Abstract
Ureaplasma parvum and Ureaplasma urealyticum are sexually transmitted, opportunistic pathogens of the human urogenital tract. There are 14 known serovars distributed between the two species. For decades, it has been postulated based upon limited data that virulence is related to serotype specificity. The results were often inconclusive due to the small sample size and extensive cross-reactivity between certain serovars. We developed real-time quantitative PCRs that allow reliable differentiation of the two species and type strains of each of the 14 serovars. To investigate species and serovar distributions, we typed 1,061 clinical isolates of human ureaplasmas from diverse patient populations. There was only a tenuous association between individual Ureaplasma serovars and certain patient populations. This may in part be explained by the fact that almost 40% of the isolates were genetic mosaics, apparently arising from the recombination of multiple serovars. This explains the extensive cross-reactivity based upon serotyping and the lack of consistent association of given serotypes with disease.
INTRODUCTION
Ureaplasmas were first described in the human urogenital tract by Shepard in 1954 (45). They belong to the class Mollicutes and are one of the smallest self-replicating prokaryotes, lacking cell walls and hydrolyzing urea to generate ATP. Thus far, in humans there are at least 14 known Ureaplasma serovars determined by the metabolism inhibition test and colony indirect epi-immunofluorescence using rabbit antisera (40). These 14 serovars were established when it was believed there was only one species of ureaplasma that occurred in humans, U. urealyticum. More recently the 14 serovars have been divided between two species by these and other criteria. U. parvum contains serovars 1, 3, 6, and 14, and U. urealyticum contains the remaining 10 serovars (42). Although ureaplasmas are common commensals in the urogenital tract, they have been associated in many invasive infections, such as nongonococcal urethritis (NGU), chorioamnionitis, endometritis, and arthritis in adults and bacteremia, meningitis, pneumonia, and chronic lung disease of prematurity (bronchopulmonary dysplasia [BPD]) in infants (52).
It has been speculated for many years that individual Ureaplasma species or serovars might be associated with certain diseases more than others, as is the case for bacteria such as Streptococcus pneumoniae and Haemophilus influenzae (2, 14). Although several studies reported that U. urealyticum is more pathogenic than U. parvum (1, 13, 26, 30, 34, 36), conflicting results have been found by others (6, 18), so it is possible that differential pathogenicity might exist at the serovar level rather than at the species level. Inconsistent results implicating specific serovars (or serovar groups) with various clinical conditions (10, 12, 29, 33, 46, 50, 58) coupled with frequent detection of Ureaplasma isolates comprised of more than one serovar have also been reported (10, 17, 29, 32, 50). Differing results among investigations could be related to inadequate or imprecise typing methods. The possibility that individual Ureaplasma isolates may express multiple serovar specificities has been suggested, but never investigated in a systematic manner due to the technical difficulty of separating all of the 14 serovars (17, 50).
Two major methodologies to classify Ureaplasma isolates to the species and serovar levels have been described. Antibody-based phenotyping methods included growth/metabolism inhibition tests (3, 41, 46), antibody-linked epi-immunofluorescence, or color reaction assays (15, 37, 44, 49). These methods yielded inconclusive results because of multiple cross-reactions and poor discriminating capacity. Cross-reactions have been observed even in certain serovar reference strains (40). Molecular genotyping methods were more rapid and accurate, readily separating the two Ureaplasma species (9, 27, 35, 43, 56). However, due to limited sequence variation in the PCR targets, only partial separation of serovars was achieved (8, 20-23). Recently, we developed a set of Ureaplasma species- and serovar-specific real-time PCR assays that separate completely all 14 ATCC serovars type strains without cross-reactions (55).
Horizontal gene transfer (HGT) plays an important role in microbial adaptation, speciation, and evolution (48). Although mycoplasmas and ureaplasmas are characterized by their minimal genomes and are thought to have undergone regressive evolution, which usually is not favorable for active DNA acquisition (54), evidence has shown that HGT occurs among phylogenetically distinct mycoplasmal species sharing the same ecological niche (47) or within the same species (51). Comparative genomic analyses have indicated possible HGT between U. parvum and Mycoplasma hominis (31). Ureaplasma spp. have also been shown to form biofilm (16), structures thought to promote DNA exchange among other bacteria. Furthermore, recombinases, transposases and putative conjugative transposon mobilization proteins have been identified in genomes of the 14 Ureaplasma serovars (V. Paralanov, unpublished observations). These findings raise the question of whether HGT occurs among Ureaplasma serovars and its potential implications for the hypothesis of differential pathogenicity at the serovars level.
Using Ureaplasma species- and serovar-specific real-time PCR assays that we recently developed and validated (55), we analyzed a large number of clinical isolates from different geographic regions and from different patient populations to classify them to species and serovar in order to investigate differential pathogenicity at these two levels. Initial evaluation of the results suggested the likelihood of HGT between species and among serovars and prompted additional investigations.
MATERIALS AND METHODS
Bacterial isolates.
Reference strains for the 14 Ureaplasma serovars were obtained from the American Type Culture Collection (ATCC) and used as the controls in serovar-specific PCR assays. The 1,061 unique clinical isolates evaluated for species and serovar distributions were obtained from cultures collected and stored frozen at −80°C or in lyophilized form dating from the late 1970s to 2009. Isolates originated from Alabama and various other states within the continental United States and from Alberta, Canada, in the patient groups described in Table 1. Ureaplasmas recovered from clinical specimens were initially identified to genus level by standard methods including colony morphology and urease production on A8 agar.
Table 1.
Description of Ureaplasma clinical isolates
| Specimen type | Group description | No. of isolates |
|---|---|---|
| Control groups | ||
| Vaginal swabs | Healthy pregnant females | 169 |
| Placental tissue collected at cesarean section in women with intact fetal membranes | Females without histologic chorioamnionitis | 42 |
| Catheterized urine | Males with neurogenic bladder without urethritis | 25 |
| Endotracheal aspirate | Preterm infants without bronchopulmonary dysplasia | 108 |
| Diseased groups | ||
| Endometrial biopsy tissue | Females with pelvic inflammatory disease and/or postpartum endometritis | 85 |
| Placental tissue collected at cesarean section in women with intact fetal membranes | Females with histologic chorioamnionitis | 18 |
| Endotracheal aspirate | Preterm infants with bronchopulmonary dysplasia | 88 |
| Urethral swab or urine | Males with nongonococcal urethritis from Canada | 421 |
| Urethral swab or urine | Males with nongonococcal urethritis from the United States | 81 |
| Blood, cerebrospinal fluid, synovial fluid, pleural fluid, lung tissue | Invasive isolates from various patient groups, including adults and children | 24 |
| Total | 1,061 |
DNA preparation.
