Abstract
HIV superinfection, which occurs when a previously infected individual acquires a new distinct HIV strain, has been described in a number of populations. Previous methods to detect superinfection have involved a combination of labor-intensive assays with various rates of success. We designed and tested a next-generation sequencing (NGS) protocol to identify HIV superinfection by targeting two regions of the HIV viral genome, p24 and gp41. The method was validated by mixing control samples infected with HIV subtype A or D at different ratios to determine the inter- and intrasubtype sensitivity by NGS. This amplicon-based NGS protocol was able to consistently identify distinct intersubtype strains at ratios of 1% and intrasubtype variants at ratios of 5%. By using stored samples from the Rakai Community Cohort Study (RCCS) in Uganda, 11 individuals who were HIV seroconcordant but virally unlinked from their spouses were then tested by this method to detect superinfection between 2002 and 2005. Two female cases of HIV intersubtype superinfection (18.2%) were identified. These results are consistent with other African studies and support the hypothesis that HIV superinfection occurs at a relatively high rate. Our results indicate that NGS can be used for detection of HIV superinfection within large cohorts, which could assist in determining the incidence and the epidemiologic, virologic, and immunological correlates of this phenomenon.
INTRODUCTION
HIV superinfection occurs when a known HIV-infected individual is subsequently infected with a new phylogenetically distinct viral strain or strains. The first documented cases of HIV superinfection were found in individuals with various modes of transmission and included inter- and intrasubtype cases (1, 9, 17). Subsequently, multiple studies have documented superinfection in small populations of high-risk individuals (2, 3, 7, 8, 11, 13, 17, 20, 22, 23, 26, 29). The rate of HIV superinfection in these high-risk groups was relatively frequent and was comparable to the incidence rate in similar populations from the same regions, especially if multiple viral genes were examined (3, 13, 14, 21, 24). In contrast, other researchers have found no evidence of superinfection in large-scale population studies (6, 15). One possible reason for this discrepancy may be due to differences in techniques and criteria used to identify superinfection (16). Initial studies designed to examine the frequency of superinfection utilized heteroduplex mobility assays (HMAs) or multiregion hybridization assays (MHAs) followed by selective clonal analysis of those samples that demonstrated the presence of new viral variants (3, 11, 15). MHA screening is limited in that it can only identify intersubtype superinfection, while possibly missing intrasubtype superinfection. Although HMA is sensitive enough to detect samples with >1.5% differences in pairwise distance, it is susceptible to false positives due to the presence of insertions or deletions (16). Additionally, both the HMA and MHA methods require verification using in-depth cloning and Sanger sequencing (13, 16). The sensitivity of these screening/cloning techniques is dependent on the number of clones amplified and the number of genes examined (12–14). To detect a minor variant approaching 1%, over 100 clones would need to be examined per sample, preferably from multiple PCRs to increase the amount and diversity of viral strains sequenced (14, 16). With the need to examine multiple regions of the viral genome to ensure accurate phylotyping and identification of superinfecting strains, in-depth cloning and Sanger sequencing are prohibitively labor-intensive for large-scale studies (12, 14, 16).
Newly developed next-generation sequencing (NGS) techniques provide unprecedented sequencing depth, offer the ability to multiplex samples, and are quicker, more cost-effective, and less labor-intensive than cloning and Sanger-based sequencing (12). Using several genomic targets and high sequence volume, NGS should be able to distinguish minor variants that arose spontaneously either through recombination, within-host viral evolution, or from newly introduced strains or subtypes (18, 30).
We designed and tested an NGS protocol and sequence analysis pipeline that focuses on amplification and sequencing of the p24 region of the viral capsid and the gp41 region of the viral envelope. These genomic regions were chosen for examination because they are relatively genetically stable and of sufficient length, are suitable for phylotyping, were previously used in PCR cloning and Sanger sequencing studies, and are not high in polymeric regions. We further tested the protocol with 11 individuals from virally unlinked HIV seroconcordant couples from Rakai District, Uganda, to detect the occurrence of HIV superinfection.
MATERIALS AND METHODS
Ethics statement.
All subjects provided written informed consent for their samples to be stored and used for future unspecified HIV-related research. The study was approved by the Science and Ethics Committee of the Uganda Virus Research Institute, the Western Institutional Review Board, and the Committee on Human Research at the Johns Hopkins Bloomberg School of Public Health.
Study population and subjects.
Serum samples were retrospectively selected from individuals in the Rakai Community Cohort Study (RCCS), a rural, community-based open cohort consisting of persons aged 15 to 49 years in Rakai District, southwestern Uganda (27). Since 1994, interviews and venous blood samples have been obtained annually from approximately 14,000 consenting adults living in 50 villages. As part of the routine interview, consenting individuals in stable sexual partnerships are linked as couples.
Control serum samples were selected from HIV-infected individuals who were previously identified as being infected with either subtype A (n = 4) or D (n = 6) in the 2002 community survey. Identification of subtypes was performed by Sanger sequencing of cloned PCR products of the p24 and gp41 target regions.
Using stored sera from 2002, we identified 18 HIV-infected individuals in 9 HIV-seroconcordant couples whose viruses were phylogenetically unlinked to their partner's virus, as determined by previous Sanger sequencing for either the gp41 or p24 regions (4). The individual's samples were labeled with their gender, couple number, and year of sample draw (e.g.,. female_1_C1_2002). Of the 18 individuals, 11 had serum samples available in 2005, and these were examined for HIV superinfection in this population. Four of the 11 individuals were from two couples (couples 1 and 2) of which both members had serum samples available from 2002 and 2005; however, for this analysis, each individual was analyzed independently. The remaining seven individuals only had serum samples available in 2002 but were included in this study to search for the source of any new superinfecting HIV strains found in their partner's 2005 samples.
