Abstract
Autotransporter biogenesis is dependent upon BamA, a central component of the β-barrel assembly machinery (BAM) complex. In this report, we detail the role of the other BAM components (BamB-E). We identify the importance of BamD in autotransporter biogenesis and show that BamB, BamC, and BamE are not required.
TEXT
Gram-negative bacteria possess a double membrane barrier that is instrumental in protecting the bacteria against the external environment and maintaining viability (4, 24). However, this unique feature poses a formidable obstacle to both the insertion of proteins into the outer membrane and the secretion of proteins across it. Most integral outer membrane proteins (OMPs) form a β-barrel structure (38), and the recently described β-barrel assembly machinery (BAM) complex (40) has been implicated in the insertion and/or folding of nearly all known OMPs (7, 20, 39). BAM has now been well characterized as a hetero-oligomeric complex composed of five proteins, one integral β-barrel membrane protein termed BamA and four outer membrane-associated lipoproteins, BamB-E (22). BamA is the central component and forms a pore with a diameter of approximately 2.5 nm in the outer membrane (30). Although other components are crucial to the functional integrity of the complex (2, 19, 34, 40), only BamA and BamD are universally essential for both bacterial cell viability and OMP biogenesis (27). Why this is, however, remains unclear.
Autotransporters (ATs) represent a large family of secreted proteins that are widespread among Gram-negative bacteria (5, 14, 16). Their name is derived from the supposition that translocation across both membranes occurs independently of accessory factors and is attributable to their conserved architecture (11). Superficially, all ATs have a three-domain architecture with an N-terminal signal sequence that mediates inner membrane translocation, a central passenger domain representing the secreted effector protein, and a C-terminal β-barrel domain that inserts into the outer membrane and facilitates the translocation of the passenger domain into the external milieu (1, 6, 28). There is now a significant body of evidence demonstrating that the biogenesis of ATs requires BamA (18, 25, 31, 33), although how this occurs remains speculative. Furthermore, the precise role of the other BAM components is not known. In order to define their functions, we used Pet, an archetypal AT that is a functionally well-characterized cytotoxin from the human pathogen enteroaggregative Escherichia coli (EAEC) strain 042 (8), the Sat cytotoxin from uropathogenic E. coli (UPEC) strain CFT073 (9), and the phase-variable OMP antigen 43 (Ag43) from laboratory E. coli strain MG1655 (17, 36). This is the first report detailing the role of each component of the BAM complex in AT biogenesis.
BamA and BamD are required for AT biogenesis.
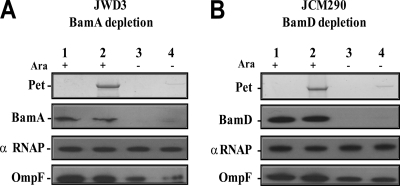
We first examined the role of the essential BamA and BamD proteins in Pet biogenesis. Full-length Pet protein was expressed under the control of its native promoter in BamA (25) and BamD (27) depletion strains by transforming them with the pet-containing recombinant plasmids pCEFN1 (8) and pACYC184/pet, respectively. In the respective depletion strains, expression of bamA or bamD is under the control of the PBAD promoter, such that expression is induced in the presence of arabinose but repressed in its absence. To construct pACYC184/pet, the pet gene, together with its native promoter, was PCR amplified from pCEFN1 using primers GGGATCCGGAAGACGGTTGTTGCG and CGGGATCCGGTTAGCTCTGAATTAAG and the product was cloned into the BamHI site of pACYC184. After growth under both replete (with arabinose) and depletion (without arabinose) conditions, the culture supernatant fractions were harvested from the BamA and BamD depletion strains. Secreted proteins were precipitated using trichloroacetic acid (26, 31), resolved by sodium dodecyl sulfate-polyacrylamide electrophoresis (SDS-PAGE), and detected by Coomassie staining (32). Analysis of proteins in whole-cell extracts followed established protocols (19). Results in Fig. 1 illustrate that when both BamA and BamD are depleted from the cells, secretion of Pet into the culture medium is severely diminished. These results confirm previous observations that BamA is required for AT biogenesis and demonstrate for the first time an essential role for BamD in AT biogenesis.
