Abstract
We analyzed 212 group B streptococci (GBS) from newborns with invasive infections in the area of Barcelona, Spain, between 1992 and 2009, with the aim of documenting changes in the prevalences of serotypes, antimicrobial resistance, and genetic lineages and evaluating their associations with either early-onset disease (EOD) or late-onset disease (LOD). Serotypes III (n = 118) and Ia (n = 47) together accounted for nearly 78% of the isolates. All isolates carried an alpha or alpha-like protein gene, and specific associations between genes and serotypes, such as serotype Ib and bca, serotype II and bca, serotype III and rib, and serotype V and alp3, reflected the presence of particular genetic lineages. Macrolide resistance (14.2%) was significantly associated with serotype V. Pulsed-field gel electrophoresis (PFGE) clustering was an excellent predictor of serotype and antibiotic resistance. The combination of PFGE and multilocus sequence typing revealed a large number of genetically distinct lineages. Still, specific lineages were dominant in our collection, particularly the serotype III/ST17/rib lineage, which had enhanced potential to cause LOD. Serotype Ia was concentrated in a single PFGE cluster composed of two genetic lineages: ST23/eps and ST24/bca. The ST24/bca sublineage of serotype Ia, which is found infrequently elsewhere, may be emerging as an important cause of neonatal invasive infections in the Mediterranean region. In spite of the introduction of prophylaxis, resulting in a pronounced decline in the frequency of EOD, the study revealed a remarkably stable clonal structure of GBS causing neonatal infections in Barcelona over a period of 18 years.
INTRODUCTION
Streptococcus agalactiae, or group B streptococcus (GBS), is well established as a leading cause of neonatal sepsis and meningitis (25, 45). In neonates, early-onset disease (EOD) is defined as occurring within the first 7 days and late-onset disease (LOD) as occurring from day 8 to 90 (12). While vertical transmission is commonly accepted to be the cause of EOD (24, 37, 51), the source of bacterial strains causing LOD is less well understood (45). In 1996, guidelines for the prevention of neonatal GBS infections by antimicrobial prophylaxis were published in the United States (12). Whereas a mixed risk-based and screening-based approach was initially suggested in the guidelines, the universal screening of pregnant women for vaginal GBS colonization at 35 to 37 weeks of gestation and the administration of intrapartum antimicrobial prophylaxis to carriers was proposed shortly afterwards (11). As a consequence, the incidence of EOD has fallen significantly over the past decade where these guidelines have been followed, yet this strategy is raising concerns that the widespread use of intrapartum antimicrobials might delay, rather than prevent, the onset of GBS disease (10, 16). In the area of Barcelona, Spain, after the local implementation of the intrapartum antimicrobial prophylaxis guidelines, the incidence of EOD declined by 86%, from 1.92 cases per 1,000 live births in 1994 to 0.26 in 2001 (P < 0.001) (2), and has remained at low levels since (ranging from 0.47 cases per 1,000 live births in 2007 to 0.18 in 2009). In the same area, the incidence of LOD increased from 0.11 cases per 1,000 live births in 1996 to 0.81 in 2009, but in spite of the difference between these values, these changes did not reflect a significant trend.
Furthermore, the increase in the use of antimicrobials due to intrapartum antibiotic prophylaxis can lead to the emergence of resistant bacteria (46), a concern that has been strengthened by the recent description of GBS strains showing reduced susceptibility to beta-lactams (28).
These considerations are driving the search for alternative prevention strategies. Studies evaluating the potential impact of vaccines in the management of GBS disease suggest that vaccination may provide additional benefits over antimicrobial prophylaxis, especially due to the expected reduction in LOD incidence (49). The vaccine formulations currently on trial are based on GBS capsular polysaccharides; however, they are not expected to provide optimal coverage in different regions, due to geographical differences in serotype distribution. In order to overcome serotype specificity, whole-genome-based approaches have been directed toward identifying protein antigens, which hold promise as components of globally effective vaccines (26, 33).
In addition to the capsular polysaccharides, multiple virulence factors have been recognized and extensively characterized in recent decades. These virulence factors may be unevenly distributed within a particular serotype and may contribute significantly to the invasive potential of a particular lineage, independently of its capsular polysaccharide. Molecular epidemiology has been used to distinguish genetic lineages in order to probe for associations between specific GBS genotypes and disease. Most of these studies, using multilocus sequence typing (MLST), have identified a lineage with enhanced invasive capacity expressing serotype III and defined by sequence type 17 (ST17) (6, 15, 31). Moreover, in a study of carriage and invasive isolates from Portugal, we found that serotype Ia presented enhanced invasive disease potential and was particularly associated with EOD (36). In that study, serotype Ia was associated mostly with a single pulsed-field gel electrophoresis (PFGE) cluster and with two sequence types (ST23 and ST24), again pointing to the possible existence of particular genetic lineages with enhanced invasive disease potential.
We undertook the analysis of GBS isolates responsible for invasive infections in newborns in the Barcelona area from 1992 to 2009 with the aim of documenting changes over this 18-year period and of testing associations with EOD and LOD. To this end, we have characterized the isolates with regard to their serotypes and antimicrobial resistance patterns and have identified the genetic lineages present by PFGE profiling, MLST, and surface protein gene profiling.
MATERIALS AND METHODS
Eight hospitals located in the Barcelona metropolitan area monitored all EOD cases from 1994 on and all LOD cases from 1996 on. In these hospitals, GBS prevention policies were implemented progressively from 1994 onward. Invasive disease was defined as the presence of GBS in a normally sterile fluid (blood or cerebrospinal fluid [CSF]). During the period from 1994 to 2009, a total of 351.950 live infants were born in the 8 hospitals, and EOD was diagnosed in 243 cases (189 infants born in the 8 hospitals and 54 referred from other hospitals). During the period from 1996 to 2009, of a total of 315.576 live births, LOD was diagnosed in 131 infants.
Bacterial isolates.
We characterized the 207 isolates available from the 374 GBS cases identified between 1994 and 2009 in the 8 Barcelona-area hospitals. Additionally, we also included 5 isolates recovered from patients with invasive GBS disease in the same centers between 1992 and 1993. Only the first isolate of each case was considered. A total of 212 GBS isolates were characterized: 123 from EOD and 89 from LOD patients.