Viral RNA extraction, cDNA synthesis, and PCR target amplification.
Viral RNA was extracted from 140 μl of serum using a QIAmp viral RNA minikit (Qiagen, Valencia CA) and eluted into 50 μl of Qiagen buffer AVE. For each genomic target region (p24 and gp41), two 50-μl reverse transcription-PCRs (RT-PCRs) were performed simultaneously to maximize the amount and diversity of viral RNA genomes amplified per sample. For the gp41 region, each 50-μl RT-PCR was performed using a 40-μl master mix composed of 20 μl of double-distilled water (ddH2O), 10 μl of 5× buffer, 3 μl of deoxynucleoside triphosphates (dNTPs), and 2 μl of enzyme mixture from the Qiagen OneStep RT-PCR kit. One microliter of RNase inhibitor was also added, along with 2 μl of 20 μM dilutions of both the forward primer (GP50F1-HXB2 nt 7691→7720) and the reverse primer (GP41R1-HXB2 nt 8347←8374) (see the supplemental material). This master mix was combined with 10 μl of purified viral RNA and incubated for 30 min at 50°C and 15 min at 94°C for RT extension. PCR was then performed for 35 cycles of 30 s at 94°C, 35 s at 53.5°C, and 90 s at 72°C, followed by 72°C for 10 min. For the p24 region, the 50-μl RT and PCRs were carried out using the same master mix as described above, with one exception: forward and reverse primers specific for the p24 target were used and designated G00 (HXB2 nt 764→782) and G01 (HXB2 nt 2264←2281), respectively (see the supplemental material). For two samples that did not amplify the p24 region during the initial PCR, a reformulated 40-μl master mix containing 20 μl of ddH2O, 10 μl of 5× buffer, 3 μl of dNTPs, and 2 μl of enzyme mix from the Qiagen OneStep RT-PCR kit, as well as 2 μl of MgCl2, 1 μl of RNase inhibitor, and 1.5 μl of 20 μM dilutions of both the forward primer and reverse primers, was used. The two samples were pooled to maximize the depth of detection, and 10 μl of this pool was used in a nested 100-μl PCR using primer sets for gp41 (E55 primer set with 14 454-bar-coded variations [MID1 to MID14]) or p24 (G100 primer set with 14 454-bar-coded variations [MID1 to MID14]) (Roche, Inc., Branford, CT) (see the supplemental material). Briefly, each nested-PCR mixture for gp41 or p24 contained 90 μl of master mix composed of 50.4 μl ddH2O, 10 μl 10× reaction buffer, 20 μl MgCl2, 3 μl dNTPs, and 0.6 μl HotStarTaq DNA polymerase (Qiagen, Valencia, CA) as well as 3 μl of the forward and reverse E55 primers or 3 μl of the forward and reverse G100 primer set both at a 20 μM final concentration for both regions (see the supplemental material). The PCR amplification conditions for the 100-μl nested reactions were identical to the first-round PCR conditions described above. Successful single-band amplification of gp41 or p24 target products was verified by agarose gel electrophoresis.
Serum HIV-1 RNA concentrations (viral loads) were determined by the Amplicor v1.5 (Roche Diagnostics, Basel, Switzerland).
Generation of control samples for inter- and intrasubtype NGS threshold detection.
Control serum samples from HIV-infected individuals, previously identified via Sanger sequencing of PCR fragments as being infected with either subtype A (n = 4) or D (n = 6), were used to determine the assay's limit of detection. Phylogenetically unlinked viral isolates were mixed in inter- and intrasubtype experiments for each viral target region.
For the p24 region, viral extracts from two HIV subtype A-infected control individuals (A1 and A2) and four subtype D-infected individuals (D1 to D4) were amplified separately in the first-round PCR. Aliquots were collected and set aside for pure sample analysis, while aliquots of each control sample were also mixed at a variety of ratios. The following ratios were tested for the p24 target region: 50:50 A2-D1, 95:5 A2-D1, 99:1 A2-D1, 99.9:0.1 D3-A2, 95:5 A1-A2, 95:5 D1-D2, and 95:5 D3-D4. Nested PCRs were performed with these samples as described above.
For the gp41 region, viral extracts from two HIV subtype A-infected individuals (A3 and A4) and two subtype D-infected control individuals (D5 and D6) were amplified separately in the first-round PCR. Aliquots of the first-round PCR were collected and set aside for pure sample analysis, while aliquots of each were mixed at a variety of ratios. The following ratios were tested for the gp41 target region: 50:50 A4-D5, 95:5 A4-D5, 99:1 A4-D5, 99.9:0.1 A4-D5, 95:5 A1-A2, and 95:5 D5-D6. Nested PCRs were performed with these samples as described above.
PCR product purification.
The amplicon library preparation method was performed as recommended by the manufacturer (Roche, Branford, CT), and all PCR products were purified with the following minor alterations. In an effort to eliminate excess primers, the bead/target ratio was reduced by incubation of 30 μl of AMPure XP beads (Agencourt, Beckman Coulter Genomics, Danvers, MA) with 25 μl of PCR product diluted in 25 μl of water. Purified PCR products were quantified using PicoGreen (Invitrogen, Carlsbad, CA), and each template was diluted to 1 × 109 molecules/μl stock. The amplicon pools were made by combining 5 μl of each diluted barcoded template to make a final 1 × 109 molecules/μl stock containing 14 bar-coded amplicons.
DNA sequencing.
Preparation of templated beads for NGS followed the emPCR Method Manual—Lib-L-MV (17a). The library pools containing 1 × 109 molecules/μl were diluted to 1 × 105 molecules/μl for a target addition of 0.175 copies per bead to the DNA capture beads. The live amplification mixture was based on the reagent volumes for paired-end libraries to reduce the amount of amplification primer in the reactions and thereby reduce the bead signal intensity during sequencing. Enriched DNA capture beads were sequenced on the Roche 454 system (Roche, Branford, CT) per the manufacturer's instructions, using a four-region gasket when indicated.