Fig. 1.
BamA and BamD are required for Pet secretion. Shown are Coomassie blue-stained SDS-PAGE of trichloroacetic acid-precipitated supernatant proteins and Western blot analysis of whole-cell extracts from cultures of BamA depletion strain E. coli JWD3 (A) and BamD depletion strain E. coli JCM290 (B). In panel A, E. coli JWD3, carrying either the empty pSPORT1 vector (lanes 1 and 3) or pCEFN1 (lanes 2 and 4), was grown in LB medium containing 100 μg/ml ampicillin supplemented with either 0.2% l-arabinose (Ara) (+) or 0.2% d-fructose (−). In panel B, E. coli JCM290, carrying either pACYC184/pet (lanes 2 and 4) or the empty vector, was grown in LB medium containing 30 μg/ml chloramphenicol supplemented with either 0.05% l-arabinose (+) or 0.05% d-fructose (−). For both panels A and B, overnight cultures were diluted into a volume of 50 ml (OD600 = 0.025) and grown at 37°C with shaking for 3 h. Cultures were harvested, and supernatant proteins were precipitated with 10% trichloroacetic acid. BamA, BamD, and OmpF were detected using antiserum raised in a rabbit, and the α subunit of RNA polymerase (αRNAP) was detected using mouse monoclonal antibodies (Neoclone). Blots were developed using the ECL Plus Western blotting detection system (GE Healthcare). Like OmpF levels, under BamA and BamD depletion conditions, Pet levels are severely diminished whereas the levels of RNAP remain constant.
BamB, BamC, and BamE are not required for AT biogenesis.
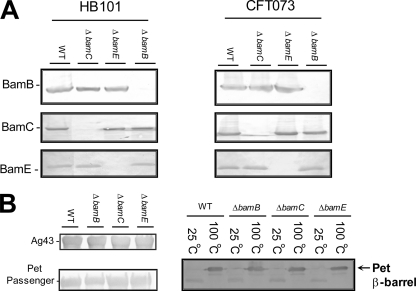
While BamA has been implicated in the biogenesis of all examined ATs (18, 33), the roles of the nonessential components of the BAM complex (BamB, BamC, and BamE) have not yet been fully investigated. In order to determine the role of these accessory factors in AT biogenesis, bamB, bamC, and bamE deletion mutants were constructed in laboratory strain E. coli HB101 and wild-type UPEC strain CFT073 using the λ Red system (3). In each mutant, the absence of the relevant endogenous protein in whole-cell lysates was confirmed by Western immunoblotting (Fig. 2). Anti-BamE antibodies were previously described (21, 23); to raise antibodies to BamB and BamC, constructs encoding these proteins but lacking their acylation site were synthesized de novo and cloned into the vector pET22b+ (Genscript). Proteins were overexpressed and purified from the periplasm of E. coli strain BL21(DE3) (Novagen) and purified by nickel affinity chromatography. These proteins were used to immunize rabbits using a standard 3-month immunization program (Eurogentec), and the antibodies were purified by affinity purification. For analysis of Pet biogenesis, E. coli HB101 and its mutant derivatives were transformed with pCEFN1 and the empty vector pSPORT1 (8). Overnight cultures were diluted 1:100 into fresh medium and grown to an optical density at 600 nm (OD600) of 1.0. The cells were pelleted, and the supernatant and outer membrane fractions were prepared as described previously (17, 26). Supernatant proteins were separated by SDS-PAGE, and Pet was detected by Western immunoblotting using a polyclonal rabbit antiserum generated toward the Pet passenger domain (8), secondary goat anti-rabbit antibodies conjugated with alkaline phosphatase (AP), and the AP substrate 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (32). Accumulation of Pet in the culture medium was unaffected by the absence of BamB, BamC, or BamE (Fig. 2). To gain a more complete picture of Pet biogenesis, we also examined the production of the C-terminal β-barrel of Pet. OMPs were purified and resolved by SDS-PAGE; Western immunoblotting was performed as described above but with a polyclonal rabbit antiserum recognizing the Pet β-barrel. To raise antibodies, the Pet β domain was cloned into the expression vector pMAL-c2X (New England BioLabs, Herts, United Kingdom) and the resulting maltose-binding protein-Pet-β-barrel