Abstract
We aimed to compare conventional ompA typing of Chlamydia trachomatis with multilocus sequence typing (MLST) and multilocus variable-number tandem-repeat (VNTR) analysis (MLVA). Previously used MLST and MLVA systems were compared to modified versions that used shorter target regions and nested PCR. Heterosexual couples were selected from among persons with urogenital C. trachomatis infections visiting the sexually transmitted infection outpatient clinic in Amsterdam, The Netherlands. We identified 30 couples with a total of 65 C. trachomatis-positive samples on which MLST and MLVA for eight target regions were performed. All regions were successfully sequenced in 52 samples, resulting in a complete profile for 18 couples and 12 individuals. Nine ompA genovars from D to K, with two variants of genovar G, were found. The numbers of sequence type and MLVA type profiles were 20 for MLST and 21 for MLVA, and a combination of MLST and MLVA yielded 28 profiles, with discriminatory indexes (D) ranging from 0.95 to 0.99. Partners in 17 couples shared identical profiles, while partners in 1 couple had completely different profiles. Three persons had infections at multiple anatomical locations, and within each of these three individuals, all profiles were identical. The discriminatory capacity of all MLST and MLVA methods is much higher than that of ompA genotyping (D = 0.78). No genotype variation was found within the samples of the same person or from heterosexual couples with a putative single transmission. This shows that the chlamydial genome in clinical specimens has an appropriate polymorphism to enable epidemiological cluster analysis using MLST and MLVA.
INTRODUCTION
In theory, urogenitally transmitted Chlamydia trachomatis seems to be an ideal target for molecular epidemiology, as it is the most prevalent sexually transmitted bacterial infection worldwide (25). C. trachomatis is present within the general population, because it is able to cause asymptomatic infections. The majority of the infections, however, occur within the transmission networks of specific risk groups, such as adolescents and men who have sex with men (MSM) (1, 15). Previous studies have shown that C. trachomatis has 17 distinct serovars, based on the antigenic properties of the major outer membrane protein (MOMP) (18), and even more genetic variants of the coding ompA gene (4). These qualities suggest that linking chlamydial types and human sexual behavior might be straightforward.
In practice however, there are difficulties. Although a considerable amount of antigenic variation between genovars exists, the genome of C. trachomatis is actually highly conserved, possibly due to its obligate intracellular life cycle (23). Within one genovar, very little variation exists within the different ompA sequences, and epidemiologically distinct risk groups have identical ompA genovars (14). More problematic is that C. trachomatis has a nearly identical distribution of genovars in most populations which seems to be independent of host risk group, geography, or calendar time (7, 14, 15, 17, 21). It has been postulated that C. trachomatis is an ancient infection that coevolved with humankind and has stabilized over time (23). This distribution is dominated by three genovars (D, E, and F), which comprise about 70% of the sexually transmitted infections (STIs), making it difficult to follow transmission patterns accurately (7, 14, 17, 21). The exception to this is the distribution of genovars among MSM, for about 85% of whom the genovars are D, G, and J (18, 22). This might be a result of different transmission dynamics, but biological differences between anogenitally transmitted and urogenitally transmitted strains are another possible explanation (10). The sensitivity of typing methods for C. trachomatis has always been problematic (18). Due to the obligate intracellular life cycle of the organism, C. trachomatis is difficult to culture, making this technique challenging for use in molecular epidemiological studies. Clinical samples might also be difficult for use for molecular typing, as they contain low bacterial loads. The samples might also suffer from interference with both human DNA and DNA from numerous other microorganisms, as C. trachomatis resides in niches with a dense microflora.
Numerous studies have shown and confirmed the stability of the distribution of ompA genovars, but very few studies have revealed the real dynamic properties of urogenital C. trachomatis strains of genovars D to K on a strain level. So far, only one clear case of a clonal outbreak has been described, being the new variant (nv) C. trachomatis outbreak in Sweden (6). The nv C. trachomatis outbreak was caused by a chlamydial strain that could not be detected by two commonly used diagnostic systems and had a prevalence among the Swedish population ranging from 10% to 65% in different counties. Later, it was identified to be a clonal outbreak by higher-resolution typing methods (6, 20). In Örebro County, Sweden, nv C. trachomatis comprised 41% of all C. trachomatis-positive samples in 2006, while no samples were found in a panel of 237 C. trachomatis-positive samples from 1999 to 2000. Although the outbreak was apparent, it was largely concealed when it was analyzed on an ompA level, as the proportion of genovar E samples (the genovar of nv C. trachomatis) in Örebro County rose from only 47% in 1999 and 2000 to 69% in 2006 (11). This shows that the data from conventional ompA typing might suggest a distribution of circulating strains that is too static, while higher-resolution typing methods might give a more dynamic view.
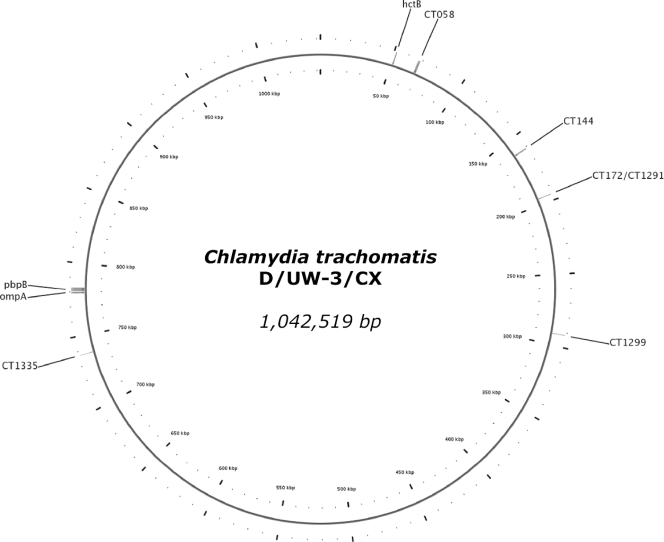
Although various genotyping methods are available, only two published methods demonstrated the degree of resolution needed for molecular epidemiological studies. In 2007, Klint et al. published a multilocus sequence typing (MLST) method for C. trachomatis that included five variable regions: hctB, CT058, CT144, CT172, and pbpB (Fig. 1) (12). A second technique, a multilocus variable-number tandem-repeat (VNTR) analysis (MLVA) published by Pedersen et al. in 2008, combined ompA with three highly variable single repeats: CT1291, CT1299, and CT1335 (Fig. 1) (20). Both techniques were evaluated by Ikryannikova et al. and showed a satisfyingly high discriminatory power for cultured samples (9). A drawback of the published techniques is that they both consist of single PCRs, which might have a decreased sensitivity with clinical samples tested directly. This can be improved by adapting the techniques to nested assays, as Lan et al. already showed in 1993 (13). Another drawback of the published MLST method is that some of the regions are over 1,500 bp in length and need multiple internal primers for sequencing. It is more convenient to use regions up to approximately 700 bp in length, because the sensitivity of the typing PCR is higher for smaller fragments, fewer PCRs and sequence reactions are needed, and the sequences are easier to assemble.
