Abstract
Rapid identification of metallo-β-lactamase-producing Gram-negative species is crucial for the timely implementation of infection control measures. We describe two pediatric cases in which colonization by VIM-1- and New Delhi metallo-beta-lactamase 1-producing Enterobacteriaceae was rapidly detected by phenotypic and genotypic methods. Phenotypic methods can be useful for routine detection of carbapenemase production.
CASE REPORT
Case 1.
In November 2010, we detected fecal carriage of health care-associated metallo-β-lactamase (MBL)-producing Klebsiella pneumoniae together with extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli, Enterobacter cloacae, and Citrobacter freundii in a 4.5-month-old infant having traveled to France from Egypt.
The child was born in Egypt in July 2010. He was treated for an occlusive syndrome in Benha Children's Hospital, Benha, Egypt, and was discharged with a rectal drain at 10 days of age. He arrived in Paris, France, 2 months later, where Hirschsprung rectosigmoid disease was diagnosed. On admission to our institution, culture on ChromID ESBL medium (bioMérieux) was used to screen his stool flora for cefpodoxime resistance. MBL and ESBL producers exhibit high-level resistance to cephalosporins and are easily detected with this medium (16, 25). A carbapenem-nonsusceptible K. pneumoniae strain was recovered from his feces at a density of 102 CFU/g. After 2 days of cefotaxime prophylaxis (100 mg/kg/24 h) for abdominal surgery, the K. pneumoniae fecal count rose to 108 CFU/g. In addition, ESBL-producing E. coli (3 different strains), E. cloacae, and C. freundii were recovered from his stool at densities of 109 CFU/g, 108 CFU/g, 107 CFU/g, 109 CFU/g, and 106 CFU/g, respectively. He was discharged from the hospital 10 days later.
Case 2.
In February 2011, we detected community-acquired fecal carriage of MBL-producing E. coli together with ESBL-producing E. coli in a 21-month-old boy having returned to France from India who was admitted to our institution for asthma exacerbation. He was born in France to parents originating from Sri Lanka.
Ten days prior to his admission, he had returned to France after a 1-month stay in India (Chennai), where no health problems or hospitalizations were reported. On admission, he had respiratory distress related to asthma but no other symptoms. Carbapenem-nonsusceptible E. coli and ESBL-producing E. coli were recovered on ChromID ESBL culture medium (bioMérieux) from an admission stool sample, both at a density of 108 CFU/g (16). He was discharged from the hospital after 7 days.
The carbapenem-nonsusceptible K. pneumoniae and E. coli isolates were resistant to all beta-lactams agents, with the exception of aztreonam for the E. coli isolate by the disk diffusion method. Both isolates were also resistant to aminoglycosides, quinolones, nitrofurantoin, and cotrimoxazole. They were susceptible to colistin and tigecycline, with respective MICs determined by Etest of 0.125 μg/ml and 1.5 μg/ml (K. pneumoniae) and 0.094 μg/ml and 0.094 μg/ml (E. coli), and also susceptible to fosfomycin (5, 6, 7). The MICs of ertapenem, meropenem, and imipenem were >32 μg/ml, >32 μg/ml, and 8 μg/ml, respectively, for the K. pneumoniae isolate and 1.5 μg/ml, 0.75 μg/ml, 0.75 μg/ml for the E. coli isolate (6). The modified Hodge test (MHT) (3) was strongly positive for carbapenemase production by the K. pneumoniae isolate and weakly positive for carbapenemase production by the E. coli isolate.
To identify the resistance mechanism(s), we used combined disk assays with meropenem (10 μg) as the substrate and distinct carbapenem inhibitors, i.e., dipicolinic acid (DPA) for MBL and boronic acid (BO) for K. pneumoniae carbapenemase (Rosco, Taastrup, Denmark), in conjunction with the Etest using imipenem/imipenem-EDTA strips (bioMérieux).