Sequence segregation.
Sequencing results were analyzed using the GS Amplicon variant analyzer, version 2.5 (Roche, Branford, CT). All sequence reads were compared, and similar sequences were combined into a single consensus sequence. Generated consensus sequences that were within 10 bases from both ends of the amplicon and comprised of a cluster of 10 individual, nearly identical sequences or more were determined using the Roche Amplicon software and were classified as being consensus sequences of HIV variants. These consensus sequences were used for subsequent phylogenetic analysis.
Phylogenetic analysis.
Consensus sequences, subtype reference sequences, and a selection of subtype reference sequences collected from Rakai (see the supplemental material) were aligned using ClustalW (25). Phylogenetic trees were generated by the neighbor-joining method (19). Statistical support for a specific clade in each phylogeny was obtained by bootstrapping (1,000 replicates). The NGS consensus sequences for gp41 and p24 have been submitted to GenBank (see below) and are also available upon request (aredd2@jhmi.edu).
HIV superinfection definition and analysis.
HIV superinfection was defined in an individual whose 2005 serum sample demonstrated two or more distinct consensus sequences forming a monophyletic cluster that was phylogenetically unlinked from the individual's entire consensus sequences in the 2002 sample. In order to be considered a superinfection, the genetic distance of the new monophyletic cluster from the closest related viral sequences found at the earlier time point had to be either ≥0.55% per year for the p24 region, ≥0.98% per year for the gp41 region for subtype D and ≥0.59% per year for the p24 region, or ≥0.72% per year for the gp41 region for subtype D, which is equal to the mean plus twice the standard deviation of the intraperson viral divergence or evolutionary rate of each HIV-1 subtype in Rakai, Uganda (data not shown). All newly identified consensus sequences were phylogenetically compared to the most prominent strains of the other bar-coded samples within NGS runs to search for microcontamination, misclassification, or sequencing errors. If instances of these errors were found, these consensus sequences were eliminated. For further verification, newly identified superinfecting viral strain sequences were translated and analyzed in order to check that a functional protein sequence was encoded in the sequence. Newly discovered superinfecting consensus sequences within an individual were compared phylogenetically to their partner's consensus viral sequences in order to determine if the partner was the source of the new superinfecting virus.
Nucleotide sequence accession numbers.
The nucleotide consensus sequences for the gp41 region have been deposited in GenBank under accession no. JN153104 to JN155099, and the nucleotide consensus sequences for the p24 region have been deposited in GenBank under accession no. JN155100 to JN157600. The sequences are also available on request.
RESULTS
Genomic target regions, sequencing depth, and consensus sequence analysis.
The p24 and gp41 regions of the viral genome were chosen for NGS because they are located at opposing ends of the HIV genome and are two of the more conserved areas of the genome. Previous research has indicated that the sensitivity of NGS for HIV quasispecies detection is 0.1% (30). Therefore, estimating an approximate read volume of 10,000 reads per sample, a cutoff of 10 similar reads, as determined by the Roche segregation software, was selected to qualify as a consensus sequence for further analysis. A cutoff of five sequences was also examined and found to not affect the findings and the overall sensitivity of the assay (12). However, when the consensus cutoff was dropped to two similar sequences, small amounts of microcontaminating sequences reflecting the inherent error rate for the technology were discovered. Therefore, for the purposes of this study, 10 reads or more was the threshold for quality consensus viral sequences (see Fig. S1 in the supplemental material).
p24 inter- and intrasubtype analysis.
Previous Sanger sequencing of PCR fragments of the p24 region identified two subtype A (A1and A2) and four subtype D (D1, D2, D3, and D4) samples used in this analysis (Table 1) (5). In order to test the intra- and intersubtype viral population sensitivities of our NGS protocol, first-round PCR products targeting the p24 region from these subtype A and subtype D samples were mixed in various ratios, amplified, and sequenced on the Roche 454 system as described above (Table 1 and Fig. 1 and 2A to D; see Fig. S2A to C in the supplemental material). In order to exclude cross contamination or poor-quality reads, consensus read data sets for all mixtures were merged, and the resulting trees were constructed (Fig. 1D). These data demonstrate that reads specific for the mixed-ratio samples are segregating properly to their respective branch locations for the components of the mixture and that the NGS protocol provides good depth and quality sequence sorting during phylogenetic analysis (Fig. 1). The ratios of A2 to D1 of 95:5 and 99:1 were examined to determine if NGS would provide adequate depth and representation of the subtypes at these ratios (Fig. 2A and B). The lower frequency of the minor variant (D1 in both cases) was adequately represented in both trees, although with a slight decrease in the number of consensus reads in the 99:1 ratio (Fig. 2B).
Table 1.