Genomic DNA was extracted from all 1,061 clinical isolates by the proteinase K method as described previously (4). Inhibited samples were further purified by using a QIAamp DNA blood mini kit (Qiagen, Valencia, CA). Prepared DNA samples were stored at −80°C unless submitted immediately for PCR assay.
Genotyping of clinical isolates by PCR.
The clinical isolates were first classified to species level by using a multiplex species-specific real-time PCR assay for which the primers, probes, reagents, and PCR conditions have been previously described (55). U. parvum and U. urealyticum isolates were then typed for their corresponding serovars by a series of serovar-specific real-time PCR assays (55). In the event that isolates were negative for any serovars within their corresponding species, additional PCR assays for serovars of the other species were performed. A designated ATCC type strain control and a negative control (distilled water) were included in every PCR run. Untypeable isolates were subjected to a secondary PCR assay targeting the urease gene (35). The species- and serovar-specific PCR assays were performed using a LightCycler 2.0 (Roche, Indianapolis, IN).
Separation of isolates containing multiple serovars.
Four isolates shown to contain multiple serovars by real-time PCR were thawed and incubated in 10B broth overnight. The broth cultures were then filtered through a 0.2-μm-pore-size filter, inoculated onto A8 agar, and incubated for 24 to 48 h until colonies were readily visible under a stereomicroscope at ×126 magnification. At least 10 single colonies from each isolate were removed with a sterile needle or pipette tip for individual overnight cultures in 10B broth. DNA was then isolated from each broth culture and prepared for PCR assay as described above.
Quantification of each serovar in clinical isolates to distinguish mixtures from hybrids.
To quantify each serovar marker in isolates containing multiple serovars, a universal control plasmid, pUC19-U, which carries one copy of each serovar marker (except for markers of serovars 4 and 5, which were not able to be stably incorporated into the plasmid), was constructed and used to generate an external quantification standard curve. Serovar-specific PCRs were performed on 271 randomly selected isolates containing multiple serovars (except serovars 4 and 5), and the quantities of each serovar were calculated. When two serovars were compared, based on the largest calculated quantity difference of the pUC19-UU plasmid control between PCR runs, an arbitrary differentiation standard was made: a difference of ≤5-fold was considered a hybrid; a difference of 5- to 10-fold was considered a hybrid or mixture (Hyb/Mix); any difference >10-fold was considered a mixed culture; and, finally, if both serovars were in low quantity, it was designated as undetermined.
DNA sequencing.
A total of seven clinical isolates, each containing two serovar markers, were selected for polymorphic loci analysis by Sanger DNA sequencing. Isolates 10902 and 97078 contained serovars 1 and 6, isolates 10901 and 8510 contained serovars 3 and 6, and isolates 24318 and 25353 contained serovars 9 and 10. For each pair of serovars, 7 to 10 loci throughout the genome that contained multiple base pair polymorphisms between each serovar pair were selected. PCR primers were designed to amplify from both serovars across the region containing the polymorphism of about 700 to 800 bp. Loci were selected to be gene coding, have at least 90% identity, and 20 or more mismatched nucleotides by BLASTn analysis (see Table S1 in the supplemental material). The mba gene of U. parvum serovars 3 and 6 was sequenced by using primers flanking the whole mba gene region. Serovar-specific PCR primer regions of serovars 9 to 12 were also amplified and sequenced. PCR was performed using a Veriti 96-well thermal cycler (Applied Biosystems, California). DNA sequencing was performed in the University of Alabama at Birmingham Heflin Center Genomics Core Facility. DNA sequences were analyzed using CLC DNA workbench 5.
We sequenced the genomes of four U. urealyticum patient isolates that we could not identify on the serovar level using 454 pyrosequencing (454 Life Sciences, Branford, CT). The genomes were assembled by using a Newbler Assembler (454 Life Sciences). We then compared the four isolates to each of the ATCC reference urealyticum genomes by generating dot plots. Sequencing was done at the J. Craig Venter Institute.
Statistical analysis.
The Fisher exact test and χ2 test were performed to compare the distributions of Ureaplasma species and serovars in different patient populations. A P value of <0.05 was considered statistically significant. Analyses were conducted by using SAS software (SAS Institute, Inc., Cary, NC) and SPSS 16.0 (SPSS, Inc., Chicago, IL).
RESULTS
Species and serovar distributions of isolates from different patient populations.
Of 1,061 unique clinical isolates that were typed to species and serovar levels (see Table S2 in the supplemental material), U. parvum was detected in 508 (48%) isolates, U. urealyticum was detected in 406 (38%) isolates, and both species were detected in 140 (13%) isolates. The remaining seven isolates (1%) could not be typed to the species level by real-time PCR. However, one of them was determined to be U. urealyticum by using a PCR targeting the urease gene (35). The prevalence of U. urealyticum was significantly increased in isolates from endometrial biopsy tissues from women with pelvic inflammatory disease (PID) and/or endometritis (33% versus 14%, P ≤ 0.01) in comparison to vaginal swabs from healthy pregnant women. It was also more common in American men with NGU compared to urine samples from men without urethritis (68% versus 48%, P < 0.05). No association of either Ureaplasma species was found in the placentas of women with versus without chorioamnionitis. There was also no difference in infants with or without chronic lung disease of prematurity or among the 24 invasive isolates. Canadian men with NGU had a significantly higher percentage of mixed species than U.S. men (26% versus 6%, P=0.001); however, the difference might merely reflect methodology and/or isolation procedures.
The 14 serovars were distributed unevenly in all groups: serovar 3 was the most common (n=332) and serovar 5 was the least common (n=19) when counting the presence of any serovars in all 1,061 isolates (see Table S2 in the supplemental material). When we compared the prevalence of serovars in diseased groups with their corresponding controls, we observed no consistent patterns implicating individual serovars. The serovars with significantly increased prevalence in diseased groups included serovars 5, 8, and 11 in the PID/endometritis group; serovar 6 in the chorioamnionitis group; serovar 9 in U.S. men with NGU; and serovar 10 in neonates in the BPD group. Among the 24 invasive isolates, serovar 3 was the most common, occurring in 7 (29%) isolates. The serovar distribution between NGU isolates from U.S. and Canadian men was significantly different, indicating that geographic factors may play an important role.
