the rhizobia, p. 37–63 In Cornelis P., Andrews S. C. (ed.), Iron uptake and homeostasis in microorganisms. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 27. Plessner O., Klapatch T., Guerinot M. L. 1993. Siderophore utilization by Bradyrhizobium japonicum. Appl. Environ. Microbiol. 59:1688–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Robinson N. J., Procter C. M., Connolly E. L., Guerinot M. L. 1999. A ferric-chelate reductase for iron uptake from soils. Nature 397:694–697 [DOI] [PubMed] [Google Scholar]
- 29. Rudolph G., et al. 2006. The iron control element, acting in positive and negative control of iron-regulated Bradyrhizobium japonicum genes, is a target for the Irr protein. J. Bacteriol. 188:733–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sangwan I., Small S. K., O'Brian M. R. 2008. The Bradyrhizobium japonicum Irr protein is a transcriptional repressor with high-affinity DNA-binding activity. J. Bacteriol. 190:5172–5177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schroder I., Johnson E., de Vries S. 2003. Microbial ferric iron reductases. FEMS Microbiol. Rev. 27:427–447 [DOI] [PubMed] [Google Scholar]
- 32. Sedlacek V., van Spanning R. J., Kucera I. 2009. Ferric reductase A is essential for effective iron acquisition in Paracoccus denitrificans. Microbiology 155:1294–1301 [DOI] [PubMed] [Google Scholar]
- 33. Small S. K., Puri S., O'Brian M. R. 2009. Heme-dependent metalloregulation by the iron response regulator (Irr) protein in Rhizobium and other Alpha-proteobacteria. Biometals 22:89–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Small S. K., Puri S., Sangwan I., O'Brian M. R. 2009. Positive control of ferric siderophore receptor gene expression by the Irr protein in Bradyrhizobium japonicum. J. Bacteriol. 191:1361–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Smith K. M. 1975. Porphyrins and metalloporphyrins. Elsevier Scientific, Amsterdam, The Netherlands [Google Scholar]
- 36. Su D., Asard H. 2006. Three mammalian cytochromes b are ascorbate-dependent ferrireductases. FEBS J. 273:3722–3734 [DOI] [PubMed] [Google Scholar]
- 37. Tusnady G. E., Simon I. 2001. The HMMTOP transmembrane topology prediction server. Bioinformatics 17:849–850 [DOI] [PubMed] [Google Scholar]
- 38. Visca P., Imperi F., Lamont I. L. 2007. Pyoverdine siderophores: from biogenesis to biosignificance. Trends Microbiol. 15:22–30 [DOI] [PubMed] [Google Scholar]
- 39. Wang J., Budde A. D., Leong S. A. 1989. Analysis of ferrichrome biosynthesis in the phytopathogenic fungus Ustilago maydis: cloning of an ornithine-N5-oxygenase gene. J. Bacteriol. 171:2811–2818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang S., Wu Y., Outten F. W. 2011. Fur and the novel regulator YqjI control transcription of the ferric reductase gene yqjH in Escherichia coli. J. Bacteriol. 193:563–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yang J., et al. 2006. Bradyrhizobium japonicum senses iron through the status of haem to regulate iron homeostasis and metabolism. Mol. Microbiol. 60:427–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang D. L., Su D., Berczi A., Vargas A., Asard H. 2006. An ascorbate-reducible cytochrome b561 is localized in macrophage lysosomes. Biochim. Biophys. Acta 1760:1903–1913 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.