Fig. 1.
Graphical representation of the circular genome of Chlamydia trachomatis (reference strain D/UW-3/CX). Indicated are the locations of the regions used by Klint et al. (12) (hctB, CT058, CT144, CT172, pbpB, and ompA) and Pedersen et al. (20) (CT1291, CT1299, CT1335, and ompA). As can be seen, CT172 and CT1291 concern the same region.
In this study, we aimed to adapt the published MLST and MLVA to nested assays of more appropriate lengths to be more suitable for clinical samples. We assessed both the minimal variation and the resolution of these novel high-resolution typing methods compared with ompA sequencing. We also aimed to compare our modified MLST with the original method published by Klint et al. (12), as that method contains some sequence information that is missed by ours. To test whether the novel typing methods are useful for molecular epidemiological research, a panel of samples from C. trachomatis-infected heterosexual couples was selected from the STI outpatient clinic in Amsterdam, The Netherlands. The study investigated which of the methods is most suitable for molecular epidemiological analysis of C. trachomatis transmission patterns in sexual networks.
MATERIALS AND METHODS
Selection of couples.
Putative heterosexual couples were selected retrospectively from people who visited the STI outpatient clinic of the Public Health Service of Amsterdam between January 2008 and April 2009 and who were diagnosed with a urogenital C. trachomatis infection. Next to urogenital samples, additional samples were occasionally taken from the patients, depending on sexual risk behavior, clinical symptoms, and complaints associated with chlamydial infections and other STIs. For inclusion, the partners within the couples had to have a concomitant urogenital C. trachomatis infection, have had heterosexual contacts in the last 6 months, share a postal address, differ by not more than 20 years in age, have different family names to avoid selecting, e.g., brother/sister couples, have visited the outpatient clinic within 5 weeks of their partner's visit, and not to have had high-risk sexual behavior (being paid for sex or having had more than two sexual partners in the preceding 6 months). Thirty putative heterosexual couples that met these criteria were selected.
All patients attending the Amsterdam STI clinic are notified during their visit that routinely collected data and samples may be used, after they are made anonymous, for scientific research. This study used only routinely collected data and samples, and no specific consent was requested. All identifer patient data (including name, address, date of birth, date of visit, and clinic registration number) were handled by qualified nurses and data managers, and none of the researchers had access to these data.
Selection of Chlamydia-positive samples.
All samples of the selected couples were tested by routine testing with the Aptima Combo 2 assay (Gen-Probe, San Diego, CA). The included C. trachomatis-positive samples consisted of a urine sample (males) and a cervical or vaginal swab sample (females) for every couple. Three persons had two infected anatomical locations (cervix and rectum) and one person had three infected locations (urethra and both eyes), which resulted in a total of 65 C. trachomatis-positive samples. The samples were stored at −20°C until use.
DNA extraction.
DNA was extracted at the Public Health Service from 200 μl swab- or urine-containing transport medium (Gen-Probe) by adding 500 μl lysis buffer (bioMérieux, Boxtel, The Netherlands), 1 μl glycogen (20 mg/ml; Roche Diagnostics, Almere, The Netherlands), and 700 μl ice-cold isopropanol. The precipitate was washed twice with 70% ethanol and dissolved in 50 μl 10 mM Tris buffer (pH 8.0). These DNA isolates were stored at −20°C until use.
Primer selection for modification of MLST and MLVA.
MLST regions were analyzed using the Uppsala, Sweden, C. trachomatis MLST database (http://mlstdb.bmc.uu.se/3) and the full genomes accessed in GenBank (genovar A, GenBank accession no. CP000051; genovar B, GenBank accession nos. FM872308 and FM872307; genovar D, GenBank accession nos. AE001273 and ACFJ01000001 genovar F, GenBank accession no. ABYF01000001; genovar J, GenBank accession no. ABYD01000001; genovar L2, GenBank accession no. AM884176; and genovar L2b, GenBank accession no. AM884177) between March 2009 and May 2010. The locations of the regions used are shown in Fig. 1.
Nested PCR and sequencing for MLST and MLVA regions (modified protocol).
The DNA isolates were amplified by a nested PCR for the regions ompA (variable domains 1 and 2), hctB, CT058, CT144, CT172, pbpB, CT1299, and CT1335 using a C1000 PCR machine (Bio-Rad, Veenendaal, The Netherlands). The outer PCR was performed in a volume of 25 μl containing 2 μl of extracted DNA, 0.63 U GoTaq polymerase (Promega, Leiden, The Netherlands), 2 mM MgCl2, 25 μM each deoxynucleoside triphosphate, and 800 ng/μl of each specific outer primer (Table 1). The inner PCR was also performed in a volume of 25 μl containing the same components as the outer assay but with 2 μl of the outer amplicon as the target. Cycling conditions were an initial step at 94°C for 3 min, followed by 35 cycles for the outer PCR and 30 cycles for the inner PCR and a final step at 72°C for 5 min. The cycles consisted of 30 s at 93°C, 30 s at 57°C, and 1 min at 72°C. The amplified DNA was cleaned with ExoSAP-IT reagent (USB, Staufen, Germany) and sequenced in both directions with an ABI BigDye Terminator (version 1.1) kit (Applied Biosystems, Nieuwerkerk a.d. IJssel, The Netherlands), using the primers from the inner PCRs. Finally, the labeled DNA was purified using an ABI BigDye XTerminator kit (Applied Biosystems) and analyzed in an ABI 3130 genetic analyzer (Applied Biosystems).