Serotyping, antimicrobial susceptibility testing, and macrolide resistance phenotype.
Capsular serotyping was carried out by a latex agglutination assay with a GBS serotyping kit (Essum, Umeå, Sweden) according to the manufacturer's instructions.
All GBS isolates were tested for susceptibility to erythromycin, clindamycin, tetracycline, chloramphenicol, levofloxacin, and penicillin by using the disk diffusion method according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (13). The macrolide resistance phenotype was determined according to a double-disk test as described previously (38).
PCR determination of the macrolide resistance genotype.
Total bacterial DNA was isolated by treatment of the cells with mutanolysin and boiling. Multiplex PCR was performed to detect the presence of the erm(B), erm(A) [erm(TR) subclass], and mef genes, as described elsewhere (17).
Pulsed-field gel profiling and MLST.
The total bacterial DNAs of the strains were isolated, digested with SmaI, and separated by PFGE as described previously (36). Whenever a complete digestion with SmaI was not achieved, the isoschizomer Cfr9I was used (48). PFGE patterns were compared by using Bionumerics software (Applied Maths, Sint-Martens-Latem, Belgium) to create dendrograms by the unweighted-pair group method with arithmetic averages (UPGMA). The Dice similarity coefficient was used with optimization and position tolerance settings of 1.0 and 1.5, respectively. PFGE-based clusters were defined as isolates with ≥80% relatedness on the dendrogram (36). MLST was performed by sequencing seven housekeeping genes as described previously (27), and sequence types (STs) were identified by using the S. agalactiae MLST database (http://pubmlst.org/sagalactiae) and were analyzed using the entire database and goeBURST (20). Alleles and sequence types not previously described were deposited at the S. agalactiae MLST database. DNA sequences were analyzed by using Bionumerics software.
Surface protein gene profile.
Total bacterial DNA was isolated by treatment of the cells with mutanolysin and boiling. A multiplex PCR assay was performed for direct identification of GBS alpha-like protein genes, as described elsewhere (14). This assay allowed the direct determination of the following GBS surface protein genes by analysis of the amplicon size: the alpha-C protein gene (bca), the epsilon protein gene (eps), and the rib, alp2 or alp3, and alp4 genes. A previously described assay was performed to differentiate the alp2 and alp3 protein antigen genes (34).
Typing concordance and statistics.
The Wallace (W) and adjusted Rand (AR) coefficients were calculated to determine the concordance between the different typing methods (9, 40, 47). The AR coefficient provides a measurement of the overall concordance between the results of two methods, whereas the W coefficient provides a directional measurement of clustering concordance between different typing methods, i.e., if the results of one typing method can predict the results of another method. Simpson's index of diversity (SID) was calculated to evaluate the diversity found among the isolates studied (9). All these calculations were performed at the Comparing Partitions website (www.comparingpartitions.info). The Fisher exact test was used to evaluate associations. Odds ratios (OR) with 95% Wald confidence intervals (CI95) (1) were calculated against all other serotypes or PFGE clusters and were used to identify particular serotypes or PFGE clusters associated with certain characteristics, controlling for a false discovery rate (FDR) under or equal to 0.05 (3). Spearman's nonparametric test was used to evaluate correlations (1).
RESULTS
Capsular serotyping.
The results of serotyping of the 212 invasive GBS isolates from neonates are summarized in Table 1. Serotypes III (n = 118) and Ia (n = 47) were the most frequent among the population, together accounting for 77.8% of the isolates. The serotypes were also differently distributed in EOD and LOD (P = 0.0067 by Fisher's exact test). Serotypes III and Ia were found in 44% and 26% of EOD cases, respectively, and in 72% and 17% of LOD cases, respectively. In fact, the number of serotype III isolates found in LOD (n = 64) was more than double the sum of all the other serotypes (n = 25), and this was the only serotype that showed a significant association with disease presentation (OR, 2.980 [CI95, 1.581 to 5.734] for an association with LOD). Although more isolates were recovered from the CSF in LOD cases (n = 25) than in EOD cases (n = 21), in agreement with a previous report (36), we did not find any association between the serotype and the biological product from which the isolate was recovered.
Table 1.
Serotype distribution among invasive GBS isolates causing EOD and LOD
| Serotype | No. of isolatesa |
||||||
|---|---|---|---|---|---|---|---|
| EOD |
LOD |
Total | |||||
| Blood | CSF | Total | Blood | CSF | Total | ||
| Ia | 27 | 5 | 32 (26) | 11 | 4 | 15 (17) | 47 |
| Ib | 8 | 0 | 8 (7) | 4 | 1 | 5 (6) | 13 |
| II | 9 | 1 | 10 (8) | 1 | 0 | 1 (1) | 11 |
| III | 41 | 13 | 54 (44) | 46 | 18 | 64 (72) | 118 |
| IV | 2 | 0 | 2 (2) | 0 | 1 | 1 (1) | 3 |
| V | 11 | 1 | 12 (10) | 2 | 0 | 2 (2) | 14 |
| NTb | 4 | 1 | 5 (4) | 0 | 1 | 1 (1) | 6 |
| Total | 102 | 21 | 123 (100) | 64 | 25 | 89 (100) | 212 |
Numbers in parentheses are percentages.
NT, nontypeable.
PFGE cluster analysis and MLST.