fusion was purified in accordance with the manufacturer's instructions before immunization of rabbits (Eurogentec) and subsequent harvesting of serum. In each case, the levels of the Pet β-barrel were unaffected by the absence of the BAM components, and the β-barrel remained heat modifiable (Fig. 2), indicating that it was inserted in the outer membrane in its native conformation. To ensure that these effects were not specific to Pet, we examined the influence of these mutations on another AT, namely, biofilm-promoting Ag43. E. coli HB101 and its knockout derivatives were transformed with agn43-containing plasmid pCO2 (35). Expression levels were monitored using a previously described heat release assay which detects Ag43 release from the bacterial cell surface (17, 35). The resulting samples were analyzed by SDS-PAGE and Western immunoblotting with anti-Ag43 antibodies (Fig. 2). The levels of Ag43 were unaffected, indicating that BamB, BamC, and BamE are not required for the translocation of Ag43 to the exterior of the cell. These observations were confirmed for production of the Sat AT cytotoxin in the wild-type strain and mutant versions of UPEC CFT073 (data not shown).
Fig. 2.
BamB, BamC, and BamE are not required for AT secretion. (A) Construction of bamB, bamC, and bamE null mutations in E. coli HB101 and E. coli CFT073. Whole-cell lysate of each mutant was prepared by spinning down 1 ml of an overnight culture and resuspending it in Laemmli buffer. Samples were separated by SDS-PAGE and Western immunoblotted with antibodies raised to BamB, BamC, and BamE, confirming the absence of the relevant protein from each mutant. (B) Western immunoblotting was performed on the BamB, BamC, and BamE null mutant strains expressing Pet and Ag43. The Pet passenger domain was prepared from supernatant fractions after filter sterilization and precipitation with a final volume of 10% trichloroacetic acid. No difference in the accumulation of Pet in culture supernatant was noted. The Pet β-barrel was detected by Western immunoblotting of outer membrane fractions with anti-Pet β-domain antibodies after heating to either 100°C or 25°C; no differences in β-domain levels or heat modifiability were detected. For analysis of Ag43 biogenesis, the Ag43 passenger domain was released from the cell surface using the standard heat release assay (60°C for 2 min) and detected by Western immunoblotting with Ag43-specific antiserum. No differences in the levels of Ag43 were detected. These data indicate that AT secretion is not significantly influenced by BamB, -C, or -E. WT, wild type.
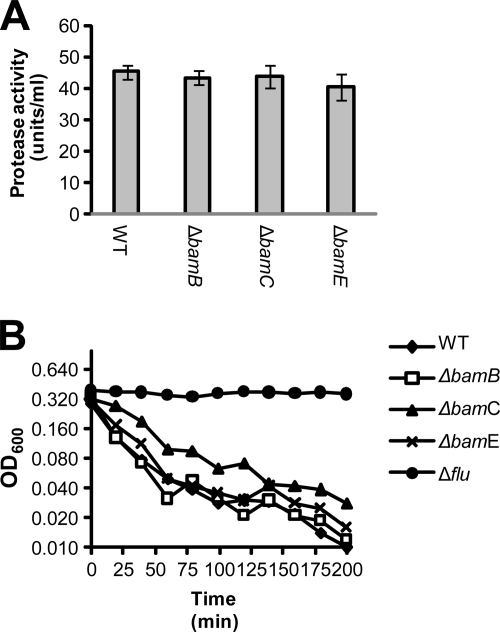
While secretion is unaffected, it remains possible that folding of the passenger domain is abnormal. To test this hypothesis, we quantified the functional activity of Pet and Ag43 as a direct indicator of protein folding. To calculate the enzymatic activity of Pet, supernatant fractions from E. coli HB101 and isogenic bamB, bamC, and bamE deletion strains expressing Pet were harvested at an OD600 of 1.0 and concentrated 100-fold using Vivaspin 20 columns (Sartorius Stedim Biotech) with a molecular weight cutoff of 50,000 as previously described (13). Pet isolated from culture supernatants was buffer exchanged into 50 mM Tris-HCl (pH 7.5), and 30 μg of protein was assayed for protease activity using azocasein as the substrate as described previously (29). One (arbitrary) unit of protease activity was defined as the protease activity resulting in an increase in A440 of 0.001 in 16 h at 37°C. The Ag43 cell-cell aggregation assay was performed as previously described (15, 17). In all of the mutants, the enzymatic activity of Pet and the ability of Ag43 to mediate cell-cell aggregation were indistinguishable from those of the wild-type organism (Fig. 3), indicating that BamB, BamC, and BamE are not required for folding of the passenger domain.