Isolates that were negative for all serovar-specific assays.
There were 67 (6%) isolates that could not be assigned to any of the 14 known serovars by PCR. To ascertain why these isolates were negative in all of our serovar-specific assays, we performed whole-genome shotgun sequencing of four of these isolates. Genome analysis showed that isolates 2033 and 2608 were most closely related to serovars 12 and 4. ATCC serovars 12 and 4 were the closest related serovars among the urealyticum group. Isolate 4155 was most similar to serovar 11, whereas isolate 4318 was most similar to serovar 2. Relative to the ATCC reference strains, all of the isolates' genomes had some minor genome rearrangements, areas that were deleted, and some areas that were inserted and are new for the urealyticum group. Analysis of the target areas for the serovar-specific PCR assays showed that the target was either missing completely, or some of the target was missing or modified so that one of the primers would not bind. However, it is clear that these isolates have changes in other areas of the genome as well. Whether we can assign new serovar numbers to any of the unidentifiable isolates is a matter of clarifying the requirements for a ureaplasma to be considered a specific serovar.
Isolates containing multiple serovars and conflicting serovars.
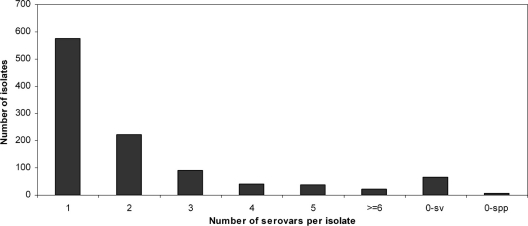
Multiple serovar markers were detected in 413 (39%) isolates, predominantly U. urealyticum (n=201) and mixed species (n=124) (Fig. 1). Among them, 223 (21%) isolates contained two serovar markers, 91 (9%) isolates contained three serovar markers, and 99 isolates (9%) contained four or more serovar markers. The maximum number of serovar markers detected in a single isolate was 10. The distribution of serovar marker numbers per isolate did not show significant differences between control and diseased groups, except for U.S. men with NGU.
Fig. 1.
Clinical isolates containing multiple serovars and untypeable serovars. A total of 1061 clinical isolates were typed to species and then serovar level by real-time PCR. In the event isolates were determined to be negative for any serovars within their designated species, additional PCR assays for serovars of the other species were performed. Untypeable isolates (0-sv, negative for any serovar; 0-spp, negative for any species) were subjected to a secondary PCR assay targeting the urease gene to confirm the species.
Discordant typing results were observed in 42 isolates. Three isolates were typed as U. parvum; however, their serovar markers belonged to U. urealyticum. On the other hand, 15 isolates were typed as U. urealyticum but contained U. parvum serovar markers. One U. urealyticum isolate showed serovar markers of both species. One isolate negative for the species-specific real-time PCR contained two different U. urealyticum serovar markers, whereas 22 isolates positive for both species showed serovar markers from only one.
Separation of isolates with multiple serovars.
In an effort to isolate pure cultures containing a single serovar from clinical isolates containing apparent mixtures of multiple serovars, we found that four isolates could not be purified into a single serovar after filtering and selecting single colonies for subculture. Two of them contained loci expressing serovars 3 and 14; one isolate expressed serovars 1 and 14, and one expressed serovars 3 and 6. As a control, a mixture of equal amounts of ATCC type strains of serovars 1 and 6 was made, and these two serovars were completely separated after the same procedures. We therefore suspected that these isolates and many of the others containing what were initially believed to be comprised of multiple serovars might not be true mixtures, but hybrid organisms carrying multiple serovar markers.
Verifying hybrid isolates by sequencing multiple gene-coding, polymorphic loci.
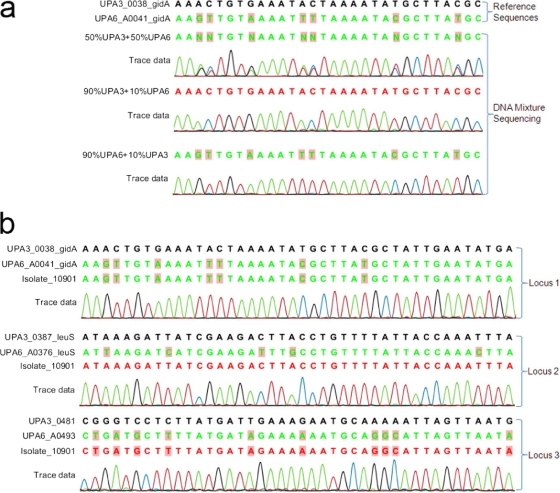
To simplify the test to determine whether the isolates were mixtures or hybrids, we focused on isolates that were positive for two serovar markers and analyzed them by DNA sequencing (primers are listed in Table S1 in the supplemental material). We chose to sequence 7 to 10 gene-coding loci that are widely distributed around the two genomes and contain multiple base pair polymorphisms. Control mixtures of equimolar genomic DNA concentration of ATCC type serovars 3 and 6 had visible double nucleotide peaks in the sequencing chromatograms as shown in Fig. 2 a. Differentiating mixed versus pure isolates was limited when the DNA concentration ratio reached 9:1, and clean single peaks representing the predominant DNA type appeared (Fig. 2a). A hybrid would be expected to show single peaks at polymorphic sites representing the sequence of one serovar in some loci and the other serovar in the rest of the loci (Fig. 2b). In all tested loci, ATCC type strains of corresponding serovars were amplified and sequenced as controls.
Fig. 2.
DNA sequencing of multiple polymorphic loci throughout the genome. DNA sequencing trace files were aligned by using CLC genomic workbench. (a) Sequencing of DNA obtained from artificial mixtures of U. parvum serovar 3 and U. parvum serovar 6. Reference sequences of gidA orthologous genes of U. parvum serovars 3 and 6 are shown on the first two lines. The type of DNA mixture for each reaction is provided on the left of the sequences. Polymorphisms are marked with shaded boxes. Trace data indicated double peaks in the 1:1 mixture and single peaks in 9:1 mixtures, representing the predominant DNA. (b) Sequencing of multiple polymorphic loci in isolate 10901. A representative window of three of seven sequenced loci is shown. Polymorphic sites are marked with shaded boxes. Isolate 10901 showed single peaks at the polymorphic loci. Loci 1 and 3 showed characteristics of serovar 6, while locus 2 was the same as serovar 3.