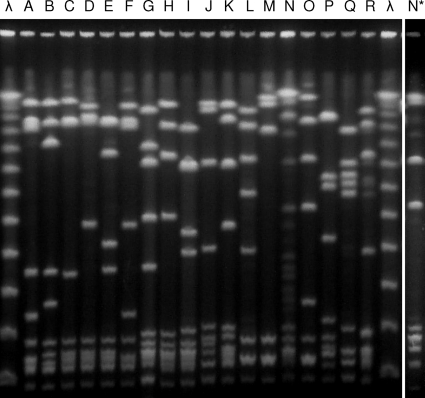
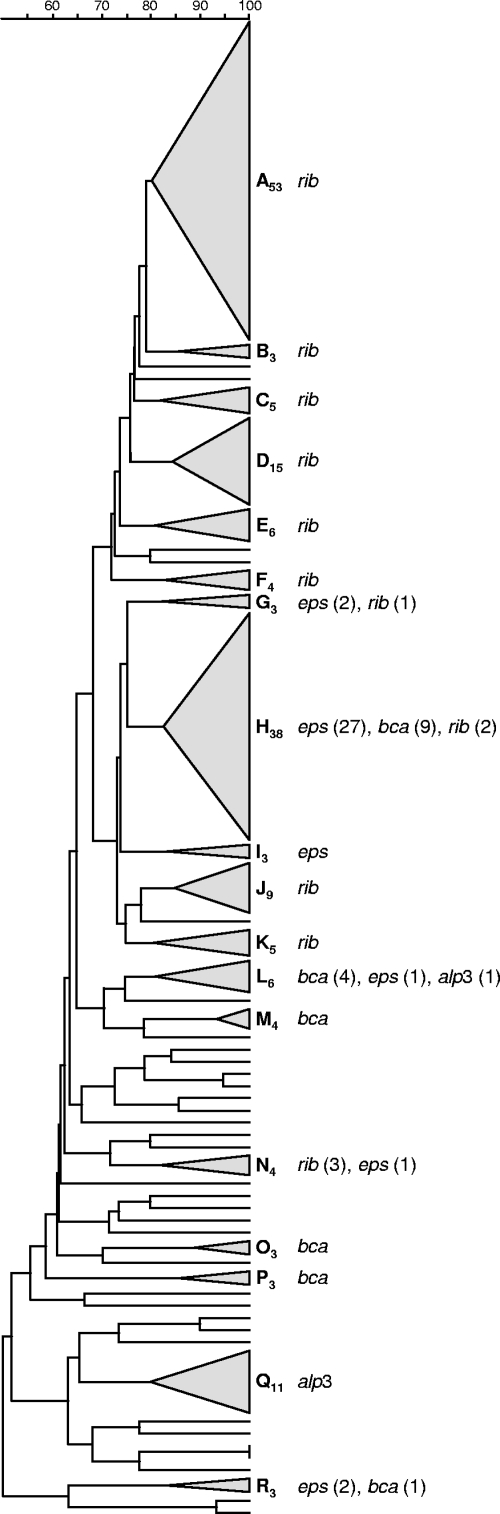
All isolates were analyzed by PFGE, and 43 different profiles were identified and were grouped into 18 different PFGE clusters (≥3 isolates), of which the major 5 accounted for nearly 60% of the isolates (Fig. 1). The remaining isolates (n = 34) were included in minor PFGE groups (≤2 isolates) or had unique profiles. The dendrogram depicting the relationship between these PFGE clusters is shown in Fig. 2. The SID for the classification of the isolates into PFGE clusters was 0.894 (CI95, 0.867 to 0.921), indicating that the collection analyzed is genetically very diverse. Each PFGE cluster was composed almost exclusively of isolates of the same serotype (W, 0.955 [CI95, 0.910 to 1.000]), indicating a very good predictive power of the PFGE-based genotypes for the serotype. The converse was not so; inspection of Fig. 2 and Table 2 reveals that each serotype was subdivided into several PFGE clusters, as expected from the existence of several genetic lineages sharing the same serotype.
Fig. 1.
Representative SmaI macrorestriction profiles of each PFGE cluster. Capital letters above the lanes correspond to cluster designations. Lane N, incomplete digestion with SmaI; lane N*, the same strain as in lane N but now digested with Cfr9I (see the text); lane λ, Lambda ladder PFG marker (New England Biolabs, Beverly, MA).
Fig. 2.
Dendrogram of the PFGE profiles of 212 GBS isolates and distribution of the surface protein genes. The dendrogram was constructed using UPGMA. Dice coefficients (percentages) are indicated in the scale above the dendrogram. Each cluster (defined as a group of three or more isolates with a Dice coefficient of ≥80%) is represented by a triangle proportional to the number of isolates included in the cluster. Clusters are designated by capital letters with a subscript number indicating the number of isolates included in the cluster. The surface protein genes found in each particular cluster are given on the right. If more than one gene was found in strains of the same PFGE cluster, the number of isolates carrying each gene is given in parentheses.
Table 2.
Properties of the genetic lineages found among the 212 invasive isolates
| PFGE clustera | Serotype | No. of isolates associated with EOD/LOD | STs in PFGE cluster with the same serotype (n)b | No. of isolates not susceptible toc: |
No. of susceptible isolatese | ||
|---|---|---|---|---|---|---|---|
| ERYd | CHL | TET | |||||
| A53 | III | 23/30 | [ST17 (48), ST147 (1), ST148 (1), ST542 (1), ST543 (1), ST550 (1)] | 1 | 0 | 51 | 2 |
| B3 | III | 2/1 | [ST17 (2), ST180 (1)] | 0 | 0 | 3 | 0 |
| C5 | III | 3/2 | [ST17 (4), ST148 (1)] | 0 | 0 | 5 | 0 |
| D15 | III | 3/12 | ST17 (15) | 0 | 0 | 15 | 0 |
| E6 | III | 2/4 | [ST17 (4), ST469 (2)] | 0 | 1 | 6 | 0 |
| F4 | III | 1/3 | ST17 (4) | 0 | 0 | 4 | 0 |
| G3 | Ia | 2/0 | ST23 (2) | 0 | 0 | 2 | 0 |
| III | 1/0 | ST19 (1) | 1 | 1 | 1 | 0 | |
| H38 | Ia | 22/14 | [ST23 (25), ST24 (9), ST223 (1), ST545 (1)] | 0 | 1 | 34 | 3 |
| III | 0/2 | ST17 (1), ST27 (1) | 0 | 0 | 1 | 1 | |
| I3 | Ia | 3/0 | ST23 (3) | 0 | 0 | 3 | 0 |
| J9 | III | 6/2 | [ST19 (5), ST456 (1), ST471 (1), ST547 (1)]* | 3f | 0 | 6 | 1 |
| II | 1/0 | ST28 (1)* | 0 | 0 | 1 | 0 | |
| K5 | III | 0/3 | ST17 (3) | 0 | 0 | 3 | 0 |
| NTg | 1/1 | ST28 (2) | 0 | 0 | 2 | 0 | |
| L6 | II | 3/0 | [ST2 (1), ST9 (1), ST12 (1)]* | 1 | 0 | 0 | 2 |
| Ia | 2/0 | ST7 (2)* | 2h | 0 | 1 | 0 | |
| Ib | 0/1 | ST1 (1)* | 0 | 0 | 1 | 0 | |
| M4 | Ib | 3/1 | [ST8 (1), ST9 (2) ST10 (1)] | 0 | 0 | 2 | 2 |
| N4 | III | 2/1 | [ST19 (2), ST27 (1)]* | 0 | 0 | 2 | 0 |
| V | 1/0 | ST19 (1)* | 0 | 0 | 2 | 0 | |
| O3 | Ib | 2/1 | ST12 (3) | 0 | 0 | 3 | 0 |
| P3 | II | 3/0 | ST22 (3) | 3f | 0 | 3 | 0 |
| Q11 | V | 9/2 | ST1 (11) | 8 | 0 | 11 | 0 |
| R3 | Ia | 2/0 | ST23 (2) | 0 | 0 | 2 | 0 |
| NTg | 1/0 | ST19 (1) | 0 | 0 | 0 | 1 | |
| Other34 | III | 11/4 | ST17 (4), ST19 (6), ST24 (1), ST106 (2), ST470 (1), ST546 (1) | 6 | 3 | 14 | 1 |
| Ib | 3/2 | ST2 (1), ST9 (1), ST10 (1), ST12 (1), ST548 (1) | 2 | 0 | 3 | 2 | |
| II | 3/1 | ST1 (1), ST12 (1), ST22 (1), ST544 (1) | 1f | 0 | 3 | 1 | |
| IV | 2/1 | ST2 (1), ST196 (1), ST549 (1) | 0 | 0 | 2 | 1 | |
| NTg | 3/0 | ST19 (1), ST27 (1), ST130 (1) | 1f | 0 | 1 | 1 | |
