Abstract
Phosphoglucosamine mutase (PNGM) is an evolutionarily conserved bacterial enzyme that participates in the cytoplasmic steps of peptidoglycan biosynthesis. As peptidoglycan is essential for bacterial survival and is absent in humans, enzymes in this pathway have been the focus of intensive inhibitor design efforts. Many aspects of the structural biology of the peptidoglycan pathway have been elucidated, with the exception of the PNGM structure. We present here the crystal structure of PNGM from the human pathogen and bioterrorism agent Bacillus anthracis. The structure reveals key residues in the large active site cleft of the enzyme which likely have roles in catalysis and specificity. A large conformational change of the C-terminal domain of PNGM is observed when comparing two independent molecules in the crystal, shedding light on both the apo- and ligand-bound conformers of the enzyme. Crystal packing analyses and dynamic light scattering studies suggest that the enzyme is a dimer in solution. Multiple sequence alignments show that residues in the dimer interface are conserved, suggesting that many PNGM enzymes adopt this oligomeric state. This work lays the foundation for the development of inhibitors for PNGM enzymes from human pathogens.
INTRODUCTION
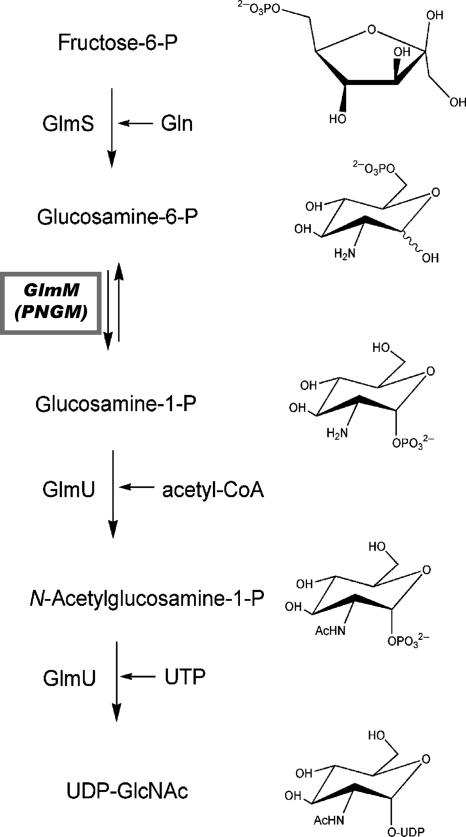
Peptidoglycan is a major component of the cell wall of both Gram-negative and Gram-positive bacteria. The molecule UDP-N-acetylglucosamine (UDP-GlcNAc) is one of the key building blocks in peptidoglycan biosynthesis (1). In bacteria, the second step in the formation of UDP-GlcNAc is the interconversion of glucosamine-6-phosphate to glucosamine-1-phosphate, which is catalyzed in the cytoplasm by the enzyme phosphoglucosamine mutase (PNGM). The glucosamine-1-phosphate product is subsequently converted to N-acetylglucosamine-1-phosphate and finally to UDP-GlcNAc, which is then further modified for incorporation into the peptidoglycan heteropolymer (Fig. 1). Bacterial enzymes that participate in the biosynthesis of UDP-GlcNAc are potential targets for antibacterial compounds, as the biosynthetic pathway of UDP-GlcNAc differs between prokaryotes and eukaryotes. To date, crystal structures have been reported for each of the bacterial proteins involved in the cytoplasmic steps of UDP-GlcNAc synthesis, with the exception of PNGM (1, 7).
Fig. 1.
An overview of the early steps of the peptidoglycan biosynthetic pathway, highlighting the role of the glmM gene product (phosphoglucosamine mutase) in the production of the critical precursor UDP-N-acetylglucosamine. PNGM catalyzes the reversible conversion of glucosamine-6-phosphate to glucosamine-1-phosphate in this pathway, via a bis-phosphorylated intermediate glucosamine-1,6-bis-phosphate (not shown). Adapted from reference 1.
Phosphoglucosamine mutases (E.C. 5.4.2.10) from various bacteria, including Escherichia coli, Helicobacter pylori, Pseudomonas aeruginosa, Salmonella enterica serovar Typhimurium, and Staphylococcus aureus, have been characterized functionally and/or kinetically. In Gram-negative organisms, these enzymes appear to be essential for viability (5, 17, 23, 31). In Gram-positive organisms, reports to date suggest that this enzyme is not required for viability, but gene deletion studies show reduced virulence, increased susceptibility to antibiotics, and decreased biofilm formation (10, 13, 30, 35, 37). The role of PNGM in the virulence of Bacillus anthracis infections has not been examined, but the peptidoglycan of this organism is known to stimulate the host inflammatory response (12).