Fig. 3.
BamB, BamC, and BamE are not required for folding of the passenger domains. If the AT passenger domains do not fold correctly, they will not be able to perform their extracellular functions. To determine if BamB, BamC, or BamE influences the folding of the passenger domain, the functional activity of the Pet and Ag43 ATs was characterized in wild-type (WT) E. coli HB101 and isogenic derivatives. (A) The enzymatic activity of Pet was calculated using an azocasein assay. Protease activity was expressed in arbitrary units. There was no difference in the functional activity of Pet harvested from the mutant strains and that of Pet harvested from the wild-type organism, indicating that Pet produced from the mutants is not aberrantly folded. (B) The ability of Ag43 to mediate cell-cell aggregation was quantified in wild-type E. coli HB101 and isogenic derivatives. No difference in the rate of sedimentation between the mutants and the parent strain was observed. In contrast, the E. coli HB101 strain lacking the gene encoding Ag43 (Δflu) did not aggregate at all. These data indicate that Ag43 is not aberrantly folded.
Concluding remarks.
In agreement with other studies, our results confirm that BamA is required for AT biogenesis (18, 33). However, this study has highlighted the additional requirement of BamD for AT biogenesis. This observation is consistent with all previous studies which have demonstrated the essential nature of BamD in OMP assembly and reinforce the concept that BamA and BamD work in concert (10, 27). Previous investigations have demonstrated that different OMPs are assembled into the outer membrane via different routes. Thus, porins such as OmpF and LamB, as well as the omptin family member OmpT, have severe biogenic defects in the absence of BamB, while, in contrast, levels of TolC increase in a BamB mutant (2, 10). Similarly, TolC levels are unaffected by loss of BamE whereas biogenesis of porins is marginally affected (2, 34, 37). We conclude from our data that, like TolC, the AT biogenesis pathway does not require the nonessential lipoproteins BamB, BamC, and BamE. However, in contrast to TolC, AT levels do not increase in the absence of BamB. These studies suggest that there are at least two distinct pathways for OMP assembly in E. coli, one which is dependent on BamB, -C, and -E (the porins) and one which is independent of these other factors (the ATs and TolC), both of which converge on the core of the BAM complex formed by the BamA and BamD subunits.
Acknowledgments
We thank Thomas Silhavy for donating the BamD depletion strain, Anthony Scott Tucker for help in preparing the anti-Pet β-domain antibodies, and Rajeev Misra for donating OmpF antibodies.
We thank the BBSRC and MRC for support.
Footnotes
Published ahead of print on 10 June 2011.