| Ia | 1/1 | ST7 (1), ST196 (1) | 0 | 0 | 1 | 1 | |
| V | 2/0 | ST26 (1), ST551 (1) | 1f | 0 | 1 | 0 | |
| Total | 123/89 | 30 | 6 | 189 | 19 | ||
PFGE clusters are identified as indicated in Fig. 1. Clusters are designated by capital letters with a subscript number indicating the number of isolates included in the cluster.
Brackets indicate STs that were grouped into the same PFGE cluster, belonged to the same clonal complex by goeBURST, and expressed the same serotype. Asterisks indicate STs or groups of STs that were found in the same PFGE cluster and belonged to the same clonal complex by goeBURST but expressed different serotypes.
ERY, erythromycin; CLI, clindamycin; CHL, chloramphenicol; TET, tetracycline.
All isolates were simultaneously resistant to erythromycin and clindamycin, presenting with the cMLSB phenotype associated with the erm(B) gene, except where otherwise indicated.
Isolates susceptible to all antimicrobials tested (erythromycin, clindamycin, chloramphenicol, and tetracycline).
One isolate carried the erm(A) gene.
NT, nontypeable.
Two isolates carried the erm(A) gene.
To further identify the genetic lineages associated with each PFGE clone, all isolates were characterized by MLST. Thirteen novel alleles and 13 novel STs (ST469 to ST471 and ST542 to ST551) were identified among the isolates studied. The detailed characterization of the clusters depicted in Fig. 1 and 2 is summarized in Table 2.
The SID for the classification of the isolates according to their MLST-based sequence types was 0.805 (CI95, 0.758 to 0.852), also indicating a genetically diverse collection but less diverse than the classification into PFGE profiles. Overall, the isolates sharing the same ST also shared the same serotype (W, 0.978 [CI95, 0.961 to 0.997]), indicating that MLST is a good predictor of serotype, a finding similar to that for the PFGE profiles.
In agreement with these observations, serotype III isolates representing ST17 were found mostly in PFGE clusters A53 and D15. Likewise, most serotype Ia isolates (of which the majority belonged to ST23 but a significant proportion belonged to ST24) and serotype V isolates (ST1) grouped into particular clusters, H38 and Q11, respectively.
Interestingly, most of the new STs were identified in serotype III isolates. Whereas ST469, ST542, ST543, and ST550 are single-locus variants (SLVs) of ST17, ST470 is a double-locus variant (DLV) of ST17 (exhibiting two novel alleles). On the other hand, ST471 and ST547 are SLVs of ST19, a sequence type previously associated with colonization (27, 31, 36). ST19 was found to be poorly represented in our collection; the largest PFGE cluster where this ST was found was J9. This was expected, because the collection analyzed included only isolates that had caused invasive neonatal infections.
Six of the newly described STs (ST470, ST544, ST546, ST548, ST549, and ST551) were found in isolates included in minor PFGE clusters or showing unique profiles (Table 2). The combination of rare PFGE profiles and STs suggests that the GBS populations causing invasive neonatal disease may reflect the larger genetic diversity found in the overall GBS population to a limited extent.
In testing for an association between the largest PFGE clusters (n ≥ 10) and the timing of disease presentation, both the major clusters of serotype III (A53 and D15) presented significant OR for an association with LOD (OR, 2.202 [CI95, 1.121 to 4.376] and 6.182 [CI95, 1.598 to 35.220], respectively), but only the latter association was significant when controlled for the FDR (P = 0.018). In agreement to what was found for the serotypes, none of these largest PFGE clusters was associated with recovery from blood or CSF.
Among sequence types, ST17 was the only genetic lineage defined by MLST that was overrepresented in LOD (OR, 4.972 [CI95, 2.659 to 9.488]). Of the two major serotype III/ST17 clusters defined by PFGE (A53 and D15), only the latter was significantly associated with LOD, suggesting that PFGE could distinguish between two ST17 lineages that are otherwise indistinguishable by their serotype, ST, and surface protein, but that could have other significant differences. This hypothesis remains to be studied further.
Antimicrobial susceptibility testing.