The PNGMs comprise one subgroup of a large and diverse enzyme superfamily, known collectively as the α-d-phosphohexomutases (29, 34). All enzymes in this superfamily catalyze the reversible conversion of 1- and 6-phosphosugars but differ in their specificity for the hexose. The PNGMs have a preference for glucosamine-based phosphosugars, while enzymes in the other subgroups utilize N-acetylglucosamine, exclusively glucose, or both glucose and mannose. Despite significant sequence diversity (typically ∼25% identity between subgroups), the α-d-phosphohexomutases share key catalytic residues and a common reaction mechanism, whereby a monophosphorylated sugar is converted into a bis-phosphorylated intermediate, which is subsequently dephosphorylated to form the product. The enzymes characterized to date require Mg2+ for full activity, and a highly conserved phosphoserine residue in the active site participates in the phosphoryl transfer reaction.
Here we report on the structure determination of PNGM from the human pathogen and category A bioterrorism agent Bacillus anthracis. This is the first report of a PNGM structure, completing structural characterization of bacterial enzymes in the UDP-GlcNAc pathway. Sequence-structure analyses reveal conserved residues in the active site and their likely roles in catalysis and substrate specificity. Conformational variability between independent molecules in the crystal shows a large domain rotation relevant to ligand binding. A conserved oligomeric interface suggests that many enzymes in the PNGM subgroup exist as dimers. These studies have implications for understanding the function of this enzyme and open the door for inhibitor design efforts for PNGMs from various pathogenic bacteria.
MATERIALS AND METHODS
Structure solution and refinement.
Expression, purification, and crystallization of native Bacillus anthracis PNGM (BaPNGM) have been previously described (16). The structure of BaPNGM was solved by molecular replacement using MOLREP (32), as outlined previously (16). However, difficulties during the early stages of refinement led us to seek additional phasing information from selenomethionine (SeMet)-substituted crystals (see the supplemental material). Although a MAD data set (data not shown) was collected from these crystals, the phasing information (from individual wavelengths or combinations thereof) was insufficient for de novo structure determination. We therefore used the SeMet data set (see Table S1 in the supplemental material) to construct an anomalous difference Fourier map, using phases from the preliminary molecular replacement solution. This map clearly showed peaks for 34 of the possible 36 SeMet residues, confirming the space group and molecular replacement solution. The SeMet peaks were then used as guideposts during model building, and they proved invaluable for placing the amino acid sequence onto the polypeptide backbone. Additional efforts for refinement, including the use of translation, libration, and serew rotation (TLS) parameters, improved the maps and allowed refinement to progress. These improved maps also revealed that there was a significant conformational difference for one domain of the protein in the two monomers in the asymmetric unit. In retrospect, we attribute the initial difficulties in refinement to the medium resolution of the data (2.7 Å), the low sequence identity of the search model available for molecular replacement (∼30%), and the conformational variability between monomers in the asymmetric unit.
Refinement was performed with Refmac 5.0 (19). The structures were refined to convergence through iterative cycles of refinement and manual rebuilding with Coot (6). Progress of the refinement was monitored by following Rfree; 5% of each data set was set aside for cross validation prior to refinement. Water molecules were placed automatically by Coot in peaks of >3.0 σ in Fo − Fc maps and within reasonable hydrogen bonding distance of oxygen or nitrogen atoms. Due to the moderate resolution of the structure, only a minority of potential water molecules retained appropriate positions and electron density after refinement, and just 17 are included in the final model. The B-factor model used during refinement consisted of an isotropic B-factor for each non-hydrogen atom and four TLS groups per chain. Additional details of the refinement are shown in Table 1.
Table 1.
Refinement statistics for B. anthracis phosphoglucosamine mutase
| Parameter | Value(s) for BaPNGM |
|---|---|
| Resolution range (Å) | 43.0-2.70 |
| Rworkb/Rfreec | 0.21/0.28 (0.36/0.45)a |
| No. of: | |
| Protein residues (chain A/B) | 445/444 |
| Protein atoms | 6,435 |
| Water molecules | 17 |
| Phosphate ions | 8 |
| RMSD bonds/angles (Å/°) | 0.017/1.865 |
| Ramachandran plot (%)d | 92.1/6.1/1.8 |
| Avg B-factors (Å2) | |
| Wilson | 93.0 |
| Protein | 89.5 |
| Water | 83.3 |
| Phosphate ions | 129.5 |
Numbers in parentheses indicate value for highest resolution shell (2.77 to 2.70 Å).
Rwork = ∑hklW||Fo| − k|Fc ||/∑hklW|Fo|, where W is the working set and hkl are the lattice points of the crystal.
Rfree = ∑hklT||Fo| − k|Fc||/∑hklT|Fo|, where T is the test set obtained by randomly selecting 5% of the data.
Residues in favored/allowed/outlier regions of the Ramachandran plot were calculated using RAMPAGE (14).
Although all PNGM enzymes are expected to require a bound Mg2+ ion for activity, no metal was located in the binding site of either monomer. This is likely due to the low pH of the crystallization buffer (4.5) (16), which presumably protonated the three aspartates that coordinate the Mg2+ ion. Related proteins, such as P. aeruginosa phosphomannomutase/phosphoglucomutase (PGM) with an identical metal-binding site, are known to lose affinity for Mg2+ at pH values below 7.0 (20). Eight phosphate ions are included in the final model of this structure; in each monomer, one of these phosphate ions is bound in the active site (see Results and Discussion). Structural figures were prepared with PyMOL (4).