Seven clinical isolates suspected to contain a hybrid organism of two serovars were analyzed. Six isolates (10902, 97078, 10901, 8510, 24318, and 25353) clearly showed some polymorphic loci characteristic of one serovar and some characteristic of another (Table 2). Seven polymorphic loci were examined in two isolates containing serovars 1 and 6. Isolate 10902 showed an obvious hybrid pattern: four loci were from UPA6, and three loci were from UPA1. In isolate 97078, six of seven loci were UPA6 specific, and one locus was closest to UPA14 (BLAST analysis showed 99.55% identity to UPA14). In order to examine the reason for having serovar-specific real-time PCR signal for both UPA1 and UPA6, we proceeded to sequence the two serovars' real-time PCR targets. Both PCR assays targeted the mba gene of each serovar. The mba sequencing chromatogram depicted only MBA1. Since real-time PCR is more sensitive than DNA sequencing, and DNA sequencing failed to recognize mixtures of 9:1 or lower ratios, we hypothesized that isolate 97078 contained two hybrids in a ratio of at least: 90% or more of a UPA6 with a UPA1 specific MBA hybrid, and 10% or less of a UPA1 with a UPA6 specific MBA hybrid. Another seven loci were tested for serovars 3 and 6, and hybrid patterns were observed in isolates 10901 and 8510. Isolates 24318 and 25353 were serial isolates from the same patient. They were positive by real-time PCR for serovars 9 and 10 and had 10 polymorphic loci from UUR10 and two specific loci from UUR9. Sequencing of the real-time PCR targets indicated that the two isolates contained serovar markers of both UUR9 and UUR10. Taken together, all of the data suggested that these two isolates are hybrids.
Table 2.
DNA sequencing results of polymorphic loci, mba gene, and serovar marker regions
| Compared serovar | Isolate | Polymorphic locus | Polymorphic character | MBA/serovar markera | Conclusion |
|---|---|---|---|---|---|
| UPA1_6 | 10902 | UPA1_43 | UPA6 | NA | Hybrid |
| UPA1_98 | UPA6 | NA | Hybrid | ||
| UPA1_100 | UPA1 | NA | Hybrid | ||
| UPA1_101 | UPA1 | NA | Hybrid | ||
| UPA1_114 | UPA6 | NA | Hybrid | ||
| UPA1_293 | UPA1 | NA | Hybrid | ||
| UPA1_359 | UPA6 | NA | Hybrid | ||
| UPA1_6 | 97078 | UPA1_43 | UPA6 | MBA1 | Hybrid |
| UPA1_98 | UPA6 | MBA1 | Hybrid | ||
| UPA1_100 | UPA6 | MBA1 | Hybrid | ||
| UPA1_101 | UPA6 | MBA1 | Hybrid | ||
| UPA1_114 | UPA6 | MBA1 | Hybrid | ||
| UPA1_293 | UPA14? | MBA1 | Hybrid | ||
| UPA1_359 | UPA6 | MBA1 | Hybrid | ||
| UPA3_6 | 10901 | UPA3_37 | UPA6 | MBA3 | Hybrid |
| UPA3_38 | UPA6 | MBA3 | Hybrid | ||
| UPA3_39 | UPA6 | MBA3 | Hybrid | ||
| UPA3_98 | UPA3 | MBA3 | Hybrid | ||
| UPA3_378 | UPA3 | MBA3 | Hybrid | ||
| UPA3_481 | UPA6 | MBA3 | Hybrid | ||
| UPA3_512 | UPA3 | MBA3 | Hybrid | ||
| UPA3_6 | 8510 | UPA3_37 | UPA3/6 | NA | Hybrid |
| UPA3_38 | UPA3* (UUR2/UPA14?) | NA | Hybrid | ||
| UPA3_39 | UPA3 | NA | Hybrid | ||
| UPA3_98 | UPA6 | NA | Hybrid | ||
| UPA3_378 | UPA3* (UPA14?) | NA | Hybrid | ||
| UPA3_481 | UPA3 | NA | Hybrid | ||
| UPA3_512 | UPA3 | NA | Hybrid | ||
| UUR9_10 | 24318 | UUR10_0043 | UUR10 | Both | Hybrid |
| UUR10_0072 | UUR10 | Both | Hybrid | ||
| UUR10_0138 | UUR10 | Both | Hybrid | ||
| UUR10_0141 | UUR10 | Both | Hybrid | ||
| UUR10_0329 | UUR10 | Both | Hybrid | ||
| UUR10_0364 | UUR10 | Both | Hybrid | ||
| UUR10_0376 | UUR10 | Both | Hybrid | ||
| UUR10_0371 | UUR10 | Both | Hybrid | ||
| UUR10_0653 | UUR10 | Both | Hybrid | ||
| UUR10_0654 | UUR10 | Both | Hybrid | ||
| UUR9_ORF01470 (UUR9 only) | UUR9 | Both | Hybrid | ||
| UUR9_ORF01469 (UUR9 only) | UUR9 | Both | Hybrid | ||
| UUR9/10_ORF00475 | One copy of UUR10 and one or two copies of UUR9 | Both | Hybrid | ||
| UUR9_10 | 25353 | UUR10_0043 | UUR10 | Both | Hybrid |
| UUR10_0072 | UUR10 | Both | Hybrid | ||
| UUR10_0138 | UUR10 | Both | Hybrid | ||
| UUR10_0141 | UUR10 | Both | Hybrid | ||
| UUR10_0329 | UUR10 | Both | Hybrid | ||
| UUR10_0364 | UUR10 | Both | Hybrid | ||
| UUR10_0376 | UUR10 | Both | Hybrid | ||
| UUR10_0371 | UUR10 | Both | Hybrid | ||
| UUR10_0653 | UUR10 | Both | Hybrid | ||
| UUR10_0654 | UUR10 | Both | Hybrid | ||
| UUR9_ORF01470 (UUR9 only) | UUR9 | Both | Hybrid | ||
| UUR9_ORF01469 (UUR9 only) | UUR9 | Both | Hybrid | ||
| UUR9/10_ORF00475 | UUR10 | Both | Hybrid |
NA, not available; Both, serovar markers of both UUR9 and UUR10.