Table 1.
Primers used for MLST and MLVA methods for Chlamydia trachomatisa
| Region | Format | Direction | Primer | Sequence (5′ to 3′) | 5′ position | Fragment length (bp)h |
|---|---|---|---|---|---|---|
| ompA (modified) | Outer | Forward | ompA OF | ATGAAAAAACTCTTGAAATCGGT | 780060 | 575–584 |
| Outer | Reverse | ompA OR | TTAGAAGCGGAATTGTGCAT | 778879 | ||
| Inner + Seq | Forward | ompA NF | CGCTTTGAGTTCTGCTTCCT | 780025 | ||
| Inner + Seq | Reverse | OMP6ASb | TGAGCGTATTGGAAAGAAGC | 779411 | ||
| hctB (modified) | Outer | Forward | Hctb39Fc | CTCGAAGACAATCCAGTAGCAT | 51219 | 624–733 |
| Outer | Reverse | Hctb794Rc | CACCAGAAGCAGCTACACGT | 52014 | ||
| Inner + Seq | Forward | CT046 NF | AACTCCAGCTTTTACTGCTA | 51959 | ||
| Inner + Seq | Reverse | CT046 NR3 | CCCCAAATATGCAACAGGAT | 51296 | ||
| CT058 (modified) | Outer | Forward | CT222 Fc | CTTTTCTGAGGCTGAGTATGATTT | 68713 | 600–601 |
| Outer | Reverse | CT1678 Rc | CCGATTCTTACTGGGAGGGT | 67215 | ||
| Inner + Seq | Forward | CT058 NF | AGGTGGCTGCGTTAAGATAACT | 68614 | ||
| Inner + Seq | Reverse | CT058 NR | AAATTGGCCTGAAGTAGAGACA | 67992 | ||
| CT058 (nested/original) | Outer | Forward | CT058 OF | TGTGGGACTTGCGTGATT | 68763 | 1455–1456 |
| Outer | Reverse | CT058 OR | TAACCGTTCCATCCACATCT | 67132 | ||
| Inner + Seq | Forward | CT222 Fc | CTTTTCTGAGGCTGAGTATGATTT | 68713 | ||
| Inner + Seq | Reverse | CT1678 Rc | CCGATTCTTACTGGGAGGGT | 67215 | ||
| Seq | Forward | CT811Fc,e | CGATAAGACAGATGCCGTTTTT | 68122 | ||
| Seq | Reverse | CT1022Rc, e | TAAGCACAGCAGGGAATGCA | 67871 | ||
| Seq | Forward | CT058 IFe | TGTCTCTACTTCAGGCCAATTT | 68013 | ||
| Seq | Reverse | CT058 IRe | AATCCTCCTTGGCCTCTCTT | 67903 | ||
| CT144 (modified) | Outer | Forward | CT144:248Fc | ATGATTAACGTGATTTGGTTTCCTT | 160641 | 443 |
| Outer | Reverse | CT144:1046Rc | GCGCACCAAAACATAGGTACT | 161439 | ||
| Inner + Seq | Forward | CT144 NF | CGAAATCGGATATCTCTTTT | 160920 | ||
| Inner + Seq | Reverse | CT144 NR | CCTAAACATACGGCTATTCC | 161400 | ||
| CT144 (nested/original) | Outer | Forward | CT144 OF | TCTATTGGGAATGAGCATCCT | 160575 | 753 |
| Outer | Reverse | CT144 OR | TTCGCTCTCCCACAATCA | 161495 | ||
| Inner + Seq | Forward | CT144:248Fc | ATGATTAACGTGATTTGGTTTCCTT | 160641 | ||
| Inner + Seq | Reverse | CT144:1046Rc | GCGCACCAAAACATAGGTACT | 161439 | ||
| CT172 (modified) | Outer | Forward | Four610 Rc | CGTCATTGCTTGCTCGGCTT | 195301 | 358–666 |
| Outer | Reverse | CT172 OR | GATCAAGCCATCTTAGACATGC | 195798 | ||
| Inner + Seq | Forward | CT172 NF | AGGTCGCCCAAATTCCATGT | 195376 | ||
| Inner + Seq | Reverse | CT172 NR | GCTCCGGCTATTTTGTTTAGGA | 195778 | ||
| pbpB (modified) | Outer | Forward | pbpB 1 Fc | TATATGAAAAGAAAACGACGCACC | 780665 | 602 |
| Outer | Reverse | pbpB 2366 Rc | TGGTCAGAAAGATGCTGCACA | 783030 | ||
| Inner + Seq | Forward | CT682 NF | TCATCACTTTGCGTATATGGCA | 780750 | ||
| Inner + Seq | Reverse | CT682 NR | AAAAGCTTGCGTACTTGATCGA | 781395 | ||
| pbpB (nested/original) | Outer | Forward | pbpB1 OF | TTGTGTTTGGAATAGCTCGAA | 780508 | 789 + 875 |
| Outer | Reverse | pbpB1 OR | AAGAACCTTCCATCTCCTGAAT | 781550 | ||
| Outer | Forward | pbpB2 OF | GGAACGATCGAGCAGCTT | 782060 | ||
| Outer | Reverse | pbpB2 OR | AGAAGCAATAGGAGAGCCGT | 783136 | ||
| Inner + Seq | Forward | pbpB 1 Fc | TATATGAAAAGAAAACGACGCACC | 780665 | ||
| Inner + Seq | Reverse | pbpB 823 Rc | CAGCATAGATCGCTTGCCTAT | 781507 | ||
| Inner + Seq | Forward | pbpB 1455 Fc | GGTCTCGTTTTTGATGTTCTATTC | 782111 | ||
| Inner + Seq | Reverse | pbpB 2366 Rc | TGGTCAGAAAGATGCTGCACA | 783030 | ||
| Seq | Forward | pbpB2 IFf | CCCAACCCCTTATGTGGA | 782545 | ||
| Seq | Reverse | pbpB2 IRf | TGGCGCCAACAAGCAAT | 782700 | ||
| CT1299 (modified) | Outer | Forward | CT1299 OF2 | CAACAATCATCACGCCCTCT | 291598 | 240–248g |
| Outer | Reverse | CT1299 OR | AGCCGCTTTCTCGTTCTAAA | 292017 | ||
| Inner + Seq | Forward | CT1299 OF | CGCTTAAGATTCTCGGAGGTA | 291656 | ||
| Inner + Seq | Reverse | CT1299 reversed | AAGTCCACGTTGTCATTGTACG | 291945 | ||
| CT1335 (modified) | Outer | Forward | CT1335 OF2 | AGTGGGTGTGAAGAACCGTA | 737151 | 191-195g |
| Outer | Reverse | CT1335 OR2 | ACCAAACCCTTTTGCAGGAA | 737463 | ||
| Inner + Seq | Forward | CT1335 OF | CGTCCTCTGGAAGGGAATAA | 737202 | ||
| Inner + Seq | Reverse | CT1335 OR | TATGCCCCAAGGAAGAGTCA | 737434 |
The regions given in boldface were used in the modified methods. For the original MLST, different PCRs were used for the regions CT058 and CT144, which are given in italics. Nested PCRs were performed for each region. The primers used for the outer PCRs are indicated with Outer, those used for the inner PCRs are indicated with Inner, and those used for sequencing are indicated with Seq. The original MLST made use only of the inner PCRs in the targets CT058, CT144, and pbpB. Unless indicated otherwise, the primers were newly designed. Positions are given in base pairs relative to the sequence of reference strain D/UW-3/CX (GenBank accession no. AE001273).