Dynamic light scattering.
Dynamic light scattering measurements were done at 25°C on a Protein Solutions DynaPro 99 instrument at a wavelength of 8,363 Å. The protein sample (concentration, 1 mg/ml) was 0.22-μm filtered prior to data collection. At least 20 measurements were taken on the sample at 5-s intervals. The standard deviation of the hydrodynamic radius measurements was less than 25%, indicating a monodisperse sample.
Protein structure accession number.
Atomic coordinates and structure factor amplitudes have been deposited in the Protein Data Bank (PDB) under accession code 3PDK.
RESULTS AND DISCUSSION
Tertiary structure of BaPNGM.
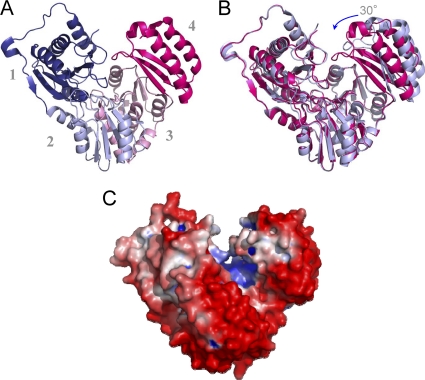
The BaPNGM monomer is a 448-residue protein with four structural domains arranged in an overall heart shape (Fig. 2 A). Domains 1 to 3 share a fold, consisting of a mixed α/β core. The β-sheet of the core consists of four antiparallel β-strands, arranged in a 2-1-3-4 pattern. In each domain, an α-helix is found between strands 1 to 2 and 2 to 3. Along with the conserved core of domains 1 to 3, additional strands and helices are also found, typically located on the periphery of the molecule. Domain 4, by contrast, is topologically distinct, and consists of a 3-stranded, antiparallel β-sheet, flanked by two α-helices. Overall, BaPNGM is structurally quite similar to other enzymes in the α-d-phosphohexomutase superfamily, in particular the PMM/PGM subgroup, which is of a similar sequence length (see Fig. S1A in the supplemental material). An exception to this is the topology of domain 4, which typically comprises a 5-stranded β-sheet (26), rather than the 3-stranded version seen in BaPNGM.
Fig. 2.
(A) A ribbon diagram of the BaPNGM monomer (chain B), showing its four domains in the following colors: domain 1 (dark blue; residues 1 to 152), domain 2 (light blue; residues 153 to 256), domain 3 (pink; residues 257 to 369), and domain 4 (magenta; residues 370 to 446). (B) A superposition of the two monomers in the asymmetric unit of the crystals. Monomer B (closed) is shown as magenta, and monomer A (open) is light blue. The rotation of domain 4 is indicated by an arrow. (C) The electrostatic potential of BaPNGM shown on a surface rendering. Negative charge is red, and positive charge is blue. The figure was made by using PMV, MSMS, and Chimera (22, 27, 28).
The active site of BaPNGM can be identified based on the location of highly conserved catalytic residues in the α-d-phosphohexomutases. It is found in the center of the molecule, in a large cleft with a surface area of ∼1,800 Å2. More than 70 residues from all four domains of the protein contribute to the active site cleft, which is highly hydrophilic and has an overall positive electrostatic potential (Fig. 2C), consistent with its negatively charged phosphosugar substrate and product.
During refinement of the BaPNGM structure, it was noted that domain 4 of the protein adopts different conformations in the two monomers of the asymmetric unit of the crystal, presumably due to crystal packing constraints. A superposition of these two monomers (Fig. 2B) shows that domain 4 rotates by ∼30°. One result of this domain rotation is that the active site environment changes from an open, solvent accessible cleft to a deep, nearly enclosed pocket. In the closed conformer, contacts between residues in domains 1 and 4 of the protein form a lid over the active site, which is absent in the open conformer. The interdomain rotation of BaPNGM is notable for its magnitude, which is significantly larger than that seen for related proteins (typically 10 to 15°) (15, 18, 24). A discussion of the functional significance of the conformational variability of domain 4 can be found in this paper (see below).
Quaternary structure of BaPNGM and conservation in the enzyme family.
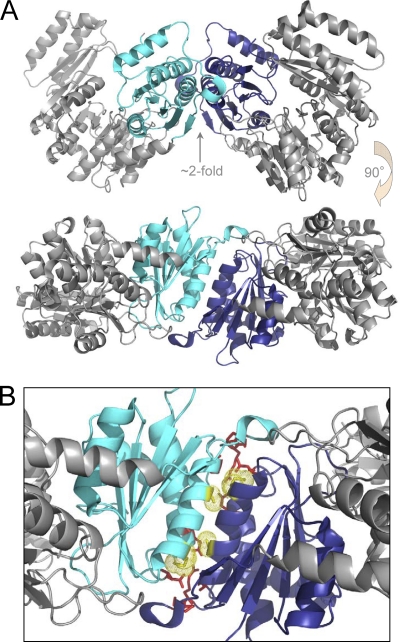
The two molecules found in the asymmetric unit of the BaPNGM crystals pack tightly against each other and are related by a pseudo-2-fold axis, which is imperfect due to the variable orientation of domain 4 (Fig. 3). The packing of all protein molecules in the crystal lattice was examined by PISA (11), which uses chemical thermodynamic calculations to assess complex stability from crystal structures and has proven highly reliable in many systems (36). This analysis predicts that the dimer observed in the asymmetric unit of the BaPNGM crystals is stable in solution. The formation of a dimer of BaPNGM in solution is also supported by measurements from dynamic light scattering, which indicate an average hydrodynamic radius of 4.2 nm, consistent with a molecular mass of 93.1 kDa, nearly twice that of the BaPNGM monomer (51.0 kDa).