Prevalence of hybrid ureaplasmas.
To quickly estimate the prevalence of hybrid ureaplasmas in a large number of clinical isolates containing multiple serovars, quantitative real-time PCR assays were performed using a universal quantification standard, plasmid pUC19-UU, which contained one copy of each serovar-specific PCR target (except for serovars 4 and 5). The rationale for this test is based on the observation and hypothesis that mixtures of different serovars will still occur as mixtures in different ratios after regrowth because of different growth rates of serovars and different mixing ratio at starting point. On the other hand, a hybrid, as a single organism will contain the same ratio of different serovar markers at the starting point and after regrowth. Thus, by quantification of serovar markers in clinical isolates, we should be able to differentiate the hybrids and mixtures.
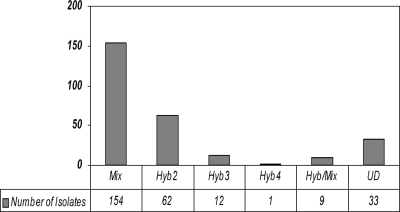
Among 271 randomly selected isolates out of 413 containing multiple serovars (except serovars 4 and 5), 75 (28%) were hybrids (Fig. 3). Among the hybrid isolates, 62 (23%) were hybrids of two serovars, 12 (4%) were hybrids of three serovars, and 1 (<1%) was a hybrid of four serovars. Most of the hybrids contained markers of the same species (Table 3). There were 10 U. parvum hybrids and 59 U. urealyticum hybrids. Interspecies hybrids were found in six isolates. The most common hybrid types that we encountered were hybrids of serovars 11 and 12. A clinical subculture could be a pure hybrid strain, a mixture of a hybrid strain and/or nonhybrid strains, or even a mixture of different hybrid strains, such as isolate 12599, which was a mixture of two hybrids—a hybrid of serovar 2/11 and another hybrid of serovar 7/12, and a nonhybrid strain of serovar 13.
Fig. 3.
Prevalence of hybrid serovars in clinical isolates. A total of 271 randomly selected clinical isolates (except serovars 4 and 5) containing multiple serovars were tested by quantitative real-time PCR, and the quantity of each serovar was calculated. To determine whether an isolate was a hybrid or mixed culture, an arbitrary standard was made based on the largest calculated deviation of the pUC19-UU control between PCR runs. A deviation of ≤5-fold among serovars was considered a hybrid strain (Hyb; the numbers following the term are the number of serovars detected in one); a deviation of 5- to 10-fold was considered a hybrid or mixture (Hyb/Mix); any deviation of >10-fold was considered to represent a mixed culture (Mix); and, finally, if both serovars were in low quantity, it was designated as undetermined (UD).
Table 3.
Types of Ureaplasma hybrids
| Hybrid type | Hybrid serovar | No. of isolates |
|---|---|---|
| Up hybrid | 1,3 | 1 |
| 1,6 | 2 | |
| 1,14 | 2 | |
| 1,3,6 | 1 | |
| 3,14 | 1 | |
| 3,6 | 2 | |
| 6,14 | 1 | |
| Uu hybrid | 2,7 | 2 |
| 2,7,12 | 1 | |
| 2,10 | 2 | |
| 2,10,12 | 2 | |
| 2,11 | 4 | |
| 7,9 | 1 | |
| 7,11 | 4 | |
| 7,12 | 6 | |
| 8,9 | 1 | |
| 8,10 | 1 | |
| 8,10,11,12 | 1 | |
| 8,11 | 7 | |
| 9,10 | 2 | |
| 9,11,12 | 1 | |
| 10,11 | 7 | |
| 10,11,12 | 6 | |
| 10,12 | 1 | |
| 11,12 | 10 | |
| Up+Uu hybrid | 1,2 | 1 |
| 3,9 | 1 | |
| 3,4,11 | 1 | |
| 6,8 | 1 | |
| 6,10 | 1 | |
| 6,11 | 1 | |
| Total | 75 |
DISCUSSION
To address the question whether differential pathogenicity exists at the Ureaplasma serovar level, an accurate typing method and a large number of clinical isolates are needed. The 14 serovar-specific real-time PCR assays have been proven to separate all 14 serovar type strains without cross-reactions (55). We applied these assays to type 1,061 clinical isolates from different patient populations. Thus far, this is the largest collection of clinical isolates that have been typed to species and serovar level by any method. The results indicated that U. urealyticum was significantly increased in men with NGU and in women with PID and/or endometritis, an observation which agrees with previous reports (1, 13, 26, 30, 34, 36). However, no agreement in association of particular serovars with diseases was achieved among the different patient groups, including the invasive isolates recovered from usually sterile sites. Previous studies also showed no consistent data correlating individual serovars and pathogenic outcome, even among invasive isolates (10, 12, 29, 33, 46, 50, 58, 59). This suggests that serovar designation is not a reliable subspecies marker for determining the differential pathogenicity of Ureaplasma.
We utilized the clinical isolates that were available to us for study, and we acknowledge there are some potential limitations in the study populations and comparator groups. One such limitation is the comparison of vaginal swabs from healthy women to serve as controls for comparison with endometrial tissue in the subjects with disease. A second is the relatively small number of control urine specimens (25) available for analysis from men without NGU. All of the clinical isolates were low-passage organisms, so it is unlikely that there would be selection of individual serovars that grow more rapidly in laboratory culture than others. However, unless one tests the original clinical specimens before any in vitro cultivation, it is not possible to know with certainty which serovars were there before the specimen is subjected to laboratory cultivation conditions. Furthermore, some serovars present in mixtures might not have survived prolonged storage and reconstitution. However, even with these potential limitations, our conclusions are not likely to be affected in the view of our finding hybrid serovars as a result of HGT. Thus, we must question the utility of serovar determination in the assessment of pathogenicity.