Sequence read totals and consensus distribution for pure subtype samples and mixture analysis
| Sample | Region | Total no. of reads | No. of consensus sequences (≥10) | No. of plate divisions | Minor variant/no. of consensus sequences detected (≥10 reads) | % minor variant |
|---|---|---|---|---|---|---|
| A1 | p24 | 20,808 | 162 | 1 | NAa | NA |
| A2 | p24 | 21,222 | 104 | 1 | NA | NA |
| D1 | p24 | 19,596 | 163 | 1 | NA | NA |
| D2 | p24 | 17,446 | 139 | 1 | NA | NA |
| D3 | p24 | 13,137 | 73 | 4 | NA | NA |
| D4 | p24 | 11,442 | 81 | 4 | NA | NA |
| A2/D1 ratio, 50:50 | p24 | 18,702 | 146 | 1 | D1/115 | 78.8 |
| A2/D1 ratio, 95:5 | p24 | 18,694 | 133 | 1 | D1/86 | 64.7 |
| A2/D1 ratio, 99:1 | p24 | 19,216 | 117 | 1 | D1/63 | 53.8 |
| D3/A2 ratio, 99.9:0.1 | p24 | 11,299 | 70 | 4 | A2/0 | 0 |
| A1/A2 ratio, 95:5 | p24 | 18,855 | 138 | 1 | A2/20 | 14.5 |
| D1/D2 ratio, 95:5 | p24 | 17,783 | 167 | 1 | D2/0 | 0 |
| D3/D4 ratio, 95:5 | p24 | 7,461 | 44 | 4 | D4/11 | 25 |
| A3 | gp41 | 9,328 | 55 | 4 | NA | NA |
| A4 | gp41 | 7,678 | 58 | 4 | NA | NA |
| D5 | gp41 | 8,461 | 40 | 4 | NA | NA |
| D6 | gp41 | 6,310 | 47 | 4 | NA | NA |
| A4/D5 ratio, 50:50 | gp41 | 7,229 | 58 | 4 | D5/19 | 32.8 |
| A4/D5 ratio, 95:5 | gp41 | 7,561 | 51 | 4 | D5/4 | 7.8 |
| A4/D5 ratio, 99:1 | gp41 | 7,990 | 64 | 4 | D5/4 | 6.3 |
| A4/D5 ratio, 99.9:0.1 | gp41 | 7,137 | 54 | 4 | D5/1 | 1.9 |
| A3/A4 ratio, 95:5 | gp41 | 9,531 | 59 | 4 | A4/14 | 23.7 |
| D5/D6 ratio, 95:5 | gp41 | 7,532 | 59 | 4 | D6/2 | 3.4 |
NA, not applicable.
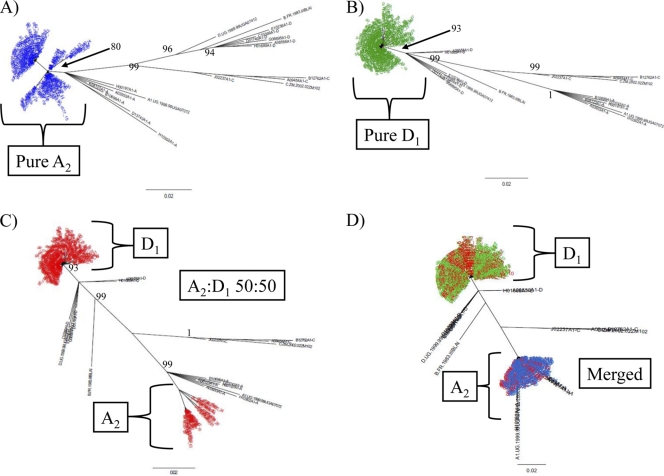
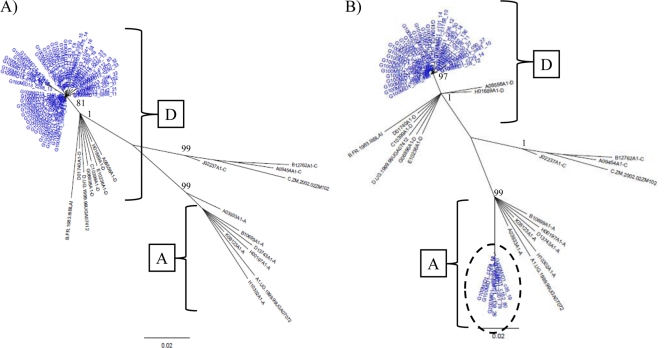
Fig. 1.
Mixture analysis of intersubtype detection by NGS. Neighbor-joining trees of p24 next-generation consensus sequences (≥10 identical reads) of control samples of A2 (A; blue), subtypes D1 (B; green), a mixture of A2 and D1 at a 50:50 ratio (C; red), and a merged tree of all three sample runs (D) are shown. The trees were constructed with a selection of subtype reference sequences and random sequences from individuals in Rakai shown in black. Brackets demonstrate the source of different clades within the merged trees. Bootstrap values higher than 80% are shown for nonmerged trees (1,000 replicates).
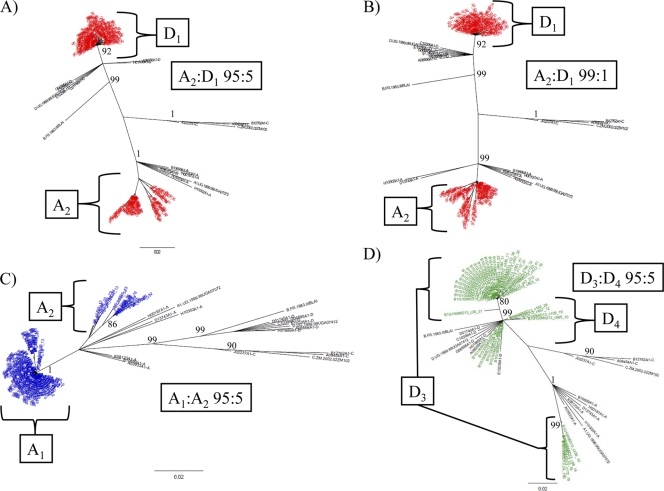
Fig. 2.
Inter- and intrasubtype detection of the p24 region by NGS. Neighbor-joining trees of p24 next-generation consensus sequences (≥10 identical reads) of intersubtype mixtures of A2 and D1 (red) at 95:5 (A) and 99:1 (B) ratios are shown. Intrasubtype mixtures of A2 and A1 (C; blue) and D3 and D4 (D; green) at the ratio of 95:5 are shown. The trees were constructed with a selection of subtype reference sequences and random sequences from individuals in Rakai shown in black. Brackets demonstrate the source of different clades within the merged trees. Bootstrap values higher than 80% are shown (1,000 replicates).