Fig. 3.
(A) The dimer of BaPNGM found in the asymmetric unit of the crystals, shown in two orientations related by a 90° rotation. Domain 1 of monomer A is cyan; domain 1 of monomer B is dark blue. (B) A close up of the dimer interface between domains 1 of each monomer. Residues involved in hydrogen bonds or ion pairs are shown as red. Residues Met55 and Ala61, which are conserved and unique to the PNGM enzymes (see the text), are shown as yellow and highlighted with a dotted surface.
The dimer interface of BaPNGM involves primarily residues from domain 1 of each molecule, and comprises an interfacial area of ∼1,200 Å2. The interface of the dimer is largely hydrophobic (∼55% of the residues), although a number of hydrogen bonds and an ion pair are also present (Fig. 3 and Table 2). Many of the packing interactions are formed by residues in the helix found between strands 1 and 2 of the α/β core of domain 1. This dimerization helix packs against its counterpart from the other monomer in an antiparallel fashion, creating a tightly fitting, solvent inaccessible interface.
Table 2.
Residues that form hydrogen bonds/ion pairs in the BaPNGM dimer interfacea
| Chain A |
Chain B |
Distance (Å) | ||
|---|---|---|---|---|
| Residue | Atom | Residue | Atom | |
| Asn15 | ND2 | Pro141 | O | 2.63 |
| Ser66 | O | 3.31 | ||
| O | Arg140 | NE | 3.34 | |
| Glu21 | OE2 | Arg140 | NH1 | 3.00 |
| Arg140 | NH2 | 2.74 | ||
| Ser66 | O | Asn15 | ND2 | 3.27 |
| Arg140 | NH1 | Glu21 | OE1 | 2.90 |
| Asn15 | OD1 | 3.13 | ||
| NH2 | Glu21 | OE2 | 2.71 | |
| Pro141 | O | Asn15 | ND2 | 2.90 |
Contacts were calculated by DIMPLOT (33). Only contacts found between corresponding residues in both chain A and chain B were included.
During preparation of the manuscript, the structure of a PNGM enzyme from the pathogen Francisella tularensis was deposited in the PDB (3I3W), although never described in a publication. This protein shares 39% sequence identity with BaPNGM, and structural comparisons show that both proteins crystallize as nearly identical dimers (Cα root mean square deviation [RMSD]=2.5 Å for 728 residues), mediated by highly similar interactions between domain 1 (see Fig. S1B in the supplemental material). The structural conservation of the dimer in these two proteins led us to examine multiple sequence alignments of PNGM proteins (see Fig. S2 and S3 in the supplemental material for selected and large alignments, respectively). Although many of the residues in the helix-helix interface are somewhat conserved, no obvious dimerization sequence motif can be elucidated. Upon closer examination, however, we find several conserved residues in the dimerization helix that distinguish the PNGM sequences from those of related monomeric proteins (e.g., P. aeruginosa PMM/PGM) (see Fig. S2). These include Met55 and Ala62 of BaPNGM (Fig. 3B). In the large PNGM alignment (see Fig. S3), Met55 is a conserved hydrophobic residue (75% methionine, 7% phenylalanine, 7% leucine, etc.), while Ala62 is nearly always an alanine (90%) or serine (8%). In contrast, in an alignment of PMM/PGM proteins (not shown), where these residues are found in a surface-exposed helix and not in a dimer interface, they are typically charged/polar residues. The high sequence conservation of these residues in the PNGMs shows that this interface may be found in a wide variety of bacterial species, suggesting that most of these proteins are also likely dimeric. Based on gel filtration, a previous report suggested that E. coli PNGM is a trimer in solution (9), but this result could also be consistent with a dimeric species given the elongated shape of the PNGM dimer (∼130 by 75 Å).
Although most enzymes in the α-d-phosphohexomutase superfamily that have been studied to date appear to be monomeric, recent characterization of a bacterial phosphoglucomutase (PGM) from S. Typhimurium shows that this enzyme is also dimeric (15). For both BaPNGM and S. Typhimurium PGM, the dimer interface involves primarily residues in domain 1. However, the overall spatial arrangement of the two protein chains in the two dimers is quite different, suggesting that the oligomers of these proteins are recent in their evolutionary development, relative to the emergence of the enzyme superfamily. As is also the case with S. Typhimurium PGM, the location of the dimer interface of BaPNGM is distant from domain 4, allowing this domain to exhibit conformational variability in the context of a dimeric species. The dimeric state of PNGM offers potential functional advantages, including the opportunity for allosteric regulation of activity.