Multiple serovars were detected in ca. 40% (413/1,061) of the clinical isolates, while 6% (67/1,061) were not typeable and 4% (42/1,061) showed conflicting species and serovar results. It has been observed since the earliest Ureaplasma typing studies that many clinical isolates contain multiple serovars (10, 17, 29, 32, 50). Although cross-reactive typing reagents and mixed cultures were generally accepted as plausible explanations, it has never been completely clear whether certain strains can contain multiple serovar specificities and how this may occur. Failure to separate multiple serovars from some clinical isolates suggests the occurrence of hybrids. Our studies indicate there are pure organisms carrying multiple serovar markers. DNA sequencing of multiple loci with multiple base pair polymorphisms provided evidence of HGT and explains why some isolates were positive in more than one serovar-specific PCR assay. Because serovar-specific markers are exchanged between ureaplasmas, some organisms might acquire multiple markers, and some might completely lose all markers. Therefore, a likely consequence of HGT is the emergence of hybrid ureaplasmas containing multiple serovar markers from one or more species and untypeable strains. Alternatively, the untypeable strains may also represent new serovars that have never been characterized. The 14-serovar classification scheme was expected to be expanded at the time it was established (40), and several later studies using antibody-based or PCR methods have reported a certain number of untypeable isolates (19, 50, 58). To determine whether those untypeable isolates represent new serovars or loss of markers, additional analysis such as whole-genome sequencing of such isolates would be instructive.
HGT between U. parvum and M. hominis, which both localize to the mucosal surface of the human urogenital tract, was recently reported (31). Five clusters of genes encoding type I and III restriction/modification systems, transposases, and cell surface proteins were predicted to undergo HGT between the two phylogenetically distinct species.
Another possible explanation for the observation of clinical isolates that apparently express markers for multiple serovars could be high-frequency mutations. Although we cannot completely discount that this may indeed occur to some extent, it is unlikely to be the primary mechanism responsible for these observations of hybrid serovars. In our experimental design we selected 7 to 10 coding loci with at least 20 polymorphisms, with some of them being consecutive bases that were spread through the genome. The patterns we observed would be difficult if not impossible to produce by high-frequency mutation.
In the present study, we have reported that HGT occurs within Ureaplasma spp. Serovar markers and other sequences throughout the genome were exchanged among serovars, including the MBA gene, which was thought to be serovar specific (53). This finding questions the definition of serovars, because the phenotypic epitopes on which the serovars are based may change, combine, or be lost after HGT. The mechanisms involved in HGT in ureaplasmas are still not clear. Mobile genetic elements, such as the conjugative transposons Tn916 and Tn1545, and plasmids have been identified in ureaplasmas (11, 25, 39). Ureaplasmas may also form biofilms in vitro (16), dense structures that enhance gene transfer (28). Recombination may occur subsequently to transposition of the DNA into the recipient genome. Sequencing hybrid isolates may help to identify possible hot spots of recombination. Furthermore, two sets of major surface antigen proteins MBA and UU376 and UU171 and UU172 of serovar 3 are phase variable due to DNA inversion events (60, 61). One of these phase variants would be undetectable using a serological assay based on the serovar 3 MBA. On the other hand, different serotyping results have been reported to occur in the same strain following subculturing (24, 50).
Genes involved in pathogenicity have not been identified conclusively in Ureaplasma spp., and we have shown that individual serovars are unlikely to be associated with differential pathogenicity. Therefore, bacterial load and different host immune responses may be alternative explanations for varied clinical findings. It has been reported that increased bacterial load is associated with NGU (5, 7, 57). A study using an animal model of urinary tract infection showed that complications associated with U. parvum infection are primarily dependent on host-specific factors (38). Further studies are needed to elucidate mechanisms of specific host response in these and other conditions that may be associated with these organisms.
In conclusion, the present study demonstrates that HGT occurs naturally among human Ureaplasma species and serovars, and Ureaplasma pathogenicity is unlikely to be associated with individual serovars. Our data suggest that “serotyping” is impractical and of limited value for the assessment of pathogenicity. To clarify the mechanisms related to pathogenicity, future studies should focus on the specific immune response to Ureaplasma infections, although there is still the possibility that a gene or group of genes might be present in pathogenic ureaplasmas and absent in commensal ureaplasmas which yet have not been distinguished by examination at the species or serovar level.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by Federal funds under grants RO1A1072577 and RO1209, contract NO1-AI-30071, from the National Institute of Allergy and Infectious Diseases and grant AI 28279 from the National Institute of Child Health and Human Development.
The technical assistance of Donna Crabb and Amy Ratliff is gratefully acknowledged.
Footnotes
Supplemental material for this article may be found at http://jcm.asm.org/.
Published ahead of print on 22 June 2011.
REFERENCES
- 1. Abele-Horn M., Wolff C., Dressel P., Pfaff F., Zimmermann A. 1997. Association of Ureaplasma urealyticum biovars with clinical outcome for neonates, obstetric patients, and gynecological patients with pelvic inflammatory disease. J. Clin. Microbiol. 35: 1199–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anonymous. 2008. Invasive pneumococcal disease in children 5 years after conjugate vaccine introduction—eight states, 1998-2005. MMWR Morb. Mortal. Wkly. Rep. 57: 144–148 [PubMed] [Google Scholar]