REFERENCES
- 1. Barnard T. J., Dautin N., Lukacik P., Bernstein H. D., Buchanan S. K. 2007. Autotransporter structure reveals intra-barrel cleavage followed by conformational changes. Nat. Struct. Mol. Biol. 14:1214–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Charlson E. S., Werner J. N., Misra R. 2006. Differential effects of yfgL mutation on Escherichia coli outer membrane proteins and lipopolysaccharide. J. Bacteriol. 188:7186–7194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Datsenko K. A., Wanner B. L. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Delcour A. H. 2009. Outer membrane permeability and antibiotic resistance. Biochim. Biophys. Acta 1794:808–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Desvaux M., Khan A., Beatson S. A., Scott-Tucker A., Henderson I. R. 2005. Protein secretion systems in Fusobacterium nucleatum: genomic identification of type 4 piliation and complete type V pathways brings new insight into mechanisms of pathogenesis. Biochim. Biophys. Acta 1713:92–112 [DOI] [PubMed] [Google Scholar]
- 6. Desvaux M., Parham N. J., Henderson I. R. 2004. Type V protein secretion: simplicity gone awry? Curr. Issues Mol. Biol. 6:111–124 [PubMed] [Google Scholar]
- 7. Doerrler W. T., Raetz C. R. 2005. Loss of outer membrane proteins without inhibition of lipid export in an Escherichia coli yaeT mutant. J. Biol. Chem. 280:27679–27687 [DOI] [PubMed] [Google Scholar]
- 8. Eslava C., et al. 1998. Pet, an autotransporter enterotoxin from enteroaggregative Escherichia coli. Infect. Immun. 66:3155–3163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guyer D. M., Henderson I. R., Nataro J. P., Mobley H. L. 2000. Identification of sat, an autotransporter toxin produced by uropathogenic Escherichia coli. Mol. Microbiol. 38:53–66 [DOI] [PubMed] [Google Scholar]
- 10. Hagan C. L., Kim S., Kahne D. 2010. Reconstitution of outer membrane protein assembly from purified components. Science 328:890–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Halter R., Pohlner J., Meyer T. F. 1984. IgA protease of Neisseria gonorrhoeae: isolation and characterization of the gene and its extracellular product. EMBO J. 3:1595–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Reference deleted.
- 13. Henderson I. R., et al. 1999. Involvement of the enteroaggregative Escherichia coli plasmid-encoded toxin in causing human intestinal damage. Infect. Immun. 67:5338–5344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Henderson I. R., Lam A. C. 2001. Polymorphic proteins of Chlamydia spp.—autotransporters beyond the Proteobacteria. Trends Microbiol. 9:573–578 [DOI] [PubMed] [Google Scholar]
- 15. Henderson I. R., Meehan M., Owen P. 1997. Antigen 43, a phase-variable bipartite outer membrane protein, determines colony morphology and autoaggregation in Escherichia coli K-12. FEMS Microbiol. Lett. 149:115–120 [DOI] [PubMed] [Google Scholar]
- 16. Henderson I. R., Navarro-Garcia F., Desvaux M., Fernandez R. C., Ala'Aldeen D. 2004. Type V protein secretion pathway: the autotransporter story. Microbiol. Mol. Biol. Rev. 68:692–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Henderson I. R., Owen P. 1999. The major phase-variable outer membrane protein of Escherichia coli structurally resembles the immunoglobulin A1 protease class of exported protein and is regulated by a novel mechanism involving Dam and oxyR. J. Bacteriol. 181:2132–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jain S., Goldberg M. B. 2007. Requirement for YaeT in the outer membrane assembly of autotransporter proteins. J. Bacteriol. 189:5393–5398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Knowles T. J., et al. 2011. Structure and function of BamE within the outer membrane and the beta-barrel assembly machine. EMBO Rep. 12:123–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Knowles T. J., et al. 2008. Fold and function of polypeptide transport-associated domains responsible for delivering unfolded proteins to membranes. Mol. Microbiol. 68:1216–1227 [DOI] [PubMed] [Google Scholar]