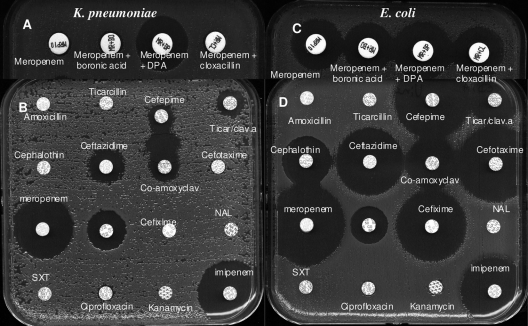
Synergy between EDTA and imipenem (Etest) and between meropenem and DPA was observed for the K. pneumoniae isolate (zone size difference, ≥5 mm), suggesting MBL production (Fig. 1A) (8, 14, 19). In contrast, the synergy between meropenem and DPA (Fig. 1C) (zone size difference, <5 mm) and between imipenem and EDTA for the E. coli isolate was equivocal. However, ertapenem could be used instead of meropenem since ertapenem showed the least activity against the New Delhi MBL 1 (NDM-1)-producing E. coli isolate.
Fig. 1.
K. pneumoniae is shown in panels A and B. E. coli is shown in panels C and D. (A and C) Phenotypic carbapenemase detection by the disk diffusion method using a combination of disks containing meropenem (10 μg), meropenem (10 μg) plus DPA, meropenem (10 μg) plus BO, and meropenem (10 μg) plus cloxacillin. (A) A zone size difference of ≥5 mm between meropenem and meropenem plus DPA points to MBL production. (C) A zone size difference of 3 mm between meropenem and meropenem plus DPA (positive if the difference is ≥5 mm). (B and D) Culture on Mueller-Hinton agar containing 5 × 10−3 M EDTA. (B) Inhibition of MBL by EDTA. Detection of an ESBL is shown. Synergy between the co-amoxiclav and cefepime disks points to ESBL production. (D) Inhibition of MBL by EDTA. Detection of plasmidic class A beta-lactamase 2b (TEM-1/-2, SHV-1) is shown. NAL, nalidixic acid; SXT, sulfamethoxazole-trimethoprim; Ticar/clav.a, ticarcillin-clavulanic acid.
On Mueller-Hinton agar containing 5 × 10−3 M EDTA, MBL production by both strains was totally inhibited. Culture on the same medium revealed synergy between co-amoxiclav and cefepime for the K. pneumoniae isolate, suggesting ESBL production (Fig. 1B) and plasmidic class A beta-lactamase 2b (TEM-1/-2, SHV-1) production by the E. coli isolate (Fig. 1D).
PCR amplification and sequencing were used to identify the beta-lactamase genes. In case 1, the carbapenem-resistant K. pneumoniae carried blaVIM-1 and blaCTX-M-14. The three E. coli isolates harbored blaTEM-1, blaCTX-M-15, and blaOXA-1; blaTEM-1 and blaCTX-M-15; and blaCTX-M-15 and blaOXA-1, respectively. The C. freundii isolate carried blaCTX-M-15 and blaOXA-1, and the E. cloacae isolate carried blaTEM-1 and blaCTX-M-15 (2, 21).
In case 2, the MBL-producing E. coli isolate harbored blaNDM-1 and blaTEM-1 and the ESBL-producing E. coli isolate carried blaCTX-M15 and blaOXA-1.
Phylogenetic analysis using a triplex PCR method targeting chuA, yjaA, and the TspE4 DNA fragment showed that the NDM-1-producing E. coli isolate belonged to commensal, less virulent group A (4). The strain was tested for virulence factor genes, revealing the presence of fuyA, papC, aer, and papGIII (1).
To our knowledge, this is the first case of imported health care-associated fecal carriage of an Egyptian VIM-1-producing K. pneumoniae strain in France, the first isolation of a community-acquired E. coli strain harboring the NDM-1 gene from a child in France, and the sixth reported isolation of NDM-1-producing bacteria in France (10, 17, 20, 23).
