Abstract
lukF-PV was present in 36% of skin and soft tissue infection (SSTI)-derived methicillin-susceptible Staphylococcus aureus (MSSA) strains and comprised six distinct clones, which contained fewer enterotoxin genes than strains without lukF-PV. Clinical presentations and outcomes of lukF-PV+ methicillin-resistant S. aureus (MRSA) and MSSA SSTIs were comparable. In multivariable analysis, the presence of lukF-PV remained a significant predictor for incision and drainage among MSSA strains.
TEXT
In the United States, Panton-Valentine leukocidin (PVL), a pore-forming cytotoxin, is investigated mostly in the emerging community-acquired methicillin-resistant Staphylococcus aureus (MRSA) clone USA300 (10). PVL contribution to virulence is disputed, as summarized by Schlievert (6). PVL is also produced by strains other than USA300, and levels vary considerably among individual strains (2, 8). Some methicillin-susceptible S. aureus (MSSA) strains produce large amounts of PVL, which has been associated with more-severe skin lesions in a murine model (8). In this study, we compared the prevalences of PVL in MSSA and MRSA strains from patients with skin and soft tissue infections (SSTIs) and compared the clinical presentations. S. aureus isolates from wound (n = 108) and blood (n = 99) were collected by the microbiology laboratory of Montefiore Medical Center as described previously (9). Detection of enterotoxin genes (staphylococcal enterotoxin A [SEA], SEB, SEC, and toxic shock syndrome toxin [TSST] genes), multilocus sequence typing (MLST), and spa typing were done in the context of a previously published study (9). For this study, detection of PVL in MSSA isolates was performed by real-time PCR targeting the lukF-PV gene (7). Also, a retrospective chart review was performed on all 104 patients from whom wound isolates had been collected and 11 patients from whom PVL-producing S. aureus strains were cultured from blood. Information on demographic characteristics, clinical presentation, antibiotic therapy, complications, and outcomes was obtained. Fever was defined as greater than 38.0°C and leukocytosis as ≥12.0 K/μl. Health care-associated (HA) risk factors included end-stage renal disease (ESRD) on dialysis, hospitalization 6 months prior to admission, history of S. aureus infection, and nursing home residence. The Wilcoxon rank-sum test was used to compare continuous variables, and the χ2 or Fisher exact test was used to compare proportions. To assess the outcome variable of incision and drainage, selected predictor variables whose P values were less than 0.20 in univariate analysis were introduced into a multivariate logistic regression model for further analysis.
Chart review determined that 101 wound isolates were derived from patients with SSTIs (46 on extremities, 55 from other body parts). Clinical data from 4 patients were not retrievable, and 3 wound isolates were excluded as they were grown from infected bone biopsy specimens and an infected vascular graft. MSSA and MRSA strains were isolated in 57% and 43% of SSTI cases, respectively. A significant percentage of SSTI-associated MSSA strains (36%) and most MRSA strains (77%) carried the lukF-PV gene (Table 1). A significantly higher proportion of S. aureus strains (both MSSA and MRSA strains) that were derived from SSTIs than were derived from blood harbored the lukF-PV gene (53% versus 11%, respectively; P < 0.001 by chi-square test). The majority (9 of 11) of lukF-PV+ blood isolates were MRSA strains derived from patients with concomitant SSTIs. Only 2 of 45 MSSA strains isolated from blood contained the lukF-PV gene.
Table 1.