Key catalytic and ligand-binding residues in the PNGM active site.
Based on sequence-structure alignments and biochemical studies of other enzymes in the α-d-phosphohexomutase family, it is possible to predict residues of BaPNGM with likely roles in catalysis and ligand binding. The active site contains four key regions: (i) the phosphoserine residue involved in the phosphoryl transfer reaction, (ii) the metal-binding site, (iii) a sugar-binding loop with residues that contact the sugar moiety of the substrate/product, and (iv) the phosphate-binding site, which makes interactions with the phosphate group of the substrate and product. Table 3 lists the residues of BaPNGM that correspond to each of these regions. In addition, based on the crystal structures of P. aeruginosa PMM/PGM in complex with glucose-6-phosphate and glucose-1-phosphate (24), models for the binding of glucosamine-6-phosphate and glucosamine-1-phosphate in the active site of BaPNGM were constructed and are shown in Fig. 4 A and B. Below we describe the key active site regions and potential enzyme-ligand interactions in detail.
Table 3.
Highly conserveda residues in the active site of PNGM enzymes and their potential functional roles
| Residueb | Proposed role (region) |
|---|---|
| Residueb3 | Proposed role (region) |
| R11 | General acid/base |
| T7 | Domain 1-to-4 latch |
| T414 | |
| S100 | Phosphoserine involved in phosphoryl transfer (i) |
| D240 | Metal-binding ligands (ii) |
| D242 | |
| D244 | |
| D308c | Specific/permissive for amino group at 2 position of sugar |
| M285c | |
| E325 | Sugar-binding loop (iii) |
| S327 | |
| R410 | Phosphate-binding site (iv) |
| S412 | |
| R419 |
All residues in this table are 100% conserved based on a multiple sequence alignment of 171 PNGM sequences (see the supplemental material).
Residue numbers refer to the BaPNGM protein.
Proposed role of D308 is specificity for binding glucosamine-6-P, while M285 appears to be permissive for binding glucosamine-1-P. The four conserved active site regions discussed in the text are indicated in parentheses.
Fig. 4.
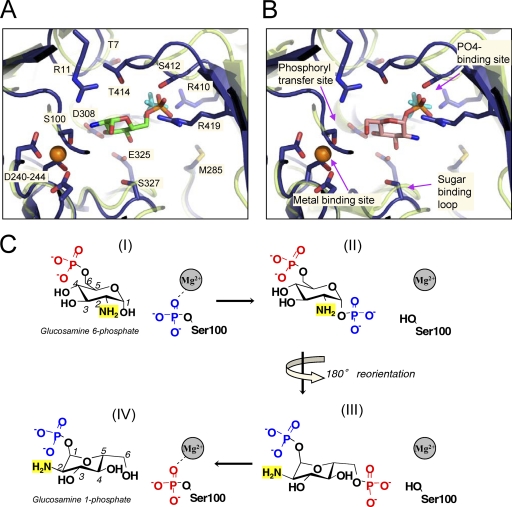
A close-up view of the active site of BaPNGM (closed monomer; blue) superposed with the structure of ligand-bound PMM/PGM from P. aeruginosa (semitransparent light green). The two proteins are only 26% identical in sequence but superpose with a Cα RMSD of 2.3 Å over 373 residues. The phosphate ion from BaPNGM is cyan; the bound metal ion from PMM/PGM is shown as an orange sphere. (Note that the backbone of the metal-binding loop is highly similar in both proteins, regardless of whether metal is bound). Key side chain residues of BaPNGM are shown as sticks. For clarity, only the backbone of PMM/PGM is shown. (A) A model of the binding of glucosamine-6-phosphate (green carbons) based on the glucose-6-phosphate complex with PMM/PGM (1P5G). (B) A model of the binding of glucosamine-1-phosphate (pink carbons) based on the glucose-1-phosphate complex with PMM/PGM (1P5D). (C) Scheme of the proposed mechanism of PNGM based on studies of related enzymes (see text). Four states (I to IV) are indicated as follows: (I) binding of substrate to the phosphorylated enzyme, (II) transfer of phosphoryl group to form a bis-phosphorylated intermediate, (III) reorientation of the intermediate to allow the second phosphoryl transfer, and (IV) formation of product and regeneration of the phosphoenzyme. States I and IV correspond to panels A and B of this figure, although the enzyme is depicted in its dephosphorylated state (as found in the crystal structure).