Primers were taken from Morré et al. (16).
Primers were taken from Klint et al. (12).
Primers were taken from Pedersen et al. (20).
For original CT058, the primers CT811F and CT1022R were used for sequencing. In the nested version, these were replaced by CT058 IF and CT058 IR.
The additional primers pbpB2 IF and pbpB2 IR were used only in the nested version.
The fragment length indicated could not always be determined directly but could be calculated using reference sequences.
Fragment length is the number of base pairs sequenced with the primers excluded.
Original MLST.
The original MLST was performed on three target regions (CT058, CT144, pbpB) that had not been fully covered by the modified MLST. Amplification and sequencing were performed as described by Jurstrand et al. (11).
Sequencing analysis.
The obtained sequences were analyzed using BioEdit software (http://www.mbio.ncsu.edu/bioedit/bioedit.html). The sequences were assembled and trimmed using the ClustalW (MEGA4) program (http://www.megasoftware.net/). Minimum-spanning trees were generated with BioNumerics software (Applied Maths, Sint-Martens-Latem, Belgium) using the generated MLST profiles.
Statistical analysis.
The discriminatory power of each typing method was calculated using Hunter and Gaston's modification of Simpson's index of diversity (8). The formula used to define Simpson's index of diversity (D) is
where N is the number of unrelated strains tested, s is the number of different types, and xj is the number of strains belonging to the jth type. The D value is between 0 and 1, and new typing methods with values of 0.95 or higher are considered highly suitable for molecular typing (24).
RESULTS
Thirty couples met the selection criteria. The 30 men had a median age of 26 years (range, 20 to 41 years), and the 30 women had a median age of 25 years (range, 19 to 42 years). We were able to obtain the complete sequences from ompA and the complete profiles for the modified MLST, modified MLVA, and the (nested) original MLST for 52 of the 65 samples (80%). For the remaining 13 samples, 4 had only partial sequences or profiles and 9 tested negative for all typing regions. Among these 9 negative samples were no complete sets of samples for couples. All 13 samples were excluded from further analyses. For 18 couples, we obtained the complete profiles for the samples from at least one of the anatomical locations tested for each partner. For the other 12 couples, we obtained complete profiles for one partner only. The samples from the 18 couples consisted of 36 urogenital samples and 4 additional samples from patients infected at more than one anatomical location. These 40 samples are further referred to as panel A.
Variation in ompA in putative single transmission.
We first analyzed panel A with ompA genotyping, as this is the reference typing technique (Table 1). In 17 of the 18 couples, the partners had identical sequences for ompA. For the three patients who contributed more than one sample, all ompA sequences within one patient were identical. The genovars in the samples from partners of one couple were discordant, with a genovar D infection in the male partner and a genovar E in the female partner. This degree of variation is impossible to have arisen by mutation during a single transmission. As these patients met our inclusion criteria, we considered that they were a couple but that the two infections must have had different origins.
Discriminating capacity of ompA sequencing.
The data obtained by ompA genotyping on panel A (together with the data of the following analyses on panel A) were used to construct a panel of epidemiologically unrelated samples. In this panel, all the couples in which the partners had concordant types all contributed one sample (n = 17) and the couple in which the partners had discordant types contributed two (n = 2). The couples for whom the sample(s) from only one of the partners could be sequenced added one sample each to this panel (n = 12). These 31 epidemiologically unrelated samples are further referred to as panel B. In this panel, we found 9 different ompA sequences: D, E, F, G variant 1 (Gvar1), Gvar2, H, Ia, Ja, and K. The genovar distribution was similar to the distributions reported in other populations, as we found 74% (23/31) to be infected with one of the most predominant genovars, D (n = 3), E (n = 13), or F (n = 7). This resulted in a rather low Hunter-Gaston D of 0.78, indicating an unsatisfactory resolution for molecular epidemiological studies (8, 24).
Design of nested MLST and MLVA.
To increase the sensitivity of the published MLST and MLVA methods, nested assays were designed for all regions (Table 1). Inner primers for the modified MLST were designed in such a way that the eight regions varied in length between 358 bp and 733 bp and that a minimum of sequence information was lost compared to the original regions of the MLST (Table 1; Fig. 2; see the data in the supplemental material). From the 96 sequence types (STs) in the Uppsala C. trachomatis MLST database that were published by September 2009, 89 could still be distinguished in silico. The MLVA regions were extended for better reads and assembling, so that the repeat region was at least 70 bp away from the start of the repeat sequence. The outer primers used in the assays were either the original primers from the papers by Klint et al. (12) and Pedersen et al. (20) or newly designed primers (Table 1). As region CT172 from the MLST and region CT1291 from the MLVA are largely overlapping, they were integrated into one assay and are referred to as CT172, from which the results for CT1291 can be derived (Fig. 2).
Fig. 2.