Baseline demographic and clinical characteristicsa
| Characteristicb | MSSA strains (n = 58) |
MRSA strains (n = 43) |
P valuec | ||
|---|---|---|---|---|---|
| PVL− | PVL+ | PVL− | PVL+ | ||
| PVL status | 37/58 (64) | 21/58 (36) | 10/43 (23) | 33/43 (77) | 0.001 |
| Median age (yr) | 39.5 (0–77) | 34 (4–65) | 68.5 (27–87) | 29.5 (0.9–87) | 0.303, 0.012* |
| Male sex | 23/37 (62) | 15/21 (71) | 3/10 (30) | 13/33 (39) | 0.005 |
| Race | |||||
| Hispanic | 17/36 (47) | 12/18 (67) | 2/10 (20) | 10/27 (37) | 0.074 |
| African-American | 14/36 (39) | 4/18 (22) | 4/10 (40) | 13/27 (48) | 0.725 |
| Caucasian | 5/36 (14) | 2/18 (11) | 4/10 (40) | 4/27 (15) | 0.420 |
| Patient risk factor | |||||
| NHR | 0/37 (0) | 0/21 (0) | 3/10 (30) | 2/33 (6) | 0.012 |
| Diabetes | 11/37 (30) | 3/21 (14) | 3/10 (30) | 6/33 (18) | 0.704 |
| ESRD/dialysis | 3/37 (8) | 0/21 (0) | 4/10 (40) | 1/33 (3) | 0.280, 0.007* |
| HIV | 3/37 (8) | 3/21 (14) | 0/10 (0) | 2/33 (6) | 0.461 |
| Malignancy | 4/37 (11) | 0/21 (0) | 2/10 (20) | 2/33 (6) | 0.720 |
| Device-related infection | 3/37 (8) | 2/21 (9) | 5/10 (50) | 1/33 (3) | 0.521, 0.001* |
| HA infection in past 6 mo | 11/36 (31) | 2/21 (10) | 9/10 (90) | 10/33 (30) | 0.031 |
| SA infection in past 6 mo | 1/36 (3) | 2/21 (9) | 1/10 (10) | 8/33 (24) | 0.027 |
| Antibiotics in last month | 8/37 (22) | 5/21 (24) | 4/10 (40) | 9/33 (27) | 0.374 |
| Presentation of SSTI | |||||
| Extremity and buttock | 20/37 (54) | 19/21 (43) | 3/10 (30) | 14/33 (42) | 0.400 |
| Fever of >100.4°F at Dx | 9/36 (25) | 2/21 (9) | 3/10 (30) | 8/33 (24) | 0.453 |
| Median WBC at Dx (range) | 8.4 (3.2–48) | 10.9 (6.3–21) | 9.5 (1.3–24) | 11.5 (3.7–30.1) | 0.485 |
| Treatment and outcome | |||||
| HA infection | 23/37 (62) | 7/21 (33) | 9/10 (90) | 18/33 (55) | 0.267, 0.035* |
| Median LOS, days (range) | 9 (0–134) | 2 (0–33) | 11.5 (1–32) | 4 (0–68) | 0.442 |
| Incision and drainage | 21/37 (57) | 17/21 (81) | 7/10 (70) | 23/33 (70) | 0.652 |
| Osteomyelitis | 4/34 (12) | 1/21 (5) | 4/10 (40) | 2/32 (6) | 0.526, 0.021* |
| Death | 1/37 (3) | 0/21 (0) | 1/10 (10) | 1/33 (3) | 0.573 |
| Appropriate Abx given | 9/36 (25) | 3/21 (14) | 10/10 (100) | 27/33 (81) | 0.000 |
| Median no. of SE (range) | 6 (2–9) | 4 (0–8) | 5 (1–9) | 3 (0–6) | 0.002, 0.008* |
| SEA+ | 3/37 (8) | 0/21 (0) | 0/13 (0) | 0/33 (0) | 0.260 |
| SEB+ | 5/37 (13) | 0/21 (0) | 0/10 (0) | 1/33 (3) | 0.233 |
| TSST+ | 5/37 (13) | 0/21 (0) | 0/13 (0) | 0/33 (0) | 0.070 |
| mecA IV | 0/37 (0) | 0/21 (0) | 13/13 (100) | 33/33 (100) | 0.000 |
Results are given as numbers (%) of strains with the characteristic, except where indicated.
NHR, nursing home residence; SA, Staphylococcus aureus; Dx, diagnosis; WBC, white blood cells; LOS, length of stay; Abx, antibiotics; SE, staphylococcal enterotoxins.
P values are for MRSA versus MSSA strains, except those marked with an asterisk, which are for PVL+ versus PVL− MRSA strains.
Patients infected with lukF-PV+ S. aureus strains were significantly younger than those infected with S. aureus strains lacking lukF-PV (33.4 and 44.7 years, respectively; P = 0.02) and had fewer hospitalizations within the prior 6 months (12 of 54 [22%] and 20 of 46 [43%], respectively; P = 0.02).