All isolates were susceptible to penicillin and levofloxacin. The overall rate of erythromycin resistance was 14.2% (n = 30). All erythromycin-resistant isolates displayed the constitutive MLSB phenotype (cMLSB), defined by cross-resistance to all macrolides, lincosamides, and streptogramin B. None of the erythromycin-resistant isolates carried the mef gene; the erm(B) gene was present in 76.7% (n = 23) and the erm(A) [erm(TR) subclass] gene in 23.3% (n = 7) of the isolates. Resistance to tetracycline was found in 89.2% (n = 189) of the isolates and resistance to chloramphenicol in 2.8% (n = 6).
While resistance to tetracycline or chloramphenicol was not clustered in particular serotypes, the same was not true for resistance to erythromycin (P < 0.001 by Fisher's exact test) (Table 2). Analysis of individual serotypes revealed that serotypes Ia (n = 2/47 [4.3%]) and III (n = 11/118 [9.3%]) presented less erythromycin resistance than expected (OR, 0.219 [CI95, 0.024 to 0.926] and 0.408 [CI95, 0.165 to 0.962], respectively), while serotypes II (n = 5/11 [45.5%]) and V (n = 9/14 [64.3%]) presented more erythromycin-resistant isolates than expected (OR, 5.788 [CI95, 1.297 to 24.7061] and 14.806 [CI95, 4.024 to 61.874], respectively), but only the finding for serotype V was significant when controlled for the FDR (P < 0.001).
Analysis of the largest PFGE clusters (n ≥ 10) revealed three clusters with significant associations with erythromycin resistance, clusters H38 (OR, 0 [CI95, 0 to 0.528]) and A53 (OR, 0.087 [CI95, 0.002 to 0.552]) with reduced resistance and cluster Q11 (OR, 21.092 [CI95, 4.645 to 131.992]) with higher resistance; all these associations remained significant when controlled for the FDR (P = 0.009 for both H38 and A53 and P < 0.001 for Q11). These results were not surprising. PFGE cluster Q11 includes the majority of serotype V isolates, all presenting with ST1, a sequence type that was also significantly associated with erythromycin resistance (OR, 12.589 [CI95, 3.303 to 53.599]). On the other hand, PFGE cluster H38 includes the majority of ST23/serotype Ia isolates, and cluster A53 represents the ST17/serotype III and related lineages. Both clusters were associated with erythromycin susceptibility, a result that had already been suggested by the serotype analysis but that had failed to reach statistical significance. However, the MLST analysis revealed that both ST23 and ST17 were significantly associated with erythromycin susceptibility (OR, 0 [CI95, 0 to 0.658] and 0.040 [CI95, 0.001 to 0.256], respectively), supporting the indication given by PFGE cluster analysis.
Even though ST19 was poorly represented in our collection, in agreement with its association with colonization (31), it was also overrepresented among erythromycin-resistant isolates (OR, 6.885 [CI95, 2.082 to 22.557]). This association was reported previously in Portugal (19).
Surface protein gene profiling.
All isolates gave positive results for the presence of only one surface protein gene, with the exception of one isolate for which we failed to amplify any of the genes tested. The surface protein gene rib was the most prevalent, followed by the eps, bca, and alp3 genes, showing variable distributions across serotypes (Table 3). No alp2 or alp4 genes were found among the isolates. There was a very high correspondence between serotypes and surface protein genes (AR, 0.789 [CI95, 0.713 to 0.867]). This was reflected in significant OR for the association of most serotypes (n ≥ 10) with particular surface protein genes: serotype Ia with eps, serotypes Ib and II with bca, serotype III with rib, and serotype V with alp3; all these associations remained significant when controlled for the FDR (P < 0.001) (Table 3).
Table 3.
Distribution of genes encoding surface proteins across serotypes
| Serotype | No. of isolates with the following surface protein genea: |
Total no. of isolates | |||
|---|---|---|---|---|---|
| bca | eps | rib | alp3 | ||
| Ia | 12 | 35 | 47 | ||
| Ib | 11 | 1 | 1 | 13 | |
| II | 7 | 1 | 2 | 1 | 11 |
| III | 2 | 116 | 118 | ||
| IV | 1 | 2 | 3 | ||
| V | 2 | 11 | 13c | ||
| NTb | 1 | 5 | 6 | ||
| Total | 34 | 41 | 123 | 13 | 211 |
Boldface numbers indicate a significant correlation between the surface protein gene and the serotype (see the text).
NT, nontypeable.
One isolate failed to amplify any of the surface proteins tested.
Surface protein genes were differently distributed across PFGE clusters (Fig. 2), correlating with the proportions of serotypes within the clusters. A major exception was serotype Ia isolates, which grouped in PFGE cluster H38, although they presented with surface protein gene eps or bca. In this cluster, an absolute association was found between the ST and the surface protein gene, with all ST23 isolates carrying the eps gene and all ST24 isolates carrying the bca gene exclusively, in support of our hypothesis that they constitute sublineages. In addition, we have also identified two isolates, one representing ST223 and the other representing the newly identified ST545, that are SLVs of ST23 but not of ST24, both harboring the eps gene, suggesting diversification of ST23 with retention of the characteristic surface protein gene.
DISCUSSION
This study comprises a considerable number of invasive GBS isolates collected over an 18-year period in the Barcelona region. In spite of the overall large number of isolates, the number of yearly infections was low, preventing detailed evaluation of the temporal changes in serotypes or PFGE clusters. Still, serotype III was present in all years of the study period, and serotype Ia was absent in only 2 years. The remaining serotypes were represented by fewer isolates than the number of years studied, but there was an overall correlation between the number of isolates and the number of years in which they were found (r = 0.964; P = 0.0028 [Spearman's test]), indicating that no significant changes in the serotypes causing neonatal infections occurred in Barcelona during the 18 years studied. A similar analysis by PFGE cluster is complicated by their larger number; still, each of the five largest clusters (n ≥ 10) was found in at least 7 years, and the two largest (A53 and H38) were found in all but 2 of the study years. Taken together, these data reveal a remarkably stable clonal structure of the GBS causing neonatal infections in Barcelona over a period of 18 years. This occurred in spite of the major epidemiological changes in neonatal GBS infections due to the introduction of prophylaxis in 1994, which resulted in a pronounced decline in EOD incidence (2).