Graphical representation of region CT172/CT1291. Indicated are the positions of regions on the genome. The upper representation is the modified version. It is designed in such a manner that it includes all significant variation of the original assay of region CT172 of Klint et al. (12). It also completely covers region CT1299 of Pedersen et al. (20). The primers are designed in such a way that the regions flanking the repeat region are at least 70 bp long. This improves the readability of the sequences. Outer primers are given in black, and inner primers are given in gray. The length of the amplified fragment is represented by the lines between the primer sets. The 5′ positions of the primers are given as in Table 1.
The regions CT058, CT144, and pbpB of the original MLST were first tested with the original single PCRs. If they tested negative, they were retested using a nested version of the original MLST method. The primers from the original MLST were used as inner primers and new outer primers were designed (see the data in the supplemental material). For original CT058, the internal primers CT811F and CT1022R were used for sequencing. In the nested version, these were replaced by CT058 IF and CT058 IR. For pbpB, the internal primers pbpB2 IF and pbpB2 IR were used only in the nested version. With these assays, we could design 4 different high-resolution typing methods: the modified MLST, the original MLST, the MLVA, and a combination of the modified MLST and MLVA (Table 2).
Table 2.
Typing methods used in this study, represented by their included regionsa
| Method | ompA (modified) | hctB (modified) | CT058 (modified) | CT058 (nested/original) | CT144 (modified) | CT144 (nested/original) | CT172 (modified) | pbpB (modified) | pbpB (nested/original) | CT1299 (modified) | CT1335 (modified) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ompA genotyping | x | ||||||||||
| Modified MLST | x | x | x | x | x | x | |||||
| Original MLST | x | x | x | x | x | x | |||||
| MLVA | x | x | x | x | |||||||
| MLST + MLVA | x | x | x | x | x | x | x | x |
The regions given in italic encode the published fragment length according to Klint et al. (12); the lengths of the regions given in boldface differ from the original fragment length. For comparison, ompA is included in the original MLST.
Variation in putative single transmission for the high-resolution typing methods.
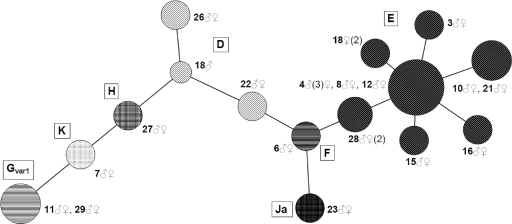
The analyses of panel A samples with the modified MLST did not reveal any differences within the 17 couples, nor did the longer reads from the (nested) original MLST. The samples from the partners within these couples had identical sequences for all of the regions in both methods and therefore had identical STs. Similarly, for the 3 persons with multiple samples from different anatomical locations, no variation was found between samples in any of the tests within one person. The exception to this was the genovar-discordant couple, couple 18, which turned out to be discordant for all 6 regions. As one or both partners of this couple could potentially have a double infection, the sequences of the three samples (two from the woman, one from the man) were checked for indications of a double infection. Also, region CT172 of these samples of both partners of couple 18 was checked on a gel, as the two alleles differ in length by 306 bp. No indications of double infections were found. These results are depicted in Fig. 3.
Fig. 3.
Minimum-spanning tree using the modified C. trachomatis MLST method with samples from 18 putative couples. The coding is by couple number and gender (♂, male; ♀, female). Genovars (ompA) are indicated with letters within the tree, and nodes with identical patterns indicate identical genovars. The sizes of the nodes indicate the number of samples included (1 to 8). The length of the branch indicates the number of alleles shared between connected sequence types (1 to 5). When multiple samples are available for one person, the number of included samples is given in parentheses. It can be seen that the partners of couple 18 do not share the same strain of Chlamydia trachomatis.
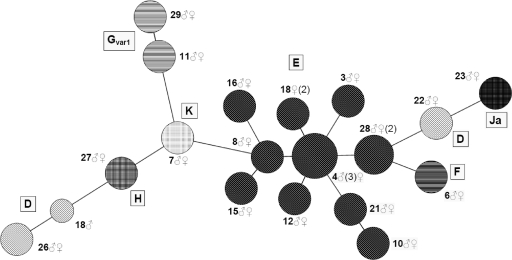
When analyzing the single nucleotide repeats of the MLVA, we found a proportion (CT172/CT1291, 10%; CT1299, 90%; CT1335, 92%) of these sequences to be ambiguous. This occurred when the repeats contained 10 or more consecutive identical nucleotides, and the ambiguity increased with length. As a result, the length of the repeat becomes difficult to determine and the 3′ flanking region becomes unreadable. Use of another high-fidelity polymerase could not correct for this phenomenon in pilot studies (data not shown), and we therefore concluded that the C. trachomatis samples could be heterogeneous for the number of single nucleotides within one repeat. As a result, samples with an identical majority repeat length could vary in the appearance of their peak patterns. The data gathered by the MLVA did show a high level of consistency, however, as between partners these patterns were always identical, again, with the exception of genovar-discordant couple 18. No variation in repeat length or appearance was seen between samples from the same person either (n = 3). A combination of the modified MLST and the MLVA is shown in Fig. 4.
Fig. 4.
Minimum-spanning tree using the modified Chlamydia trachomatis MLST and VNTR analysis MLVA method combined on 18 putative couples. The coding is by couple number and gender (♂, male; ♀, female). Genovars (ompA) are indicated with letters within the tree, and nodes with identical patterns indicate identical genovars. The sizes of the nodes indicate the number of samples included (1 to 8). The length of the branch indicates the number of alleles shared between connected sequence types (1 to 7). When multiple samples are available for one person, the number of included samples is given in parentheses. It can be seen that the partners of couple 18 do not share the same strain of Chlamydia trachomatis.
Discriminating capacity of the high-resolution typing methods.
Analysis of the 31 specimens in panel B with the modified MLST system resulted in 20 STs. Especially the more abundant ompA genovars were split, which resulted in 3 different STs for genovar D, 7 STs for genovar E, and 3 STs for genovar F. In the minimum-spanning tree generated from these data, the STs of one genovar were located in each other's proximity. This is similar to the results shown in Fig. 3. The D of the modified MLST system was 0.95. This changed when the original MLST was applied to the samples, as the cluster of couples 5, 10, 21, and 30 separated into two clusters. Also, there was a minor shift in the distribution of the STs in the minimum-spanning tree. The D of the original MLST system was 0.96. Only 8 (38%) of the 21 STs already existed in the Uppsala C. trachomatis MLST database; thus, 13 STs (62%) were types not described previously. Also, new polymorphisms were found for the regions hctB, CT058, CT172, and pbpB (Table 3).