A comparison of patients with SSTIs caused by MRSA lukF-PV+ or MSSA lukF-PV+ strains demonstrated similar lengths of hospital stay (median lengths of 6 and 4 days, respectively; P = 0.59), proportions of complications by osteomyelitis (6% and 5%, respectively; P = 0.85), and numbers of strains associated with bacteremia (3 MSSA strains and 2 MRSA strains). The percentage of patients that were treated by incision and drainage tended to be higher in those with SSTIs caused by lukF-PV+ MSSA than in those with SSTIs caused by lukF-PV-negative MSSA (81% versus 57%, respectively). To assess the outcome of incision and drainage, selected predictors, including age, sex, presence of fever at diagnosis, methicillin resistance, and presence of the lukF-PV gene, were examined for an association (Table 2). There was a trend toward increased odds for incision and drainage among all patients with lukF-PV+ infections (odds ratio [OR], 2.24; 95% confidence interval [95% CI], 0.79 to 6.35). When multivariable analysis was stratified by status of methicillin resistance, the odds for incision and drainage among patients with lukF-PV+ MSSA infections was significantly higher than among those with lukF-PV-negative MSSA infections (OR, 4.73; 95% CI, 1.14 to 19.68). In contrast, the presence of methicillin resistance was not associated with the outcome of incision and drainage (P = 0.7). In multivariable analysis, the presence of PVL remained a significant predictor for incision and drainage among MSSA strains.
Table 2.
Multivariate analysis predicting need for incision and drainage
| Characteristic | All strains (MRSA and MSSA) |
MSSA strains only |
||||
|---|---|---|---|---|---|---|
| P value | OR | 95% CI | P value | OR | 95% CI | |
| Age | 0.42 | 0.99 | 0.97–1.01 | 0.86 | 1.00 | 0.97–1.03 |
| Male sex | 0.05 | 2.57 | 0.98–6.70 | 0.06 | 3.44 | 0.95–12.50 |
| Methicillin resistance | 0.71 | 1.22 | 0.42–3.55 | |||
| PVL present | 0.13 | 2.24 | 0.79–6.35 | 0.03 | 4.73 | 1.14–19.68 |
| Fever present at Dx | 0.016 | 5.67 | 1.39–23.13 | 0.07 | 5.50 | 0.89–33.92 |
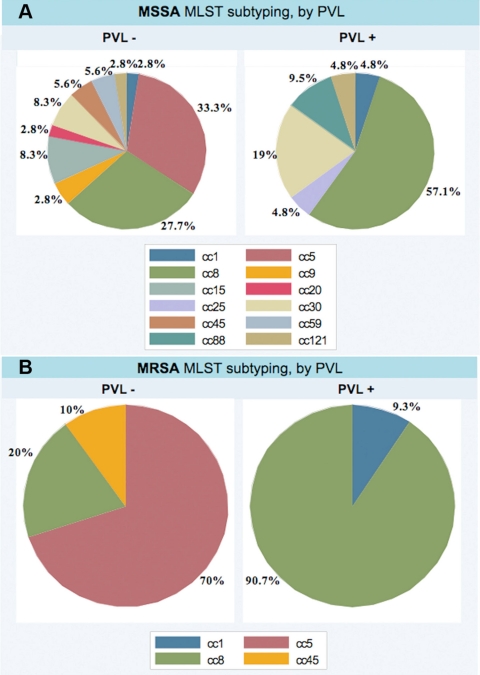
Similarly to other studies, lukF-PV+ MRSA strains were mostly the epidemic strain t008/eGenomics type 1 (USA300). The three remaining were USA400 strains. Among 58 MSSA strains, 12 distinct MLST clonal types were identified. Pattern CC8 was most common, representing 57% of PVL-positive MSSA isolates, followed by CC30, CC88, CC25, CC121, and CC1, respectively (Fig. 1A). t008/eGenomics type 1 was identified in only 7 MSSA strains. In a New York prison population, this clone is also dominant among colonizing MSSA strains, of which 32.2% carry lukF-PV (4). Of note is that 4 of the t008/eGenomics type 1 strains (2 MRSA and 2 MSSA strains) did not contain the lukF-PV gene. Similarly to USA300 strains, enterotoxin gene content was lower in lukF-PV+ MSSA strains than in lukF-PV-negative MSSA strains (4 versus 5.7 genes, respectively; P = 0.002 by t test). TSST, SEB, and SEC genes were absent in lukF-PV+ MSSA and MRSA strains. The SEA gene was present in one MSSA strain from blood; however, all 3 USA400 strains had the SEC gene.
Fig. 1.
Schemata of MLST of PVL-producing MSSA (A) and MRSA (B) strains.