We found substantial diversity among the GBS isolates causing neonatal invasive disease, not only in terms of capsular polysaccharides, but also in the genetic lineages defined by both PFGE and MLST (Fig. 2 and Table 2). However, most isolates belonged to two serotypes and to a few STs and major PFGE clusters. The serotype distribution found in the population was similar to that described in some European countries, where capsular types Ia and III prevail among isolates causing invasive neonatal infections (5, 36, 41). In contrast to some U.S. studies, where the prevalence of serotype V reached as high as 30% (42), and to the recent increase reported in the prevalence of this serotype in Scandinavia (4, 39), serotype V was found much less frequently in Barcelona, in agreement with the results of most studies across Europe.
Classification by PFGE and MLST defined groups of isolates frequently sharing the same serotype and surface protein gene, indicating that both techniques are identifying groups of closely related isolates. Strong correlations between genes encoding surface proteins and serotypes were also found (Table 3): most serotypes were associated primarily with a single surface protein gene. The exception was serotype Ia, which was associated with two proteins, although only one reached significance, with approximately one-quarter of the isolates carrying the bca gene and the remaining three-quarters carrying the eps gene. While our data are consistent with those reported elsewhere in Europe, contrasting data can be found in some studies from the United States reporting the absence of the bca surface protein gene in all serotype Ia isolates causing invasive neonatal infections (32). We believe that this is a defining characteristic of a sublineage of serotype Ia, as discussed below. Also in contrast to the data reported here, another study from the United States found the bca surface protein gene in more than 50% of serotype V isolates (50). Considering that most studies describe a strong association of serotype V with the alp3 gene (21, 29, 43), it is possible that the higher prevalence of serotype V in the United States than in Europe results from the expansion of a sublineage not found in Europe. This is despite the fact that most serotype V isolates on either continent share the same sequence type (ST1).
As more collections are analyzed by their complement of surface protein genes, a broader understanding of these genes' relationships with serotypes may be obtained. However, our data suggest that it may be naïve to expect an absolute correlation with serotype and that additional typing methods, such as PFGE or MLST, are potentially useful for identifying distinct genetic lineages within each serotype that may also differ in their surface proteins.
Of note in this context was the high prevalence of ST24 found among serotype Ia isolates, which concentrated mostly in PFGE cluster H38, together with the more widely disseminated ST23. ST24 is a DLV of ST23 that has rarely been found among large collections of GBS isolates characterized by MLST, with the exception of Portugal, where a significant prevalence of this ST was first reported in 2007 (36). Also in 2007, a study from Italy showed most serotype Ia isolates grouping together in the same PFGE cluster, presenting either the bca or the eps surface protein gene as described here, and the one representative isolate of this cluster characterized by MLST harbored the bca gene and belonged to ST24 (21). Later, ST24 was found in 3 out of 52 serotype Ia invasive isolates from neonates in the United States; these 3 isolates were reported as a rare invasive clone (8). This suggests that within serotype Ia there are two different sublineages not distinguishable by PFGE but discriminated by MLST and surface protein gene profiling. In agreement with this hypothesis is the absolute association between the ST and the surface protein gene in our collection, with all ST23 isolates carrying the eps gene and all ST24 isolates carrying the bca gene exclusively. Previous results from our laboratory had provided strong support for the circulation of both sublineages in Portugal (35). Taken together, these observations suggest that the presence of a particular alpha or alpha-like surface protein gene is a clonal property rather than a feature of the serotype. Interestingly, we have also found an additional ST24/bca isolate belonging to serotype III that may be the result of capsular switching (34).
This study, together with previous data from Portugal and Italy (21, 36), suggests that a particular sublineage of serotype Ia may be disseminated in the Mediterranean region. Although we could not find a correlation between each of the surface protein-defined sublineages and the isolate source (blood or CSF) or the timing of disease presentation (EOD or LOD), the ST24 sublineage may have other properties that could explain its success.
In spite of the recent report of penicillin nonsusceptibility among GBS (28) and the intensive use of beta-lactams in prophylaxis in Barcelona since 1994, all isolates were fully susceptible to penicillin. The macrolides can also be used in chemoprophylaxis, and in contrast to penicillin, a significant proportion of erythromycin resistance was found (14.2%), in line with previous results from a multicenter study in Spain (13.7%) (22). The phenotypes and genotypes of macrolide-resistant isolates from Barcelona were also similar to those previously identified in Spain (23). In agreement with other studies in different geographic regions, we found an association between macrolide resistance, serotype V (7, 53), and ST1 (44). Still, only 30% of erythromycin resistance was found in serotype V, while the remaining erythromycin resistance was dispersed in all other serotypes except serotype IV, a situation observed in a few previous studies (18, 30, 52). Resistance was also found to be dispersed in many genetic lineages as defined by PFGE and MLST, in agreement with the carriage of the methylase genes on mobile genetic elements. However, clonal expansion is also important in macrolide resistance; PFGE cluster Q11, expressing serotype V and ST1, was found to be significantly associated with resistance. On the other hand, cluster A53, representing the highly virulent ST17 and associated lineages, was less resistant than expected, reflecting the general observation that isolates representing ST17 are rarely macrolide resistant. The reasons why this highly virulent and successful lineage is seldom resistant are unknown.
The characterization of the population of GBS causing invasive infections in neonates in the Barcelona region revealed the existence of a large number of genetically distinct lineages that were present over a significant time span. The stability and dominance of a few lineages that are responsible for the majority of infections in spite of continuous antibiotic and immune selective pressures suggest that they are extremely well adapted to their particular niche. Although most of these lineages are widely disseminated worldwide, we have also identified seemingly regionally successful clones, raising the possibility of ongoing selection and expansion of specific virulent GBS clones. Continuous surveillance will shed further light on these processes and will determine if these clones will expand beyond their current geographical boundaries.
ACKNOWLEDGMENTS
This work was partly supported by a grant from the Fundação Calouste Gulbenkian, by the Fundação para a Ciência e a Tecnologia (POCI/SAU-ESP/57646/2004), and by an unrestricted grant from Glaxo SmithKline Portugal. E.R.M. was supported by a grant from the Fundação para a Ciência e a Tecnologia (SFRH/BD/41761/2007).