Table 3.
Sequence types of 31 epidemiologically unrelated samples (panel B) determined using the original and nested Chlamydia trachomatis MLST method and the Uppsala Chlamydia trachomatis MLST databasea
| Genovar | Sample no. | Allele |
ST | ||||
|---|---|---|---|---|---|---|---|
| hctB | CT058 | CT144 | CT172 | pbpB | |||
| D | 22 | 5 | 19 | 15 | 1 | 4 | 13 |
| 26 | 10 | 8 | 1 | 4 | 23 | 35 | |
| 18♂ | NAL 1 | NAL 1 | 1 | 3 | 23 | NST 1 | |
| E | 5, 21, 30 | 1 | 2 | 6 | 2 | 2 | 3 |
| 3 | 7 | 19 | 14 | 2 | 1 | 16 | |
| 4, 8, 12, 24 | 1 | 19 | 7 | 2 | 1 | 56 | |
| 10 | 1 | 23 | 6 | 2 | 2 | NST 2 | |
| 15 | NAL 2 | 19 | 7 | 2 | 1 | NST 3 | |
| 16 | NAL 3 | 2 | 7 | 2 | NAL 1 | NST 4 | |
| 18♀ | NAL 4 | 19 | 7 | 2 | 1 | NST 5 | |
| 28 | 5 | 19 | 7 | 2 | 2 | NST 6 | |
| F | 1, 9, 13, 17, 25 | 5 | 19 | 7 | 1 | 4 | 12 |
| 6 | 5 | 19 | 7 | 2 | 4 | 148 | |
| 14 | 5 | 2 | 7 | 2 | NAL 2 | NST 7 | |
| Gvar1 | 11, 29 | 10 | 6 | 10 | 15 | 6 | NST 8 |
| Gvar2 | 20 | 10 | NAL 2 | 22 | 4 | 6 | NST 9 |
| H | 27 | 10 | 4 | 1 | 3 | 21 | NST 10 |
| Ia | 2 | 10 | 5 | 12 | 4 | 18 | 135 |
| 19 | 10 | 5 | 12 | NAL 1 | 18 | NST 11 | |
| Ja | 23 | 37 | 15 | 7 | 1 | NAL 3 | NST 12 |
| K | 7 | 10 | 6 | 22 | 3 | 8 | NST 13 |
Sample number refers to both partners of a couple, if the complete profiles were successfully sequenced. The exception to this is couple 18, where the male (♂) and female (♀) are indicated. NST, new sequence type; NAL, new allele. Allele designations are retrieved from the Uppsala CT MLST database.
The modified MLVA system gave similar results for panel B. It gave rise to 21 MLVA types (MTs; similar to sequence types), where genovar D comprised 3 different MTs and genovar E and F gave 5 MTs each (Table 4). In the minimum-spanning tree generated from the profiles, most genovars grouped together, but both genovar D and genovar G were divided over two different locations within the tree. The distribution of the samples over the MTs was considerably different compared to the MLSTs, as different samples grouped together. The D of the MLVA system was 0.96. The majority (67%) of the 21 MTs found were not yet identified in the Århus University database, which contains the different profiles generated with the MLVA method and consists of 61 different MTs found in 161 randomly selected samples (19, 20). New variants were found for all VNTRs. These are 7C and 12C for C. CT1291, 15C and 16C for CT1299, and 8T9A and 9T9A for CT1335. Transitions (C → T) were found in CT1299 at positions 10, 11, and 12.
Table 4.
MLVA types of 31 epidemiologically unrelated samples (panel B) determined using the Chlamydia trachomatis MLVA method and the Århus University databasea
| Genovar | Sample no. | Allele |
MT | ||
|---|---|---|---|---|---|
| CT1335 | CT1299 | CT1291 | |||
| D | 22 | 8 | 5 | 2 | 8.5.2e |
| 18♂ | 3 | NV 1 (15C)b | 3 | NMT 1 | |
| 26 | 3 | NV 1 (15C)b | 4 | NMT 2 | |
| E | 12 | 6 | 5 | 1 | 6.5.1 |
| 8, 10, 15, 16 | 8 | 4 | 1 | 8.4.1 | |
| 3 | 8 | 6 | 1 | 8.6.1 | |
| 5, 24 | 6 | 4 | 1 | NMT 3 | |
| 4, 18♀, 21, 28, 30 | 8 | 5 | 1 | NMT 4 | |
| F | 13 | 8 | 5 | 2 | 8.5.2e |
| 1, 17, 25 | 8 | 6 | 2 | 8.6.2 | |
| 6 | 8 | 3 | 1 | NMT 5 | |
| 14 | 8 | 5 | 1 | NMT 6 | |
| 9 | 8 | 7 | 2 | NMT 7 | |
| Gvar1 | 29 | NV 1 (8T9A) | 7c | NV 1 (7C) | NMT 8 |
| 11 | NV 1 (8T9A) | 8d | NV 1 (7C) | NMT 9 | |
| Gvar2 | 20 | 3 | NV 2 (16C) | 4 | NMT 10 |
| H | 27 | 3 | NV 1 (15C)b | 3 | NMT 11 |
| Ia | 19 | NV 2 (9T9A) | 5 | NV 2 (12C) | NMT 12 |
| 2 | NV 2 (9T9A) | 5 | 4 | NMT 13 | |
| Ja | 23 | 8 | 5 | 2 | NMT 14e |
| K | 7 | 3 | 4 | 3 | 3.4.3 |
Sample number refers to both partners of a couple, if the complete profiles were successfully sequenced. The exception to this is couple 18, where the male (♂) and female (♀) are indicated. Allele designations are retrieved from the Århus University database. NMT, new MLVA type; NV, VNTR.
The C → T transition was found at position 12 in CT1299.
The C → T transition was found at position 10 in CT1299.
The C → T transition was found at position 11 in CT1299.
The sequence type 8.5.2 has previously been described for samples with genovars D, E, and F (19) but not for genovar Ja.