Discussion.
The emergence of MBL-producing bacteria in children is a matter of serious concern because it severely limits treatment options. The most frequent MBLs reported to date belong to the VIM and IMP types, which have been reported extensively worldwide (18). NDM-1, a novel acquired MBL, has recently started to spread (11). The genes that encode these enzymes are a source of concern, as they usually are carried by mobile genetic elements with a high capacity for horizontal dissemination (11, 22). In contrast, carbapenem resistance due to a combination of ESBL or AmpC production and porin loss has a fitness cost and spreads slowly. It is therefore important to determine the mechanism of carbapenem resistance. The MHT has been used to detect carbapenemase producers (3). In contrast to the VIM-1-producing K. pneumoniae isolate, the MHT showed only weak production of the NDM-1 carbapenemase by the E. coli isolate. However, weak or negative MHT results for NDM-1 have already been reported (15). Molecular testing is recommended to characterize the MBL genes of Enterobacteriaceae (16), but PCR is not universally available. Phenotypic methods such as those used in our study can efficiently detect the resistance mechanism.
Invasive infections by carbapenem-resistant strains are associated with high morbidity and mortality rates (24). Early identification of colonized patients on hospital admission is crucial for timely implementation of infection control measures and for rapid adaptation of antimicrobial chemotherapy in case of infection. Indeed, delayed detection of carbapenem-resistant organisms may lead to outbreaks of colonization or infection (12, 13). The French health authorities recommend the screening of all patients hospitalized abroad for carriage of multiresistant bacteria on the day of their admission to any French hospital (9). In our pediatric hospital, all patients arriving from countries where infections with these bacteria are endemic, whether or not they were hospitalized abroad, are screened on admission. Further cases of colonization or infection by the carbapenemase-producing bacteria described here were prevented by reinforcing infection control procedures and by screening all of the patients on the affected ward by rectal swabbing.
In conclusion, infections by carbapenem-resistant bacteria are difficult to treat and rapid identification of MBL-producing Gram-negative species is crucial both for appropriate treatment and for timely implementation of infection control measures. Phenotypic methods can be useful for routine detection of carbapenemase production, particularly when PCR is not immediately available.
Footnotes
Published ahead of print on 8 June 2011.
REFERENCES
- 1. Bingen-Bidois M., et al. 2002. Phylogenetic analysis and prevalence of urosepsis strains of Escherichia coli bearing pathogenicity island-like domains. Infect. Immun. 70:3216–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Branger C., et al. 2005. Genetic background of Escherichia coli and extended- spectrum beta-lactamase type. Emerg. Infect. Dis. 11:54–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Centers for Disease Control Prevention Last accessed 20 August 2010. Modified Hodge test for carbapenemase detection in Enterobacteriaceae. Centers for Disease Control and Prevention, Atlanta, GA. www.cdc.gov/ncidod/dhqp/pdf/ar/HodgeTest_carbapenemase_Enterobacteriaceae.pdf
- 4. Clermont O., Bonacorsi S., Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555–4558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Clinical Laboratory Standards Institute 2011. Performance standards for antimicrobial susceptibility testing. CLSI M100-S21. Clinical and Laboratory Standards Institute, Wayne, PA.
- 6. Clinical Laboratory Standards Institute 2010. Performance standards for antimicrobial susceptibility testing; M100-S20U. Update June 2010. Clinical and Laboratory Standards Institute, Wayne, PA.
- 7. Comité de l'Antibiogramme de la Societé Française de Microbiologie 2011. Recommandations 2011. Edition de janvier 2011. Societé Française de Microbiologie, Paris, France.