- 3. Black F. T. 1973. Modifications of the growth inhibition test and its application to human T-mycoplasmas. Appl. Microbiol. 25: 528–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Blanchard A., Hentschel J., Duffy L., Baldus K., Cassell G. H. 1993. Detection of Ureaplasma urealyticum by polymerase chain reaction in the urogenital tract of adults, in amniotic fluid, and in the respiratory tract of newborns. Clin. Infect. Dis. 17Suppl. 1: S148–S153 [DOI] [PubMed] [Google Scholar]
- 5. Bowie W. R., et al. 1977. Etiology of nongonococcal urethritis. Evidence for Chlamydia trachomatis and Ureaplasma urealyticum. J. Clin. Invest. 59: 735–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bradshaw C. S., et al. 2006. Etiologies of nongonococcal urethritis: bacteria, viruses, and the association with orogenital exposure. J. Infect. Dis. 193: 336–345 [DOI] [PubMed] [Google Scholar]
- 7. Brunner H., Weidner W., Schiefer H. G. 1983. Quantitative studies on the role of Ureaplasma urealyticum in non-gonococcal urethritis and chronic prostatitis. Yale J. Biol. Med. 56: 545–550 [PMC free article] [PubMed] [Google Scholar]
- 8. Cao X., Jiang Z., Wang Y., Gong R., Zhang C. 2007. Two multiplex real-time TaqMan PCR systems for simultaneous detecting and serotyping of Ureaplasma parvum. Diagn. Microbiol. Infect. Dis. 59: 109–111 [DOI] [PubMed] [Google Scholar]
- 9. Cao X., Wang Y., Hu X., Qing H., Wang H. 2007. Real-time TaqMan PCR assays for quantitative detection and differentiation of Ureaplasma urealyticum and Ureaplasma parvum. Diagn. Microbiol. Infect. Dis. 57: 373–378 [DOI] [PubMed] [Google Scholar]
- 10. Cracea E., Constantinescu S., Lazar M. 1985. Serotypes of Ureaplasma urealyticum isolated from patients with nongonococcal urethritis and gonorrhea and from asymptomatic urethral carriers. Sex Transm. Dis. 12: 219–223 [DOI] [PubMed] [Google Scholar]
- 11. de Barbeyrac B., Dupon M., Rodriguez P., Renaudin H., Bebear C. 1996. A Tn1545-like transposon carries the tet(M) gene in tetracycline resistant strains of Bacteroides ureolyticus as well as Ureaplasma urealyticum but not Neisseria gonorrhoeae. J. Antimicrob. Chemother. 37: 223–232 [DOI] [PubMed] [Google Scholar]
- 12. De Francesco M. A., Negrini R., Pinsi G., Peroni L., Manca N. 2009. Detection of Ureaplasma biovars and PCR-based subtyping of Ureaplasma parvum in women with or without symptoms of genital infections. Eur. J. Clin. Microbiol. Infect. Dis. 28: 641–646 [DOI] [PubMed] [Google Scholar]
- 13. Deguchi T., et al. 2004. Association of Ureaplasma urealyticum (biovar 2) with nongonococcal urethritis. Sex. Transm. Dis. 31: 192–195 [DOI] [PubMed] [Google Scholar]
- 14. Dworkin M. S., Park L., Borchardt S. M. 2007. The changing epidemiology of invasive Haemophilus influenzae disease, especially in persons > or=65 years old. Clin. Infect. Dis. 44: 810–816 [DOI] [PubMed] [Google Scholar]
- 15. Echahidi F., Muyldermans G., Lauwers S., Naessens A. 2001. Development of an enzyme-linked immunosorbent assay for serotyping Ureaplasma urealyticum strains using monoclonal antibodies. Clin. Diagn. Lab. Immunol. 8: 52–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Garcia-Castillo M., et al. 2008. Differences in biofilm development and antibiotic susceptibility among clinical Ureaplasma urealyticum and Ureaplasma parvum isolates. J. Antimicrob. Chemother. 62: 1027–1030 [DOI] [PubMed] [Google Scholar]
- 17. Horowitz S. A., et al. 1986. Can group- and serovar-specific proteins be detected in Ureaplasma urealyticum? Pediatr. Infect. Dis. 5: S325–S331 [DOI] [PubMed] [Google Scholar]
- 18. Katz B., et al. 2005. Characterization of ureaplasmas isolated from preterm infants with and without bronchopulmonary dysplasia. J. Clin. Microbiol. 43: 4852–4854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Knox C. L., et al. 2003. Ureaplasma parvum and Ureaplasma urealyticum are detected in semen after washing before assisted reproductive technology procedures. Fertil. Steril. 80: 921–929 [DOI] [PubMed] [Google Scholar]
- 20. Knox C. L., Giffard P., Timms P. 1998. The phylogeny of Ureaplasma urealyticum based on the mba gene fragment. Int. J. Syst. Bacteriol. 4: 1323–1331 [DOI] [PubMed] [Google Scholar]
- 21. Kong F., Ma Z., James G., Gordon S., Gilbert G. L. 2000. Molecular genotyping of human Ureaplasma species based on multiple-banded antigen (MBA) gene sequences. Int. J. Syst. Evol. Microbiol. 5: 1921–1929 [DOI] [PubMed] [Google Scholar]
- 22. Kong F., Ma Z., James G., Gordon S., Gilbert G. L. 2000. Species identification and subtyping of Ureaplasma parvum and Ureaplasma urealyticum using PCR-based assays. J. Clin. Microbiol. 38: 1175–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kong F., et al. 1999. Comparative analysis and serovar-specific identification of multiple-banded antigen genes of Ureaplasma urealyticum biovar 1. J. Clin. Microbiol. 37: 538–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lin J. S., Kass E. H. 1973. Serotypic heterogeneity in isolates of human genital T-mycoplasmas. Infect. Immun. 7: 499–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lu C., et al. 2010. Phenotypic and genetic characteristics of macrolide- and lincosamide-resistant Ureaplasma urealyticum isolated in Guangzhou, China. Curr. Microbiol. 61: 44–49 [DOI] [PubMed] [Google Scholar]
- 26. Maeda S., et al. 2004. Detection of Mycoplasma genitalium, Mycoplasma hominis, Ureaplasma parvum (biovar 1), and Ureaplasma urealyticum (biovar 2) in patients with non-gonococcal urethritis using PCR-microtiter plate hybridization. Int. J. Urol. 11: 750–754 [DOI] [PubMed] [Google Scholar]
- 27. Mallard K., Schopfer K., Bodmer T. 2005. Development of real-time PCR for the differential detection and quantification of Ureaplasma urealyticum and Ureaplasma parvum. J. Microbiol. Methods 60: 13–19 [DOI] [PubMed] [Google Scholar]
- 28. Molin S., Tolker-Nielsen T. 2003. Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilization of the biofilm structure. Curr. Opin. Biotechnol. 14: 255–261 [DOI] [PubMed] [Google Scholar]