To further test the sensitivity of this assay, we analyzed a mixture of D3 to A2 at a ratio of 99.9:0.1. When we merged these ratio data with the control data sets (D3 and A2), the minor variant (A2) did not appear in the data (see Fig. S2C in the supplemental material). These results suggest that for the p24 target, an intersubtype ratio of ≤0.1% cannot be reliably identified by this NGS protocol.
In order to test the protocol for its ability to adequately sequence and separate related subtypes, the following ratios were tested: 95:5 A1-A2, 95:5 D1-D2, and 95:5 D3-D4 (Table 1 and Fig. 2C and D; see Fig. S2A and B in the supplemental material). The minor viral variant population in the 95:5 A1/A2 ratio (A2) was identified as 14.5% of the total number of consensus sequences (Table 1 and Fig. 2C). The 95:5 D1/D2 ratio sample did not appear to adequately amplify the minor variant (D2) when the data were merged with the data sets for D1 and D2 (Table 1; see Fig. S2B in the supplemental material). This suggests a lower limit for D1- versus D2-related intrasubtype identification for the p24 target. To determine if this lack of detection or amplification of D2 was unique to the D1/D2 ratio of 95:5, this test was repeated using the ratio of 95:5 D3-D4. In this test, the minor variant (D4) was identified in 25% of the total number of consensus sequences (Table 1 and Fig. 2D). It was found that the consensus sequences that were expanded from the minor variant (D3) corresponded to the most prominent subtype sequences present in the pure sample for D3 (see Fig. S2A in the supplemental material).
gp41 inter- and intrasubtype analysis.
Due to limited amounts of viral RNA available for samples A1, A2, and D1 to D4, different control samples were used to test the minor intra- and intersubtype viral population sensitivities of our NGS protocol of the gp41 region (A3, A4, D5, and D6,) (Table 1 and Fig. 3). The majority of the p24 NGS reactions were performed on a full 454 slide with 14 different bar-coded samples, whereas the gp41 test samples were run on a slide that had been divided into four quadrants. The reason for this change was to increase the sample throughput per run, resulting in a lower read volume per bar-coded sample (Table 1).
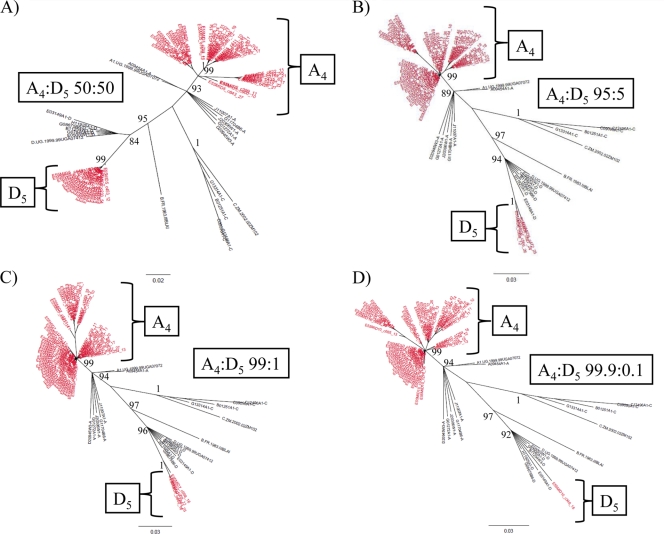
Fig. 3.
Intersubtype detection of the gp41 region by NGS. Neighbor-joining trees of gp41 next-generation consensus sequences (≥10 identical reads) of intersubtype mixtures of A4 and D5 (red) at 50:50 (A), 95:5 (B), 99:1 (C), and 99.9:0.1 (D) ratios are shown. The trees were constructed with a selection of subtype reference sequences and random sequences from individuals in Rakai shown in black. Brackets demonstrate the source of different clades within the merged trees. Bootstrap values higher than 80% are shown (1,000 replicates).
NGS analysis of all four intersubtype mixtures (A versus D) for the gp41 region demonstrated detectable consensus sequences of the minor variant (Table 1 and Fig. 3A to D). However, in the case of the 99.9:0.1 mixture, only one consensus sequence from the minority variant subtype was amplified (Table 1 and Fig. 3D). While the sensitivity for minor viral variants was increased for gp41 relative to the results for p24, the lack of two or more distinct consensus sequences means that this would not qualify as a superinfecting viral species according to the parameters described above.
NGS analysis of the two intrasubtype comparisons (A3 versus A4,or D5 versus D6) at a 95:5 ratio demonstrated that in a merged data format, the minor variants (A4 and D5) were detected (Table 1; see Fig. S3A and B in the supplemental material). These data also demonstrated that the A3 individual, who previously was identified by PCR cloning and Sanger sequencing analysis as being infected with only subtype A, was in fact infected with two distinct variants which coincided with both subtypes A and D (see Fig. S3A in the supplemental material).
HIV superinfection in Rakai, Uganda.
Eleven HIV-infected individuals from whom serum samples were collected at 2002 and 2005 were evaluated at both p24 and gp41for evidence of HIV superinfection (Table 2). In addition, for each individual, their partner's sample from 2002, or in the case of two couples (C_1 and C_2), the samples from 2002 and 2005, were amplified and sequenced by NGS to examine if superinfecting strains discovered in 2005 originated from their partner (Table 2). Serum HIV loads were calculated for each sample tested (Table 2). Each member was treated independently in this analysis.
Table 2.