The phosphoserine residue of region i is a conserved feature of the entire α-d-phosphohexomutase superfamily. The phosphorylated version of this residue provides the phosphoryl group that is transferred to substrate and subsequently accepts a phosphoryl group from the reaction intermediate to form the product (Fig. 4C). In BaPNGM, this residue is Ser100, which is observed in its dephosphorylated state in the crystal structure. The side chain oxygen (Oγ) of this residue also typically acts as a ligand for the metal ion bound to region ii, while three other ligands for the metal are provided by the side chains of aspartate residues 240, 242, and 244. The metal (typically Mg2+ for full activity) serves to withdraw electrons from the leaving group, facilitating the phosphoryl transfer reaction. However, due to crystallization at pH 4.5 (16), the aspartate carboxylates are likely protonated, and no metal is observed in the binding sites of the BaPNGM structure. Despite this, the geometry of the metal-binding loop is quite similar to that observed in metal-bound forms of related enzymes, such as P. aeruginosa PMM/PGM (Fig. 4). Rather than acting as metal ligands, the three aspartate side chains of BaPNGM make multiple hydrogen bond interactions with neighboring residues, including Ser100. Also near the site of phosphoryl transfer is Arg11, which we propose to be a general acid/base in the reaction, acting to abstract/donate a proton to/from the hydroxyl group of the phosphosugar (at C-1 or C-6) where phosphoryl transfer occurs. Mutation of this conserved arginine has been shown to greatly reduce activity in other enzymes in the α-d-phosphohexomutase superfamily (3, 25). Although arginine does not commonly serve this role in enzymes, there are a number of established examples in the literature, many of which involve reactions with hydrophilic, charged substrates (8).
Region iii of the active site is a loop containing Glu325 and Ser327. Structures of enzyme-ligand complexes for related proteins (21, 24) have shown that the side chains of these residues contact the O3 and O4 hydroxyls of the phosphosugar substrate and product. We therefore propose that they will make similar contacts to the O3 and O4 hydroxyls of glucosamine-1-phosphate and -6-phosphate. We note that the reaction mechanism of enzymes in the α-d-phosphohexomutase superfamily requires a 180° reorientation of the reaction intermediate (i.e., glucosamine-1,6-bis-phosphate) in between successive phosphoryl transfer reactions (Fig. 4C), which has been substantiated by multiple studies (21, 24). This requires that the sugar moiety of glucosamine-1-phosphate and -6-phosphate bind in different orientations in the active site (Fig. 4, compare panels A and B). However, in both orientations, the O3 and O4 hydroxyls are positioned to interact with these conserved glutamate and serine residues.
Region iv of the active site is the phosphate-binding site, which is formed by residues in/near a loop of domain 4. Although no ligands are present in the BaPNGM structure, the protein was crystallized in 2.0 M Na,K phosphate (16), and a phosphate ion is found near the expected phosphate-binding site in both monomers (Fig. 4). Residues of BaPNGM involved in direct phosphate contacts include the side chains of Arg410, Ser412, and Arg419. These enzyme-phosphate interactions directly support the proposed roles of these residues in binding the phosphate group of substrate and product (Table 3). Also near the phosphate-binding site are two conserved threonines (residues 7 and 414 of BaPNGM) that participate in the domain 1-to-4 interface found in the closed conformer of the protein. A similar, highly conserved “threonine latch” has been observed in the crystal structures of related phosphoglucomutase enzymes (15, 18).
Although described separately above, the four key regions of the PNGM active site do not function independently. To create a high-affinity ligand-binding site, these regions must be correctly positioned relative to each other. In related enzymes, the conformation of domain 4 has been shown to be critical for this (21, 24). In the BaPNGM crystal structure, the observed conformational variability of domain 4 suggests that rotation of this domain will also occur upon ligand binding for the PNGM enzymes. Based on structural superpositions, only in the closed conformer of BaPNGM are all four key regions of the active site positioned such that each can make the expected contacts with ligand. Thus, as is true for other α-d-phosphohexomutases, it appears that conformational change and ligand binding will be intimately associated in the PNGM enzymes.
Specificity of the PNGM enzymes and prospects for inhibitor design.
Modeling of substrate and product in the active site of BaPNGM (Fig. 4) provides the first insights into the structural basis of specificity of the PNGM enzyme family for glucosamine phosphosugars. Glucosamine is distinguished from glucose by the presence of an amine at the 2 position of the sugar ring, rather than a hydroxyl. Our model of the enzyme-substrate complex (Fig. 4A) predicts the 2-amino group of glucosamine-6-phosphate to be near a conserved aspartate residue (Asp308 of BaPNGM). Given a reported pKa of 7.6 for glucosamine (2), a favorable charge-charge interaction may occur between this residue and the 2-amino group of the sugar. In contrast, such an interaction would not be expected with the 2-hydroxyl of glucose-6-phosphate, and indeed aspartate is not found at the corresponding position in related enzymes (e.g., PMM/PGM, PGM) that prefer glucose-based substrates (not shown).
In the model of the enzyme-product complex for BaPNGM, the amino group of glucosamine-1-phosphate does not appear situated to make any direct interactions with the enzyme. Nearby, however, is a methionine residue (Met285 of BaPNGM) that is highly conserved in the PNGM alignment (Fig. 4B). In the related PMM/PGM proteins, this residue is a lysine that interacts with O2 of glucose-1-phosphate (24). Hence, the substitution of methionine at this position in the PNGM active site removes the positively charged lysine side chain from the vicinity of the 2-amino group and may thereby facilitate binding of glucosamine phosphosugars (rather than their glucose counterparts) in the active site.