When the modified MLST and MLVA were combined into one analysis, the resolution increased to 28 different genotypes (combination of both STs and MTs), splitting genovar D into 3 genotypes, genovar E into 12 genotypes, and genovar F into 7 genotypes. In the minimum-spanning tree generated from the profiles, most of the samples with the same genovars grouped together. Only the genovar D samples were located at different positions, which is also shown for panel A in Fig. 4. The diversity index of the combined MLST/MLVA was 0.99, as only 2 genotypes contained more than one sample and the 19 remaining genotypes were unique. In Table 5, a summary of the various typing techniques applied to panel B samples and their discriminatory indexes are presented.
Table 5.
Discriminatory power for the typing methods used in this study, based on 31 epidemiologically unrelated samples (panel B)
| Method | No. of loci | No. of variants | D |
|---|---|---|---|
| ompA genotyping | 1 | 9 | 0.78 |
| Modified MLST | 6 | 20 | 0.95 |
| Original MLSTa | 6 | 21 | 0.96 |
| MLVA | 4 | 21 | 0.96 |
| MLST + MLVA | 8 | 28 | 0.99 |
For comparison, ompA is included in the original MLST.
DISCUSSION
Using a limited panel of samples derived from putative heterosexual transmissions, we showed that we successfully modified two methods previously published by Klint et al. (12) and Pedersen et al. (20) with a very high resolution for typing of Chlamydia trachomatis. By changing them into nested PCR formats, their sensitivity was increased, while the discriminatory capacity was maintained. The maximum target length in our assay format was less than 750 bp, which allowed easy sequence analysis from single PCR fragments. Various new polymorphisms per included region were identified. Comparing original MLST results of our samples with the Uppsala C. trachomatis MLST database and the MLVA types with the Århus University database, we found that the majority (62% and 67%, respectively) of the generated profiles were not identified before.
Since the selected couples were not linked epidemiologically, the high sequence variation resulted in unique C. trachomatis profile types for almost every couple. The diversity found within Chlamydia trachomatis strains with MLST and MLVA is thus substantially higher than serotyping or ompA genotyping has indicated previously. The discriminatory power for each typing method ranged from 0.95 to 0.99 (Table 5), and such resolutions are satisfactory according to established guidelines (D ≥ 0.95) (24) for most molecular epidemiological questions. Combination of the modified MLST and MLVA typing assays resulted in the highest discriminatory power (D = 0.99). However, the method of choice for each typing purpose might not be by default. If the mutation rate is too high, inducing too much resolution, the epidemiological links might get lost.
The data from the MLVA typing should especially be analyzed with care. As elaborated on in the Results section, a large proportion of the sequences of the MLVA regions were ambiguous. This method relies on determining the number of single nucleotide repeats within one target region. In pilot studies, we used a special high-fidelity Taq polymerase enzyme, and this did not resolve the ambiguities, such as double peaks and shifts, in the MLVA target sequences. Using a defined algorithm, we were still able to assign a type in the majority of the samples. However, interpretation is thus user dependent and error prone. We therefore aim to preferentially use the modified MLST for our future studies and add MLVA to it only when a higher discriminatory power is needed.
These high-resolution typing methods were shown to be robust. No variation was found within one person or between persons who were likely to have infected each other. The samples from two individuals of one couple, number 18, were found to be highly discordant for all genomic regions, and the variation between these samples was so large that it was impossible to have arisen during a single transmission step. This was shown by studies on clonal outbreaks using high-resolution typing methods. No variation was seen previously in samples containing the nv C. trachomatis strain and lymphogranuloma venereum (LGV) strain L2b when these were typed with MLST (2, 6, 11), and variations were seen only exceptionally in MLVA (20). If the chosen MLST and MLVA regions would have been too polymorphic due to high mutation rates, these clonal outbreaks would not be recognizable as such. This confirms that when using high-resolution typing methods, a clonal outbreak can be identified with more certainty and that sexual transmission connections will remain apparent.
The sensitivity of a typing method greatly depends on the initial detection method that was used to diagnose the infection. In this study, the samples were screened using a highly sensitive detection system for Chlamydia trachomatis, the Aptima Combo 2 assay. This system utilizes 16S rRNA as an amplification target, and a few thousand copies of this target are present in every chlamydial cell. Several studies have shown a 2 to 12% higher sensitivity of Aptima Combo 2 compared to other commercial C. trachomatis detection systems, as reviewed by Cook et al. (3). Although false-positive Aptima results are always a possibility, at least one of the partners in each couple was positive in the typing PCRs, making the occurrence of false positivity unlikely. Since our typing PCRs were less sensitive than commercial C. trachomatis detection systems and about 10 to 100 times less sensitive than the Aptima Combo 2 assay, we did not expect to be able to type all Aptima C. trachomatis-positive samples, yet 80% of the clinical C. trachomatis-positive samples could successfully be sequenced for all included regions. This percentage was comparable to the percentages from similar typing studies (50% to 94%) that also used highly sensitive C. trachomatis screening diagnostics and nested PCR (7, 14, 17, 21). It should be noted that these typing studies used only 1 region (ompA), compared to the 11 different genomic targets that we amplified. We therefore conclude that using nested PCR in our modified typing methods is appropriately sensitive for typing a large majority of direct (uncultured) clinical samples.
For this study, we used C. trachomatis-positive samples from putative heterosexual couples to ensure that only a single transmission step occurred. As only a limited number of couples met all criteria, only a rather small number of samples were included. This led to a small number of epidemiologically unrelated samples (n = 31), limiting the power of this study. The calculated discriminatory indexes might therefore either increase or decrease, when using larger sample numbers. However, the discriminatory indexes that we calculated were similar to those in previous reports (9, 20). Also, the samples were considered epidemiologically unrelated, but all samples were selected from heterosexuals in the Amsterdam area. Therefore, the methods used in this study might be biased toward local Dutch types of urogenital Chlamydia trachomatis. Previous studies with the original MLST, however, found high levels of polymorphism in distinct strains of Chlamydia trachomatis, such as strains causing LGV and trachoma (2, 5). This indicates that the performance of the typing methods is probably not limited to specific risk groups or strains.
Detailed epidemiological data and larger study groups are needed to determine what behavior underlies the transmission of Chlamydia trachomatis within a population and the degree of mixing between specific risk groups. We conclude that the high-resolution typing methods described here show a satisfactory discriminatory capacity, sensitivity, and robustness and can be used in future studies. We aim to focus on these risk groups, such as men who have sex with men, adolescents, and migrant groups (studying the effects of traveling), and thus, we might find the determining factors of the chains of transmission of Chlamydia trachomatis.