Contributing members of The Microbiologist Group for the Study of Vertical Transmission Infections are M. Gimenez, Hospital Germans Trias i Pujol, Badalona; M. Sierra, Hospital Barcelona-SCIAS, Barcelona; J. Lite, Hospital Mutua Terrassa, Terrassa; I. Sanfeliu, Coorporació Parc Taulí, Sabadell; F. Sanchez, Hospital de Sant Pau, Barcelona; E. Dopico, Laboratori Clinic l'Hospitalet, Hospitalet; and C. Guardià, Laboratori Clinic Barcelones Nord i Valles Oriental, Badalona.
Footnotes
Published ahead of print on 22 June 2011.
REFERENCES
- 1. Altman D. G. 1999. Practical statistics for medical research. Chapman & Hall/CRC, Boca Raton, Fla [Google Scholar]
- 2. Andreu A., et al. 2003. Decreasing incidence of perinatal group B streptococcal disease (Barcelona 1994-2002). Relation with hospital prevention policies. Enferm. Infecc. Microbiol. Clin. 21:174–179 (In Spanish) [DOI] [PubMed] [Google Scholar]
- 3. Benjamini Y., Hochberg Y. 1995. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodological) 57: 289–300 [Google Scholar]
- 4. Bergseng H., et al. 2009. Molecular and phenotypic characterization of invasive group B streptococcus strains from infants in Norway 2006-2007. Clin. Microbiol. Infect. 15: 1182–1185 [DOI] [PubMed] [Google Scholar]
- 5. Bergseng H., Rygg M., Bevanger L., Bergh K. 2008. Invasive group B streptococcus (GBS) disease in Norway 1996-2006. Eur. J. Clin. Microbiol. Infect. Dis. 27: 1193–1199 [DOI] [PubMed] [Google Scholar]
- 6. Bidet P., Brahimi N., Chalas C., Aujard Y., Bingen E. 2003. Molecular characterization of serotype III group B-streptococcus isolates causing neonatal meningitis. J. Infect. Dis. 188: 1132–1137 [DOI] [PubMed] [Google Scholar]
- 7. Blumberg H. M., et al. 1996. Invasive group B streptococcal disease: the emergence of serotype V. J. Infect. Dis. 173: 365–373 [DOI] [PubMed] [Google Scholar]
- 8. Bohnsack J. F., et al. 2008. Population structure of invasive and colonizing strains of Streptococcus agalactiae from neonates of six U.S. academic centers from 1995 to 1999. J. Clin. Microbiol. 46: 1285–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Carriço J. A., et al. 2006. Illustration of a common framework for relating multiple typing methods by application to macrolide-resistant Streptococcus pyogenes. J. Clin. Microbiol. 44: 2524–2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Centers for Disease Control and Prevention 2005. Early-onset and late-onset neonatal group B streptococcal disease—United States, 1996-2004. MMWR Morb. Mortal. Wkly. Rep. 54: 1205–1208 [PubMed] [Google Scholar]
- 11. Centers for Disease Control and Prevention 2002. Prevention of perinatal group B streptococcal disease. Revised guidelines from CDC. MMWR Recommend. Rep. 51: 1–22 [PubMed] [Google Scholar]
- 12. Centers for Disease Control and Prevention 1996. Prevention of perinatal group B streptococcal disease: a public health perspective. MMWR Recommend. Rep. 45: 1–24 [PubMed] [Google Scholar]
- 13. Clinical and Laboratory Standards Institute 2009. Performance standards for antimicrobial susceptibility testing, nineteenth informational supplement. CLSI document M100-S19. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 14. Creti R., Fabretti F., Orefici G., von Hunolstein C. 2004. Multiplex PCR assay for direct identification of group B streptococcal alpha-protein-like protein genes. J. Clin. Microbiol. 42: 1326–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Davies H. D., et al. 2004. Multilocus sequence typing of serotype III group B streptococcus and correlation with pathogenic potential. J. Infect. Dis. 189: 1097–1102 [DOI] [PubMed] [Google Scholar]
- 16. de la Rosa Fraile M., Cabero L., Andreu A., Rao G. G. 2001. Prevention of group B streptococcal neonatal disease: a plea for a European consensus. Clin. Microbiol. Infect. 7: 25–27 [DOI] [PubMed] [Google Scholar]
- 17. Figueira-Coelho J., Ramirez M., Salgado M. J., Melo-Cristino J. 2004. Streptococcus agalactiae in a large Portuguese teaching hospital: antimicrobial susceptibility, serotype distribution, and clonal analysis of macrolide-resistant isolates. Microb. Drug Resist. 10: 31–36 [DOI] [PubMed] [Google Scholar]
- 18. Fitoussi F., et al. 2001. Mechanisms of macrolide resistance in clinical group B streptococci isolated in France. Antimicrob. Agents Chemother. 45: 1889–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Florindo C., et al. 2010. Molecular characterization and antimicrobial susceptibility profiles in Streptococcus agalactiae colonizing strains: association of erythromycin resistance with subtype III-1 genetic clone family. Clin. Microbiol. Infect. 16: 1458–1463 [DOI] [PubMed] [Google Scholar]
- 20. Francisco A. P., Bugalho M., Ramirez M., Carriço J. A. 2009. Global optimal eBURST analysis of multilocus typing data using a graphic matroid approach. BMC Bioinformatics 10: 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gherardi G., et al. 2007. Molecular epidemiology and distribution of serotypes, surface proteins, and antibiotic resistance among group B streptococci in Italy. J. Clin. Microbiol. 45: 2909–2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. González J. J., et al. 2004. Sensibilidad a antimicrobianos del estreptococo del grupo B de transmisión vertical. Estudio multicéntrico. Enferm. Infecc. Microbiol. Clin. 22: 286–291 [DOI] [PubMed] [Google Scholar]