Subject viral loads, sequence read totals, and consensus subtype distributiona
| Subject no. | Viral load (log10 viral copies/ml) | p24 |
gp41 |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Total no. of reads | No. of consensus sequences (≥10) | No. of consensus sequences by subtype: |
Total no. of reads | No. of consensus sequences (≥10) | No. of consensus sequences by subtype: |
||||||
| A | D | C | A | D | C | ||||||
| Female_C1_02 | 4.43 | 18,186 | 91 | 91 | 7,517 | 60 | 49 | 11 | |||
| Female_C1_05 | 5.31 | 17,874 | 153 | 8 | 145 | 6,812 | 56 | 47 | 9 | ||
| Male_C1_02 | 3.03 | 16,378 | 138 | 138 | 11,180 | 81 | 81 | ||||
| Male_C1_05 | 4.52 | 19,554 | 148 | 148 | 7,679 | 53 | 53 | ||||
| Female_C2_02 | 4.94 | 10,921 | 72 | 72 | 6,939 | 46 | 46 | ||||
| Female_C2_05 | 5.27 | 13,331 | 102 | 102 | 5,553 | 37 | 37 | ||||
| Male_C2_02 | 5.35 | 12,651 | 16 | 16 | 6,235 | 65 | 65 | ||||
| Male_C2_05 | 6.30 | 12,076 | 99 | 99 | 5,506 | 50 | 50 | ||||
| Female_C3_02 | 4.83 | 8,119 | 50 | 50 | 6,382 | 46 | 46 | ||||
| Female_C3_05 | 5.07 | 6,980 | 39 | 7 | 32 | 11,155 | 86 | 86 | |||
| Male_C3_02 | 5.30 | 11,779 | 93 | 93 | 8,406 | 75 | 71 | 4 | |||
| Female_C4_02 | 4.08 | 13,525 | 93 | 93 | 11,418 | 100 | 100 | ||||
| Female_C4_05 | 4.30 | 15,274 | 95 | 95 | 12,345 | 84 | 84 | ||||
| Male_C4_02 | 5.04 | 17,534 | 134 | 134 | 14,384 | 109 | 109 | ||||
| Male_C5_02 | 4.86 | 16,915 | 105 | 105 | 13,799 | 120 | 120 | ||||
| Male_C5_05 | 5.66 | 15,038 | 109 | 109 | 10,557 | 84 | 84 | ||||
| Female_C5_02 | <2.6 | 6,936 | 93 | 93 | |||||||
| Female_C6_02 | 4.10 | 20,767 | 110 | 110 | 14,437 | 90 | 90 | ||||
| Female_C6_05 | 3.80 | 17,158 | 93 | 93 | 11,959 | 101 | 101 | ||||
| Male_C6_02 | 5.69 | 19,648 | 89 | 89 | 9,450 | 75 | 20 | 55 | |||
| Female_C7_02 | 4.13 | 14,882 | 133 | 133 | 6,850 | 49 | 49 | ||||
| Female_C7_05 | 4.77 | 12,154 | 102 | 102 | 6,829 | 41 | 41 | ||||
| Male_C7_02 | <2.6 | 12,799 | 90 | 90 | 12,358 | 68 | 68 | ||||
| Male_C8_02 | 4.33 | 7,971 | 60 | 60 | 9,237 | 80 | 80 | ||||
| Male_C8_05 | 5.10 | 8,261 | 67 | 67 | 5,988 | 12 | 12 | ||||
| Female_C8_02 | 5.31 | 8,251 | 39 | 39 | 7,556 | 40 | 40 | ||||
| Male_C9_02 | 4.70 | 10,655 | 62 | 62 | 7,329 | 77 | 77 | ||||
| Male_C9_05 | 4.88 | 10,158 | 64 | 64 | 9,476 | 51 | 51 | ||||
| Female_C9_02 | 4.73 | 9,927 | 55 | 55 | 7,544 | 67 | 67 | ||||
Results for samples with superinfecting strains are shown in bold. Corresponding partner read totals and consensus sequence are indicated for the individual's samples, which are labeled with their gender, couple number, and year of sample draw.
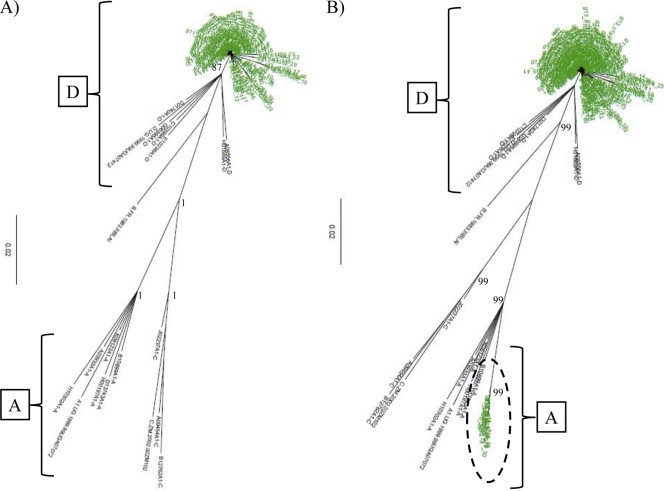
Using NGS, two of the 11 individuals (18.2%) had evidence of HIV superinfection in their 2005 sera (Table 2 and Fig. 4 and 5). The first case of superinfection was documented in female_C1, who was infected in 2002 with a viral population that grouped with subtype D in the p24 region and with subtypes D and C in the gp41 region (Table 2 and Fig. 4A; see Fig. S4 in the supplemental material). In 2005, she had multiple consensus sequences in the p24 target region which grouped with subtype A, indicating a superinfection of a new HIV species (Fig. 4B). NGS analysis of her male partner (male_C1) demonstrated that he was infected with an apparent D/C recombinant strain that was linked with his female partner's viral strains in both regions in 2002 and 2005 when examined in a merged phylogenetic tree (merged data not shown), indicating that she was superinfected by another source (Table 2).
Fig. 4.