Supplementary Material
ACKNOWLEDGEMENTS
We thank M. S. van Rooijen, N. Nassir Hajipour, and R. L. J. Heijman for their contributions.
Footnotes
Supplemental material for this article may be found at http://jcm.asm.org/.
Published ahead of print on 8 June 2011.
REFERENCES
- 1. Barnes R. C., Rompalo A. M., Stamm W. E. 1987. Comparison of Chlamydia trachomatis serovars causing rectal and cervical infections. J. Infect. Dis. 156:953–958 [DOI] [PubMed] [Google Scholar]
- 2. Christerson L., et al. 2010. Typing of lymphogranuloma venereum Chlamydia trachomatis strains. Emerg. Infect. Dis. 16:1777–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cook R. L., Hutchison S. L., Ostergaard L., Braithwaite R. S., Ness R. B. 2005. Systematic review: noninvasive testing for Chlamydia trachomatis and Neisseria gonorrhoeae. Ann. Intern. Med. 142:914–925 [DOI] [PubMed] [Google Scholar]
- 4. Dean D., Millman K. 1997. Molecular and mutation trends analyses of omp1 alleles for serovar E of Chlamydia trachomatis. Implications for the immunopathogenesis of disease. J. Clin. Invest. 99:475–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Harding-Esch E. M., et al. 2010. Multi-locus sequence typing: a useful tool for trachoma molecular epidemiology, p. 55–58 In Chlamydial Infections: Proceedings of the Twelfth International Symposium on Human Chlamydial Infections, International Chlamydia Symposium, San Francisco, CA [Google Scholar]
- 6. Herrmann B., et al. 2008. Emergence and spread of Chlamydia trachomatis variant, Sweden. Emerg. Infect. Dis. 14:1462–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hsu M. C., et al. 2006. Genotyping of Chlamydia trachomatis from clinical specimens in Taiwan. J. Med. Microbiol. 55: 301–308 [DOI] [PubMed] [Google Scholar]
- 8. Hunter P. R., Gaston M. A. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26: 2465–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ikryannikova L. N., Shkarupeta M. M., Shitikov E. A., Il'ina E. N., Govorun V. M. 2010. Comparative evaluation of new typing schemes for urogenital Chlamydia trachomatis isolates. FEMS Immunol. Med. Microbiol. 59:188–196 [DOI] [PubMed] [Google Scholar]
- 10. Jeffrey B. M., et al. 2010. Genome sequencing of recent clinical Chlamydia trachomatis strains identifies loci associated with tissue tropism and regions of apparent recombination. Infect. Immun. 78: 2544–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jurstrand M., et al. 2010. Characterisation of Chlamydia trachomatis by ompA sequencing and multilocus sequence typing in a Swedish county before and after identification of the new variant. Sex. Transm. Infect. 86: 56–60 [DOI] [PubMed] [Google Scholar]
- 12. Klint M., et al. 2007. High-resolution genotyping of Chlamydia trachomatis strains by multilocus sequence analysis. J. Clin. Microbiol. 45: 1410–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lan J., et al. 1993. Direct detection and genotyping of Chlamydia trachomatis in cervical scrapes by using PCR and restriction fragment length polymorphism analysis. J. Clin. Microbiol. 31: 1060–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lysén M., et al. 2004. Characterization of ompA genotypes by sequence analysis of DNA from all detected cases of Chlamydia trachomatis infections during 1 year of contact tracing in a Swedish county. J. Clin. Microbiol. 42: 1641–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Machado A. C., et al. 2010. Distribution of Chlamydia trachomatis genovars among youths and adults in Brazil. J. Med. Microbiol. 60: 472–476 [DOI] [PubMed] [Google Scholar]
- 16. Morré S. A., et al. 1998. Serotyping and genotyping of genital Chlamydia trachomatis isolates reveal variants of serovars Ba, G, and J as confirmed by omp1 nucleotide sequence analysis. J. Clin. Microbiol. 36: 345–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mossman D., et al. 2008. Genotyping of urogenital Chlamydia trachomatis in regional New South Wales, Australia. Sex. Transm. Dis. 35: 614–616 [DOI] [PubMed] [Google Scholar]
- 18. Pedersen L. N., Herrmann B., Moller J. K. 2009. Typing Chlamydia trachomatis: from egg yolk to nanotechnology. FEMS Immunol. Med. Microbiol. 55: 120–130 [DOI] [PubMed] [Google Scholar]
- 19. Pedersen L. N., Moller J. K. 2009. Distribution of Chlamydia trachomatis genotypes in rectal and urogenital samples, poster p3.60 Abstr Int. Soc. Sex. Trans. Dis., London, United Kingdom http://www.klinmik.dk/poster/isstdr2009_p360.pdf [Google Scholar]
- 20. Pedersen L. N., Podenphant L., Moller J. K. 2008. Highly discriminative genotyping of Chlamydia trachomatis using omp1 and a set of variable number tandem repeats. Clin. Microbiol. Infect. 14: 644–652 [DOI] [PubMed] [Google Scholar]
- 21. Pineiro L., et al. 2009. Genotyping of Chlamydia trachomatis in an area of northern Spain. Enferm. Infecc. Microbiol. Clin. 27: 462–464(In Spanish.) [DOI] [PubMed] [Google Scholar]
- 22. Quint K. D., et al. 2011. Analysis of infections with concomitant Chlamydia trachomatis genotypes among men who have sex with men in Amsterdam, The Netherlands. BMC Infect. Dis. 11: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stephens R. S. 2002. Chlamydiae and evolution: a billion years and counting, p. 3–12 In Chlamydial infections: Proceedings of the Tenth International Symposium on Human Chlamydial Infections, International Chlamydia Symposium, San Francisco, CA. [Google Scholar]
- 24. van Belkum A., et al. 2007. Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin. Microbiol. Infect. 13(Suppl. 3):1–46 [DOI] [PubMed] [Google Scholar]
- 25. World Health Organization. 2010. Sexually transmitted diseases. World Health Organization, Geneva, Switzerland. http://www.who.int/vaccine_research/diseases/soa_std/en/index1.html [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.