- 29. Naessens A., Foulon W., Breynaert J., Lauwers S. 1988. Serotypes of Ureaplasma urealyticum isolated from normal pregnant women and patients with pregnancy complications. J. Clin. Microbiol. 26: 319–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ondondo R. O., Whittington W. L., Astete S. G., Totten P. A. Differential association of ureaplasma species with non-gonococcal urethritis in heterosexual men. Sex Transm. Infect. 86: 271–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pereyre S., et al. 2009. Life on arginine for Mycoplasma hominis: clues from its minimal genome and comparison with other human urogenital mycoplasmas. PLoS Genet. 5: e1000677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Piot P. 1977. Comparison of growth inhibition and immunofluorescence tests in serotyping clinical isolates of Ureaplasma urealyticum. Br. J. Vener. Dis. 53: 186–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Piot P. 1976. Distribution of eight serotypes of Ureaplasma urealyticum in cases of non-gonococcal urethritis and of gonorrhoea and in healthy persons. Br. J. Vener. Dis. 52: 266–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Povlsen K., Bjornelius E., Lidbrink P., Lind I. 2002. Relationship of Ureaplasma urealyticum biovar 2 to nongonococcal urethritis. Eur. J. Clin. Microbiol. Infect. Dis. 21: 97–101 [DOI] [PubMed] [Google Scholar]
- 35. Povlsen K., Jensen J. S., Lind I. 1998. Detection of Ureaplasma urealyticum by PCR and biovar determination by liquid hybridization. J. Clin. Microbiol. 36: 3211–3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Povlsen K., Thorsen P., Lind I. 2001. Relationship of Ureaplasma urealyticum biovars to the presence or absence of bacterial vaginosis in pregnant women and to the time of delivery. Eur. J. Clin. Microbiol. Infect. Dis. 20: 65–67 [DOI] [PubMed] [Google Scholar]
- 37. Quinn P. A., Arshoff L. U., Li H. C. 1981. Serotyping of Ureaplasma urealyticum by immunoperoxidase assay. J. Clin. Microbiol. 13: 670–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Reyes L., Reinhard M., Brown M. B. 2009. Different inflammatory responses are associated with Ureaplasma parvum-induced UTI and urolith formation. BMC Infect. Dis. 9: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Roberts M. C., Kenny G. E. 1986. TetM tetracycline-resistant determinants in Ureaplasma urealyticum. Pediatr. Infect. Dis. 5: S338–S340 [DOI] [PubMed] [Google Scholar]
- 40. Robertson J. A., Stemke G. W. 1982. Expanded serotyping scheme for Ureaplasma urealyticum strains isolated from humans. J. Clin. Microbiol. 15: 873–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Robertson J. A., Stemke G. W. 1979. Modified metabolic inhibition test for serotyping strains of Ureaplasma urealyticum (T-strain Mycoplasma). J. Clin. Microbiol. 9: 673–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Robertson J. A., et al. 2002. Proposal of Ureaplasma parvum sp. nov. and emended description of Ureaplasma urealyticum (Shepard et al. 1974). Int. J. Syst. Evol. Microbiol. 52: 587–597 [DOI] [PubMed] [Google Scholar]
- 43. Robertson J. A., Vekris A., Bebear C., Stemke G. W. 1993. PCR using 16S rRNA gene sequences distinguishes the two biovars of Ureaplasma urealyticum. J. Clin. Microbiol. 31: 824–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rosendal S., Black F. T. 1972. Direct and indirect immunofluorescence of unfixed and fixed Mycoplasma colonies. Acta Pathol. Microbiol. Scand. B Microbiol. Immunol. 80: 615–622 [DOI] [PubMed] [Google Scholar]
- 45. Shepard M. C. 1954. The recovery of pleuropneumonia-like organisms from Negro men with and without nongonococcal urethritis. Am. J. Syph. Gonor. Vener. Dis. 38: 113–124 [PubMed] [Google Scholar]
- 46. Shepard M. C., Lunceford C. D. 1978. Serological typing of Ureaplasma urealyticum isolates from urethritis patients by an agar growth inhibition method. J. Clin. Microbiol. 8: 566–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sirand-Pugnet P., et al. 2007. Being pathogenic, plastic, and sexual while living with a nearly minimal bacterial genome. PLoS Genet. 3: e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Smets B. F., Barkay T. 2005. Horizontal gene transfer: perspectives at a crossroads of scientific disciplines. Nat. Rev. Microbiol. 3: 675–678 [DOI] [PubMed] [Google Scholar]
- 49. Stemke G. W., Robertson J. A. 1981. Modified colony indirect epifluorescence test for serotyping Ureaplasma urealyticum and an adaptation to detect common antigenic specificity. J. Clin. Microbiol. 14: 582–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stemke G. W., Robertson J. A. 1985. Problems associated with serotyping strains of Ureaplasma urealyticum. Diagn. Microbiol. Infect. Dis. 3: 311–320 [DOI] [PubMed] [Google Scholar]
- 51. Teachman A. M., French C. T., Yu H., Simmons W. L., Dybvig K. 2002. Gene transfer in Mycoplasma pulmonis. J. Bacteriol. 184: 947–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Waites K. B., Katz B., Schelonka R. L. 2005. Mycoplasmas and ureaplasmas as neonatal pathogens. Clin. Microbiol. Rev. 18: 757–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Watson H. L., Blalock D. K., Cassell G. H. 1990. Variable antigens of Ureaplasma urealyticum containing both serovar-specific and serovar-cross-reactive epitopes. Infect. Immun. 58: 3679–3688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Woese C. R., Maniloff J., Zablen L. B. 1980. Phylogenetic analysis of the mycoplasmas. Proc. Natl. Acad. Sci. U. S. A. 77: 494–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Xiao L., et al. 2010. Detection and characterization of human Ureaplasma species and serovars by real-time PCR. J. Clin. Microbiol. 48: 2715–2723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yi J., Yoon B. H., Kim E. C. 2005. Detection and biovar discrimination of Ureaplasma urealyticum by real-time PCR. Mol. Cell Probes 19: 255–260 [DOI] [PubMed] [Google Scholar]
- 57. Yoshida T., et al. 2007. Quantitative detection of Ureaplasma parvum (biovar 1) and Ureaplasma urealyticum (biovar 2) in urine specimens from men with and without urethritis by real-time PCR. Sex. Transm. Dis. 34: 416–419 [DOI] [PubMed] [Google Scholar]
- 58. Yoshida T., et al. 2005. PCR-based subtyping of Ureaplasma parvum and Ureaplasma urealyticum in first-pass urine samples from men with or without urethritis. Sex. Transm. Dis. 32: 454–457 [DOI] [PubMed] [Google Scholar]
- 59. Zheng X., Watson H. L., Waites K. B., Cassell G. H. 1992. Serotype diversity and antigen variation among invasive isolates of Ureaplasma urealyticum from neonates. Infect. Immun. 60: 3472–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zimmerman C. U., Rosengarten R., Spergser J. 2011. Ureaplasma antigenic variation beyond MBA phase variation: DNA inversions generating chimeric structures and switching in expression of the MBA N-terminal paralogue UU172. Mol. Microbiol. 79: 663–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zimmerman C. U., Stiedl T., Rosengarten R., Spergser J. 2009. Alternate phase variation in expression of two major surface membrane proteins (MBA and UU376) of Ureaplasma parvum serovar 3. FEMS Microbiol. Lett. 292: 187–193 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.