Detection of HIV superinfection in the p24 region. Neighbor-joining trees of HIV p24 next-generation consensus sequences (≥10 identical reads) from female_C1 (green) in 2002 (A) and 2005 (B) are shown. The trees were constructed with a selection of subtype reference sequences and random sequences from individuals in Rakai shown in black. Superinfecting strains are shown with a circle. Brackets demonstrate the individual's HIV subtypes within the trees. Bootstrap values higher than 80% are shown (1,000 replicates).
Fig. 5.
Detection of HIV superinfection in the p24 region. Neighbor-joining trees of HIV p24 next-generation consensus sequences (≥10 identical reads) from female_C3 (blue) in 2002 (A) and 2005 (B) are shown. The trees were constructed with a selection of subtype reference sequences and random sequences from individuals in Rakai shown in black. Superinfecting strains are shown with a circle. Brackets demonstrate the individual's HIV subtypes within the trees. Bootstrap values higher than 80% are shown (1,000 replicates).
The second case of superinfection was observed in female_C3, who was initially infected with HIV subtype D in both genomic regions (Table 2 and Fig. 5A; see Fig. S5 in the supplemental material). In her 2005 sample, she had acquired a new viral strain in the p24 region with multiple consensus sequences that clustered with subtype A (Fig. 5B). Her partner, male_C3, was infected in 2002 with a dual population of viruses that clustered with subtypes D and C in the gp41 region and subtype D in the p24 region (Table 2). Merged phylogenetic tree analysis demonstrated that her superinfecting strain was not found in her partner, suggesting she was superinfected by another source (merged data not shown). No other cases of superinfection were observed in the remaining nine individuals during merged and unmerged phylogenetic tree analysis (Table 2).
DISCUSSION
Identification of HIV superinfection in the past has been accomplished using a variety of screening techniques in conjunction with labor-intensive cloning or single-genome amplification (3, 6, 11–13, 21). This has led to a significant amount of variability in the estimated rates of HIV superinfection (3, 6, 8, 21). The data presented here describe a new NGS protocol to identify HIV superinfection with relatively high inter- and intrasubtype sensitivities. The consensus of 10 repeated sequences was chosen since it was approximately 1/1,000 of the estimated total reads and appeared to be an appropriate cutoff to identify inter- and intrasubtype minor variants while avoiding data artifacts. Using mixtures of HIV-infected samples containing subtypes A and D, the predominant viral species found in Uganda, the assay's intersubtype sensitivity in both the p24 and gp41 target regions was determined to be at least 1%. Minor viral strains were found at lower levels (0.1%) in the gp41 region, but not consistently or at high enough consensus counts to lower the threshold of detection for the protocol. Intrasubtype sensitivity was approximately 5%, although intrasubtype detection within the subtype A mixtures seemed more robust than that for the subtype D samples. We hypothesize that primer specificity and target sequence variation may be driving some of these differences and is a limitation of our protocol.
The NGS protocol was able to identify two cases of HIV superinfection in women from 11 individuals who were members of virally unlinked concordantly infected couples. In both cases, the superinfecting strain was HIV subtype A, which has been shown to be more infectious than subtype D (10). In addition, both women's viral loads increased during the period. None of the superinfecting strains were detected in the women's male partners, suggesting that the superinfecting strain was acquired from another source. It is possible that the new strains found in these two individuals were present in the earlier time points at levels that were too low to be detected in our assay. However, according to the data from our mixture analysis, the levels in the first time point would most likely be less than 1%, and therefore we feel these events should be classified as superinfections. The relatively high proportion of superinfected individuals in our population agrees with other studies of high-risk individuals in Africa (13, 14). However, given the small number of individuals examined, further investigation is needed to estimate the rate and correlates of superinfection in the Rakai population. In addition, the individuals in this study were selected based upon a high likelihood of superinfection since they were initially virally unlinked from their partners and therefore may not represent the natural rate of superinfection in the larger HIV-infected population. NGS is substantially easier and more cost-effective than previous methods used to detect superinfection, particularly for screening large numbers of subjects (12, 28). It should be noted that NGS protocols like ours require specialized equipment that somewhat limits their utility in resource-poor settings. The data presented here demonstrate that HIV superinfection can be detected in an accurate and sensitive manner, in a high-throughput environment, and suggest that future studies examining HIV superinfection rates in large cohorts should utilize these types of deep sequencing techniques. The ability to rapidly determine the nature and extent of HIV superinfection could have a profound influence on studies of HIV disease, therapeutic interventions, transmission of potential drug resistance, and viral evolution in the population.
Supplementary Material
ACKNOWLEDGMENTS
We thank all the participants of the Rakai cohort, and the staff of the Rakai Health Science Program. We especially thank Susanna Lamers for assistance with sequence submission.
All subjects provided written informed consent for their samples to be stored and used for future HIV-related research. The study was approved by the Science and Ethics Committee of the Uganda Virus Research Institute, the Western Institutional Review Board, and the Committee on Human Research at Johns Hopkins Bloomberg School of Public Health. There are no conflicts of interests for any of the study authors. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
This study was supported in part by funding from the Division of Intramural Research, NIAID, NIH, NIAID grants R01 A134826 and R01 A134265, NICHD grant 5P30HD06826, the World Bank STI Project, Uganda, the Henry M. Jackson Foundation, the Fogarty Foundation (grant 5D43TW00010), and the Bill and Melinda Gates Institute for Population and Reproductive Health at JHU.
Footnotes
Supplemental material for this article may be found at http://jcm.asm.org/.
Published ahead of print on 22 June 2011.