- 23. González J. J., Andreu A., the Spanish Group for the Study of Perinatal Infection from the Spanish Society for Clinical Microbiology and Infectious Diseases 2005. Multicenter study of the mechanisms of resistance and clonal relationships of Streptococcus agalactiae isolates resistant to macrolides, lincosamides, and ketolides in Spain. Antimicrob. Agents Chemother. 49: 2525–2527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hansen S. M., Uldbjerg N., Kilian M., Sorensen U. B. 2004. Dynamics of Streptococcus agalactiae colonization in women during and after pregnancy and in their infants. J. Clin. Microbiol. 42: 83–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Holt D. E., Halket S., de Louvois J., Harvey D. 2001. Neonatal meningitis in England and Wales: 10 years on. Arch. Dis. Child. Fetal Neonatal Ed. 84: F85–F89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Johri A. K., et al. 2006. Group B streptococcus: global incidence and vaccine development. Nat. Rev. Microbiol. 4: 932–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jones N., et al. 2003. Multilocus sequence typing system for group B streptococcus. J. Clin. Microbiol. 41: 2530–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kimura K., et al. 2008. First molecular characterization of group B streptococci with reduced penicillin susceptibility. Antimicrob. Agents Chemother. 52: 2890–2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kong F., Gowan S., Martin D., James G., Gilbert G. L. 2002. Serotype identification of group B streptococci by PCR and sequencing. J. Clin. Microbiol. 40: 216–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lin F. Y., et al. 2000. Antibiotic susceptibility profiles for group B streptococci isolated from neonates, 1995-1998. Clin. Infect. Dis. 31: 76–79 [DOI] [PubMed] [Google Scholar]
- 31. Lin F. Y., et al. 2006. Phylogenetic lineages of invasive and colonizing strains of serotype III group B streptococci from neonates: a multicenter prospective study. J. Clin. Microbiol. 44: 1257–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Manning S. D., et al. 2006. The frequency of genes encoding three putative group B streptococcal virulence factors among invasive and colonizing isolates. BMC Infect. Dis. 6: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Margarit I., et al. 2009. Preventing bacterial infections with pilus-based vaccines: the group B streptococcus paradigm. J. Infect. Dis. 199: 108–115 [DOI] [PubMed] [Google Scholar]
- 34. Martins E. R., Melo-Cristino J., Ramirez M. 2010. Evidence for rare capsular switching in Streptococcus agalactiae. J. Bacteriol. 192: 1361–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Martins E. R., Melo-Cristino J., Ramirez M. 2008. Characterization of a Streptococcus agalactiae serotype Ia lineage with enhanced invasiveness. Clin. Microbiol. Infect. 14: S244 [Google Scholar]
- 36. Martins E. R., Pessanha M. A., Ramirez M., Melo-Cristino J., the Portuguese Group for the Study of Streptococcal Infections 2007. Analysis of group B streptococcal isolates from infants and pregnant women in Portugal revealing two lineages with enhanced invasiveness. J. Clin. Microbiol. 45: 3224–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Melchers W. J., et al. 2003. Genetic analysis of Streptococcus agalactiae strains isolated from neonates and their mothers. FEMS Immunol. Med. Microbiol. 36: 111–113 [DOI] [PubMed] [Google Scholar]
- 38. Melo-Cristino J., Fernandes M. L., The Portuguese Surveillance Group for the Study of Respiratory Pathogens 1999. Streptococcus pyogenes isolated in Portugal: macrolide resistance phenotypes and correlation with T types. Microb. Drug Resist. 5: 219–225 [DOI] [PubMed] [Google Scholar]
- 39. Persson E., et al. 2004. Serotypes and clinical manifestations of invasive group B streptococcal infections in western Sweden 1998-2001. Clin. Microbiol. Infect. 10: 791–796 [DOI] [PubMed] [Google Scholar]
- 40. Pinto F. R., Melo-Cristino J., Ramirez M. 2008. A confidence interval for the Wallace coefficient of concordance and its application to microbial typing methods. PLoS One 3: e3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Poyart C., et al. 2008. Invasive group B streptococcal infections in infants, France. Emerg. Infect. Dis. 14: 1647–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Puopolo K. M., Hollingshead S. K., Carey V. J., Madoff L. C. 2001. Tandem repeat deletion in the alpha C protein of group B streptococcus is recA independent. Infect. Immun. 69: 5037–5045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Radtke A., et al. 2009. Identification of surface proteins of group B streptococci: serotyping versus genotyping. J. Microbiol. Methods 78: 363–365 [DOI] [PubMed] [Google Scholar]
- 44. Sadowy E., Matynia B., Hryniewicz W. 2010. Population structure, virulence factors and resistance determinants of invasive, non-invasive and colonizing Streptococcus agalactiae in Poland. J. Antimicrob. Chemother. 65: 1907–1914 [DOI] [PubMed] [Google Scholar]
- 45. Schuchat A. 1998. Epidemiology of group B streptococcal disease in the United States: shifting paradigms. Clin. Microbiol. Rev. 11: 497–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schuchat A. 1999. Group B streptococcus. Lancet 353: 51–56 [DOI] [PubMed] [Google Scholar]
- 47. Severiano A., Carriço J. A., Robinson D. A., Ramirez M., Pinto F. R. 2011. Evaluation of jackknife and bootstrap for defining confidence intervals for pairwise agreement measures. PLoS One 6: e19539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Silva-Costa C., Ramirez M., Melo-Cristino J. 2006. Identification of macrolide-resistant clones of Streptococcus pyogenes in Portugal. Clin. Microbiol. Infect. 12: 513–518 [DOI] [PubMed] [Google Scholar]
- 49. Sinha A., Lieu T. A., Paoletti L. C., Weinstein M. C., Platt R. 2005. The projected health benefits of maternal group B streptococcal vaccination in the era of chemoprophylaxis. Vaccine 23: 3187–3195 [DOI] [PubMed] [Google Scholar]
- 50. Smith T. C., et al. 2007. Distribution of novel and previously investigated virulence genes in colonizing and invasive isolates of Streptococcus agalactiae. Epidemiol. Infect. 135: 1046–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tsolia M., et al. 2003. Group B streptococcus colonization of Greek pregnant women and neonates: prevalence, risk factors and serotypes. Clin. Microbiol. Infect. 9: 832–838 [DOI] [PubMed] [Google Scholar]
- 52. Uh Y., et al. 2004. Serotypes and genotypes of erythromycin-resistant group B streptococci in Korea. J. Clin. Microbiol. 42: 3306–3308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. von Both U., et al. 2003. A serotype V clone is predominant among erythromycin-resistant Streptococcus agalactiae isolates in a southwestern region of Germany. J. Clin. Microbiol. 41: 2166–2169 [DOI] [PMC free article] [PubMed] [Google Scholar]