- 21. Knowles T. J., McClelland D. M., Rajesh S., Henderson I. R., Overduin M. 2009. Secondary structure and (1)H, (13)C and (15)N backbone resonance assignments of BamC, a component of the outer membrane protein assembly machinery in Escherichia coli. Biomol. NMR Assign. 3:203–206 [DOI] [PubMed] [Google Scholar]
- 22. Knowles T. J., Scott-Tucker A., Overduin M., Henderson I. R. 2009. Membrane protein architects: the role of the BAM complex in outer membrane protein assembly. Nat. Rev. Microbiol. 7:206–214 [DOI] [PubMed] [Google Scholar]
- 23. Knowles T. J., et al. 2010. Secondary structure and 1H, 13C and 15N resonance assignments of BamE, a component of the outer membrane protein assembly machinery in Escherichia coli. Biomol. NMR Assign. 4:179–181 [DOI] [PubMed] [Google Scholar]
- 24. Lee V. T., Schneewind O. 2001. Protein secretion and the pathogenesis of bacterial infections. Genes Dev. 15:1725–1752 [DOI] [PubMed] [Google Scholar]
- 25. Lehr U., et al. 2010. C-terminal amino acid residues of the trimeric autotransporter adhesin YadA of Yersinia enterocolitica are decisive for its recognition and assembly by BamA. Mol. Microbiol. 78:932–946 [DOI] [PubMed] [Google Scholar]
- 26. Leyton D. L., Sloan J., Hill R. E., Doughty S., Hartland E. L. 2003. Transfer region of pO113 from enterohemorrhagic Escherichia coli: similarity with R64 and identification of a novel plasmid-encoded autotransporter, EpeA. Infect. Immun. 71:6307–6319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Malinverni J. C., et al. 2006. YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol. Microbiol. 61:151–164 [DOI] [PubMed] [Google Scholar]
- 28. Oomen C. J., et al. 2004. Structure of the translocator domain of a bacterial autotransporter. EMBO J. 23:1257–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ridgway I. D., et al. 2008. Extracellular proteases and possible disease related virulence mechanisms of two marine bacteria implicated in an opportunistic bacterial infection of Nephrops norvegicus. J. Invertebr. Pathol. 99:14–19 [DOI] [PubMed] [Google Scholar]
- 30. Robert V., et al. 2006. Assembly factor Omp85 recognizes its outer membrane protein substrates by a species-specific C-terminal motif. PLoS Biol. 4:e377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ruiz-Perez F., et al. 2009. Roles of periplasmic chaperone proteins in the biogenesis of serine protease autotransporters of Enterobacteriaceae. J. Bacteriol. 191:6571–6583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sambrook J., Russell D. W. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring har bor, NY [Google Scholar]
- 33. Sauri A., et al. 2009. The Bam (Omp85) complex is involved in secretion of the autotransporter haemoglobin protease. Microbiology 155:3982–3991 [DOI] [PubMed] [Google Scholar]
- 34. Sklar J. G., et al. 2007. Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 104:6400–6405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ulett G. C., et al. 2007. Functional analysis of antigen 43 in uropathogenic Escherichia coli reveals a role in long-term persistence in the urinary tract. Infect. Immun. 75:3233–3244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. van der Woude M. W., Henderson I. R. 2008. Regulation and function of Ag43 (flu). Annu. Rev. Microbiol. 62:153–169 [DOI] [PubMed] [Google Scholar]
- 37. Volokhina E. B., Beckers F., Tommassen J., Bos M. P. 2009. The beta-barrel outer membrane protein assembly complex of Neisseria meningitidis. J. Bacteriol. 191:7074–7085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Walther D. M., Rapaport D., Tommassen J. 2009. Biogenesis of beta-barrel membrane proteins in bacteria and eukaryotes: evolutionary conservation and divergence. Cell. Mol. Life Sci. 66:2789–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Werner J., Misra R. 2005. YaeT (Omp85) affects the assembly of lipid-dependent and lipid-independent outer membrane proteins of Escherichia coli. Mol. Microbiol. 57:1450–1459 [DOI] [PubMed] [Google Scholar]
- 40. Wu T., et al. 2005. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell 121:235–245 [DOI] [PubMed] [Google Scholar]