- 8. Deshpande P., et al. 2010. New Delhi metallo-β lactamase (NDM-1) in Enterobacteriaceae: treatment options with carbapenems compromised. J. Assoc. Physicians India 58:147–149 [PubMed] [Google Scholar]
- 9. Haut Conseil de Santé Publique 2010. Dépistage du portage digestif des bactéries commensales multirésistantes aux antibiotiques importées en France à l'occasion du rapatriement de patients en provenance de l'étranger et maîtrise de leur diffusion. Haut Conseil de Santé Publique, Paris, France. www.hcsp.fr/explore.cgi/hcspr20100518_bmrimportees.pdf
- 10. Institut National de Veille Sanitaire 2011. Entérobactéries productrices de carbapénèmases. Situation épidémiologique au 7 janvier 2011. Institut National de Veille Sanitaire, Saint-Maurice, France. http://www.invs.sante.fr/surveillance/enterobacteries/situation_070111.htm
- 11. Kumarasamy K. K., et al. 2010. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 10:597–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mammina C., et al. 2010. Multiclonal emergence of carbapenem-resistant Klebsiella pneumoniae in Tuscany, Italy. Int. J. Antimicrob. Agents 36:576–578 [DOI] [PubMed] [Google Scholar]
- 13. Mammina C., et al. 2010. Outbreak of infection with Klebsiella pneumoniae sequence type 258 producing Klebsiella pneumoniae carbapenemase 3 in an intensive care unit in Italy. J. Clin. Microbiol. 48:1506–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Miriagou V., et al. 2010. Detecting VIM-1 production in Proteus mirabilis by an imipenem-dipicolinic acid double disk synergy test. J. Clin. Microbiol. 48:667–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mochon A. B., et al. 2011. New Delhi metallo-{beta}-lactamase (NDM-1)-producing Klebsiella pneumoniae: a case report and laboratory detection strategies. J. Clin. Microbiol. 49:1667–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nordmann P., Poirel L., Carrër A., Toleman M. A., Walsh T. R. 2011. How to detect NDM-1 producers. J. Clin. Microbiol. 49:718–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nordmann P., Poirel L., Toleman M. A., Walsh T. R. 2011. Does broad-spectrum {beta}-lactam resistance due to NDM-1 herald the end of the antibiotic era for treatment of infections caused by Gram-negative bacteria? J. Antimicrob. Chemother. 66:689–692 [DOI] [PubMed] [Google Scholar]
- 18. Overturf G. D. 2010. Carbapenemases: a brief review for pediatric infectious disease specialists. Pediatr. Infect. Dis. J. 29:68–70 [DOI] [PubMed] [Google Scholar]
- 19. Picão R. C., et al. 2008. Metallo-β-lactamase detection: comparative evaluation of double-disk synergy versus combined disk tests for IMP-, GIM-, SIM-, SPM-, or VIM-producing isolates. J. Clin. Microbiol. 46:2028–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Poirel L., Hombrouck-Alet C., Freneaux C., Bernabeu S., Nordmann P. 2010. Global spread of New Delhi metallo-β-lactamase 1. Lancet Infect. Dis. 10:832. [DOI] [PubMed] [Google Scholar]
- 21. Poirel L., Naas T., Nordmann P. 2010. Diversity, epidemiology, and genetics of class D beta-lactamases. Antimicrob. Agents Chemother. 54:24–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Poirel L., Revathi G., Bernabeu S., Nordmann P. 2011. Detection of NDM-1-producing Klebsiella pneumoniae in Kenya. Antimicrob. Agents Chemother. 55:934–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Poirel L., et al. 2011. Extremely drug-resistant Citrobacter freundii isolate producing NDM-1 and other carbapenemases identified in a patient returning from India. Antimicrob. Agents Chemother. 55:447–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Struelens M. J., et al. 2010. New Delhi metallo-beta-lactamase 1-producing Enterobacteriaceae: emergence and response in Europe. Euro Surveill. 15:19716. [DOI] [PubMed] [Google Scholar]
- 25. Yong D., et al. 2009. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53:5046–5054 [DOI] [PMC free article] [PubMed] [Google Scholar]