Determination of a PNGM structure completes the structural characterization of at least one representative of the four subgroups of the α-d-phosphohexomutase superfamily. Given the ubiquitous presence of these enzymes in organisms in all kingdoms of life, it is of considerable interest to understand the sequence/structure determinants of substrate specificity, as this should help predict the biosynthetic pathways in which they participate. Understanding specificity may also be critical for the design of selective and highly potent inhibitors for enzymes in different subgroups of the superfamily. The identification of Asp308 and Met285, which are both very highly conserved in the PNGM family but not in other subgroups of the α-d-phosphohexomutase superfamily, is a first step toward characterizing residues responsible for the specificity of the PNGMs. However, additional biochemical studies are needed to fully define the importance of PNGM residues to ligand binding and specificity.
Enzymes in peptidoglycan biosynthesis have long been considered attractive targets for inhibitor design. In the case of PNGM, recent reports indicate a growing recognition of the importance of this enzyme in bacterial virulence and infectivity (10, 13, 30, 35, 37). Therefore, inhibitors selective for the bacterial PNGMs that do not cross-react with related eukaryotic proteins, such as PGM, may find clinical utility as antimicrobial agents. The crystal structure of BaPNGM presented here sheds light on active site residues and the conformational change of this enzyme and should greatly facilitate the design of specific inhibitors for the PNGM enzymes.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jay Nix of ALS beamline 4.2.2 for assistance with data collection. We thank Manfred Weiss for helpful discussions regarding the structure solution and refinement and an anonymous reviewer for helpful comments.
This work was supported by a grant from the National Science Foundation (MCB-0918389) to L.J.B. Part of this work was performed at the Advanced Light Source. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under contract number DE-AC02-05CH11231.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 17 June 2011.
REFERENCES
- 1. Barreteau H., et al. 2008. Cytoplasmic steps of peptidoglycan biosynthesis. FEMS Microbiol. Rev. 32:168–207 [DOI] [PubMed] [Google Scholar]
- 2. Bichsel Y., Von Gunten U. 2000. Formation of iodo-trihalomethanes during disinfection and oxidation of iodide-containing waters. Environ. Sci. Technol. 34:2784–2791 [Google Scholar]
- 3. Brautaset T., Petersen S. B., Valla S. 2000. In vitro determined kinetic properties of mutant phosphoglucomutases and their effects on sugar catabolism in Escherichia coli. Metab. Eng. 2:104–114 [DOI] [PubMed] [Google Scholar]
- 4. DeLano W. L. 2002. The PyMOL Molecular Graphics System. DeLano Scientific, San Carlos, CA [Google Scholar]
- 5. De Reuse H., Labigne A., Mengin-Lecreulx D. 1997. The Helicobacter pylori ureC gene codes for a phosphoglucosamine mutase. J. Bacteriol. 179:3488–3493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Emsley P., Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 [DOI] [PubMed] [Google Scholar]
- 7. Gautam A., Vyas R., Tewari R. 22 November 2010. Peptidoglycan biosynthesis machinery: a rich source of drug targets. Crit. Rev. Biotechnol. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 8. Guillen Schlippe Y. V., Hedstrom L. 2005. A twisted base? The role of arginine in enzyme-catalyzed proton abstractions. Arch. Biochem. Biophys. 433:266–278 [DOI] [PubMed] [Google Scholar]
- 9. Jolly L., et al. 1999. Reaction mechanism of phosphoglucosamine mutase from Escherichia coli. Eur. J. Biochem. 262:202–210 [DOI] [PubMed] [Google Scholar]
- 10. Jolly L., et al. 1997. The femR315 gene from Staphylococcus aureus, the interruption of which results in reduced methicillin resistance, encodes a phosphoglucosamine mutase. J. Bacteriol. 179:5321–5325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Krissinel E., Henrick K. 2007. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372:774–797 [DOI] [PubMed] [Google Scholar]
- 12. Langer M., et al. 2008. Bacillus anthracis peptidoglycan stimulates an inflammatory response in monocytes through the p38 mitogen-activated protein kinase pathway. PLoS One 3:e3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu X. D., Duan J., Guo L. H. 2009. Role of phosphoglucosamine mutase on virulence properties of Streptococcus mutans. Oral Microbiol. Immunol. 24:272–277 [DOI] [PubMed] [Google Scholar]
- 14. Lovell S. C., et al. 2003. Structure validation by Cα geometry: φ, ψand Cβ deviation. Proteins 50:437–450 [DOI] [PubMed] [Google Scholar]