Polyclonality among lukF-PV+ MSSA strains has been reported by several studies and stands in contrast to the clonal homogeneity among lukF-PV+ MRSA strains (1, 5). This finding could also be relevant for the pathogenesis of community-acquired MRSA (CA-MRSA) disease. First, most studies report that nasal colonization by MSSA is more common than that by MRSA. However, it is unknown how many colonizing MSSA strains produce PVL. Second, colonization with PVL-secreting MRSA or MSSA strains elicits a neutralizing polyclonal immune response to PVL, which is not protective (3) but is probably boostered by each infection. Third, it has been proposed, based on murine studies, that high titers of PVL neutralizing antibodies (Abs) inhibit clearance of MRSA infection (11). Finally, it is conceivable that recurrent infection with PVL-producing MRSA or MSSA strains may promote selection of strains that secrete higher levels of PVL, which is associated with worse SSTI in mice (8).
In summary, our data indicate that lukF-PV+ MSSA strains, similarly to lukF-PV+ MRSA strains, commonly cause complicated SSTIs in patients in the Bronx. The lukF-PV gene is uncommon in MSSA strains that cause septicemia without SSTI. As the epidemiology of CA-MRSA infection evolves, careful monitoring of emergence of genes encoding PVL in both MRSA and MSSA isolates is necessary to optimize the treatment and prevention of complicated SSTIs. This may further our understanding of S. aureus microevolution and how this process is affected by the human host response to its toxins.
Acknowledgments
We thank Marilou Corpuz for assistance with obtaining institutional review board permission.
This work was funded by the NIH-funded Northeast Biodefense Center, U54-AI057158-Lipkin. This work was also supported by CTSA grant UL1 RR025750; KL2 RR025749 and TL1 RR025748 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH); and the NIH Roadmap for Medical Research.
The contents of this work are solely the responsibility of the authors and do not necessarily represent the official view of the NCRR or the NIH.
We declare no conflict of interest.
Footnotes
Published ahead of print on 8 June 2011.
REFERENCES
- 1. Chini V., et al. 2006. Spread of Staphylococcus aureus clinical isolates carrying Panton-Valentine leukocidin genes during a 3-year period in Greece. Clin. Microbiol. Infect. 12:29–34 [DOI] [PubMed] [Google Scholar]
- 2. Hamilton S. M., et al. 2007. In vitro production of Panton-Valentine leukocidin among strains of methicillin-resistant Staphylococcus aureus causing diverse infections. Clin. Infect. Dis. 45:1550–1558 [DOI] [PubMed] [Google Scholar]
- 3. Hermos C. R., Yoong P., Pier G. B. 2010. High levels of antibody to Panton-Valentine leukocidin are not associated with resistance to Staphylococcus aureus-associated skin and soft-tissue infection. Clin. Infect. Dis. 51:1138–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lowy F. D., et al. 2007. Staphylococcus aureus colonization and infection in New York State prisons. J. Infect. Dis. 196:911–918 [DOI] [PubMed] [Google Scholar]
- 5. Monecke S., Slickers P., Ellington M. J., Kearns A. M., Ehricht R. 2007. High diversity of Panton-Valentine leukocidin-positive, methicillin-susceptible isolates of Staphylococcus aureus and implications for the evolution of community-associated methicillin-resistant S. aureus. Clin. Microbiol. Infect. 13:1157–1164 [DOI] [PubMed] [Google Scholar]
- 6. Schlievert P. M. 2009. Cytolysins, superantigens, and pneumonia due to community-associated methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 200:676–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sinsimer D., et al. 2005. Use of a multiplex molecular beacon platform for rapid detection of methicillin and vancomycin resistance in Staphylococcus aureus. J. Clin. Microbiol. 43:4585–4591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Varshney A. K., et al. 2010. Augmented production of Panton-Valentine leukocidin toxin in methicillin-resistant and methicillin-susceptible Staphylococcus aureus is associated with worse outcome in a murine skin infection model. J. Infect. Dis. 201:92–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Varshney A. K., et al. 2009. Diverse enterotoxin gene profiles among clonal complexes of Staphylococcus aureus isolates from the Bronx, New York. Appl. Environ. Microbiol. 75:6839–6849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Voyich J. M., et al. 2006. Is Panton-Valentine leukocidin the major virulence determinant in community-associated methicillin-resistant Staphylococcus aureus disease? J. Infect. Dis. 194:1761–1770 [DOI] [PubMed] [Google Scholar]
- 11. Yoong P., Pier G. B. 2010. Antibody-mediated enhancement of community-acquired methicillin-resistant Staphylococcus aureus infection. Proc. Natl. Acad. Sci. U. S. A. 107:2241–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]