REFERENCES
- 1. Altfeld M., et al. 2002. HIV-1 superinfection despite broad CD8+ T-cell responses containing replication of the primary virus. Nature 420:434–439 [DOI] [PubMed] [Google Scholar]
- 2. Braibant M., et al. 2010. Disease progression due to dual infection in an HLA-B57-positive asymptomatic long-term nonprogressor infected with a nef-defective HIV-1 strain. Virology 405:81–92 [DOI] [PubMed] [Google Scholar]
- 3. Chohan B., Lavreys L., Rainwater S. M., Overbaugh J. 2005. Evidence for frequent reinfection with human immunodeficiency virus type 1 of a different subtype. J. Virol. 79:10701–10708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Collinson-Streng A. N., et al. 2009. Geographic HIV type 1 subtype distribution in Rakai District, Uganda. AIDS Res. Hum. Retroviruses 25:1045–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Conroy S. A., et al. 2010. Changes in the distribution of HIV-1 subtypes D and A in Rakai District, Uganda between 1994 and 2002. AIDS Res. Hum. Retroviruses 26:1087–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gonzales M. J., et al. 2003. Lack of detectable human immunodeficiency virus type 1 superinfection during 1072 person-years of observation. J. Infect. Dis. 188:397–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gunthard H. F., et al. 2009. HIV-1 superinfection in an HIV-2-infected woman with subsequent control of HIV-1 plasma viremia. Clin. Infect. Dis. 48:e117–e120 [DOI] [PubMed] [Google Scholar]
- 8. Herbinger K. H., et al. 2006. Frequency of HIV type 1 dual infection and HIV diversity: analysis of low- and high-risk populations in Mbeya Region, Tanzania. AIDS Res. Hum. Retroviruses 22:599–606 [DOI] [PubMed] [Google Scholar]
- 9. Jost S., et al. 2002. A patient with HIV-1 superinfection. N. Engl. J. Med. 347:731–736 [DOI] [PubMed] [Google Scholar]
- 10. Kiwanuka N., et al. 2009. HIV-1 subtypes and differences in heterosexual transmission of HIV among HIV-1 discordant couples in Rakai, Uganda. AIDS 23:2479–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McCutchan F. E., et al. 2005. In-depth analysis of a heterosexually acquired human immunodeficiency virus type 1 superinfection: evolution, temporal fluctuation, and intercompartment dynamics from the seronegative window period through 30 months postinfection. J. Virol. 79:11693–11704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pacold M., et al. 2010. Comparison of methods to detect HIV dual infection. AIDS Res. Hum. Retroviruses 26:1291–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Piantadosi A., Chohan B., Chohan V., McClelland R. S., Overbaugh J. 2007. Chronic HIV-1 infection frequently fails to protect against superinfection. PLoS Pathog. 3:e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Piantadosi A., Ngayo M. O., Chohan B., Overbaugh J. 2008. Examination of a second region of the HIV type 1 genome reveals additional cases of superinfection. AIDS Res. Hum. Retroviruses 24:1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rachinger A., et al. 2010. Absence of HIV-1 superinfection 1 year after infection between 1985 and 1997 coincides with a reduction in sexual risk behavior in the seroincident Amsterdam cohort of homosexual men. Clin. Infect. Dis. 50:1309–1315 [DOI] [PubMed] [Google Scholar]
- 16. Rachinger A., van de Ven T. D., Burger J. A., Schuitemaker H., van't Wout A. B. 2010. Evaluation of pre-screening methods for the identification of HIV-1 superinfection. J. Virol. Methods 165:311–317 [DOI] [PubMed] [Google Scholar]
- 17. Ramos A., et al. 2002. Intersubtype human immunodeficiency virus type 1 superinfection following seroconversion to primary infection in two injection drug users. J. Virol. 76:7444–7452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17a. Roche 2009. emPCR method manual—Lib-L-MV. Roche, Branford, CT [Google Scholar]
- 18. Rozera G., et al. 2009. Massively parallel pyrosequencing highlights minority variants in the HIV-1 env quasispecies deriving from lymphomonocyte sub-populations. Retrovirology 6:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Saitou N., Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 20. Smith D. M., et al. 2006. Lack of neutralizing antibody response to HIV-1 predisposes to superinfection. Virology 355:1–5 [DOI] [PubMed] [Google Scholar]
- 21. Smith D. M., et al. 2004. Incidence of HIV superinfection following primary infection. JAMA 292:1177–1178 [DOI] [PubMed] [Google Scholar]
- 22. Smith D. M., et al. 2005. HIV drug resistance acquired through superinfection. AIDS 19:1251–1256 [DOI] [PubMed] [Google Scholar]
- 23. Streeck H., et al. 2008. Immune-driven recombination and loss of control after HIV superinfection. J. Exp. Med. 205:1789–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Taylor J. E., Korber B. T. 2005. HIV-1 intra-subtype superinfection rates: estimates using a structured coalescent with recombination. Infect. Genet. Evol. 5:85–95 [DOI] [PubMed] [Google Scholar]
- 25. Thompson J. D., Higgins D. G., Gibson T. J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. van der Kuyl A. C., et al. 2010. Analysis of infectious virus clones from two HIV-1 superinfection cases suggests that the primary strains have lower fitness. Retrovirology 7:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wawer M. J., et al. 1998. A randomized, community trial of intensive sexually transmitted disease control for AIDS prevention, Rakai, Uganda. AIDS. 12:1211–1225 [DOI] [PubMed] [Google Scholar]
- 28. Willerth S. M., et al. 2010. Development of a low bias method for characterizing viral populations using next generation sequencing technology. PLoS One 5:e13564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yang O. O., et al. 2005. Human immunodeficiency virus type 1 clade B superinfection: evidence for differential immune containment of distinct clade B strains. J. Virol. 79:860–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zagordi O., Klein R., Daumer M., Beerenwinkel N. 2010. Error correction of next-generation sequencing data and reliable estimation of HIV quasispecies. Nucleic Acids Res. 38:7400–7409 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.