- 15. Mehra-Chaudhary R., Mick J., Tanner J. J., Henzl M. T., Beamer L. J. 2011. Crystal structure of a bacterial phosphoglucomutase, an enzyme involved in the virulence of multiple human pathogens. Proteins 79:1215–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mehra-Chaudhary R., Neace C. E., Beamer L. J. 2009. Crystallization and initial crystallographic analysis of phosphoglucosamine mutase from Bacillus anthracis. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 65:733–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mengin-Lecreulx D., van Heijenoort J. 1996. Characterization of the essential gene glmM encoding phosphoglucosamine mutase in Escherichia coli. J. Biol. Chem. 271:32–39 [DOI] [PubMed] [Google Scholar]
- 18. Müller S., et al. 2002. Crystal structure analysis of the exocytosis-sensitive phosphoprotein, pp63/parafusin (phosphoglucomutase), from Paramecium reveals significant conformational variability. J. Mol. Biol. 315:141–153 [DOI] [PubMed] [Google Scholar]
- 19. Murshudov G. N., Vagin A. A., Lebedev A., Wilson K. S., Dodson E. J. 1999. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr. D 55:247–255 [DOI] [PubMed] [Google Scholar]
- 20. Naught L. E., Tipton P. A. 2001. Kinetic Mechanism and pH dependence of the kinetic parameters of Pseudomonas aeruginosa phosphomannomutase/phosphoglucomutase. Arch. Biochem. Biophys. 396:111–118 [DOI] [PubMed] [Google Scholar]
- 21. Nishitani Y., et al. 2006. Crystal structures of N-acetylglucosamine-phosphate mutase, a member of the alpha-D-phosphohexomutase superfamily, and its substrate and product complexes. J. Biol. Chem. 281:19740–19747 [DOI] [PubMed] [Google Scholar]
- 22. Pettersen E. F., et al. 2004. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612 [DOI] [PubMed] [Google Scholar]
- 23. Rathi B., Sarangi A. N., Trivedi N. 2009. Genome subtraction for novel target definition in Salmonella typhi. Bioinformation 4:143–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Regni C., Naught L. E., Tipton P. A., Beamer L. J. 2004. Structural basis of diverse substrate recognition by the enzyme PMM/PGM from P. aeruginosa. Structure 12:55–63 [DOI] [PubMed] [Google Scholar]
- 25. Regni C., Schramm A. M., Beamer L. J. 2006. The reaction of phosphohexomutase from Pseudomonas aeruginosa: structural insights into a simple processive enzyme. J. Biol. Chem. 281:15564–15571 [DOI] [PubMed] [Google Scholar]
- 26. Regni C., Tipton P. A., Beamer L. J. 2002. Crystal structure of PMM/PGM: an enzyme in the biosynthetic pathway of P. aeruginosa virulence factors. Structure 10:269–279 [DOI] [PubMed] [Google Scholar]
- 27. Sanner M. F. 1999. Python: a programming language for software integration and development. J. Mol. Graph Model. 17:57–61 [PubMed] [Google Scholar]
- 28. Sanner M. F., Olson A. J., Spehner J. C. 1996. Reduced surface: an efficient way to compute molecular surfaces. Biopolymers 38:305–320 [DOI] [PubMed] [Google Scholar]
- 29. Shackelford G. S., Regni C. A., Beamer L. J. 2004. Evolutionary trace analysis of the alpha-d-phosphohexomutase superfamily. Protein Sci. 13:2130–2138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shimazu K., et al. 2008. Identification of the Streptococcus gordonii glmM gene encoding phosphoglucosamine mutase and its role in bacterial cell morphology, biofilm formation, and sensitivity to antibiotics. FEMS Immunol. Med. Microbiol. 53:166–177 [DOI] [PubMed] [Google Scholar]
- 31. Tavares I. M., Leitao J. H., Sa-Correia I. 2003. Chromosomal organization and transcription analysis of genes in the vicinity of Pseudomonas aeruginosa glmM gene encoding phosphoglucosamine mutase. Biochem. Biophys. Res. Commun. 302:363–371 [DOI] [PubMed] [Google Scholar]
- 32. Vagin A., Teplyakov A. 1997. MOLREP: an automated program for molecular replacement. J. App. Cryst. 30:1022–1025 [Google Scholar]
- 33. Wallace A. C., Laskowski R. A., Thornton J. M. 1995. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 8:127–134 [DOI] [PubMed] [Google Scholar]
- 34. Whitehouse D. B., Tomkins J., Lovegrove J. U., Hopkinson D. A., McMillan W. O. 1998. A phylogenetic approach to the identification of phosphoglucomutase genes. Mol. Biol. Evol. 15:456–462 [DOI] [PubMed] [Google Scholar]
- 35. Wu S., de Lencastre H., Sali A., Tomasz A. 1996. A phosphoglucomutase-like gene essential for the optimal expression of methicillin resistance in Staphylococcus aureus: molecular cloning and DNA sequencing. Microb. Drug Resist. 2:277–286 [DOI] [PubMed] [Google Scholar]
- 36. Xu Q., et al. 2008. Statistical analysis of interface similarity in crystals of homologous proteins. J. Mol. Biol. 381:487–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yajima A., et al. 2009. Contribution of phosphoglucosamine mutase to the resistance of Streptococcus gordonii DL1 to polymorphonuclear leukocyte killing. FEMS Microbiol. Lett. 297:196–202 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.