Abstract
The performance characteristics of four different assays for hepatitis B virus (HBV) quantification were assessed: the Abbott RealTime HBV IUO, the Roche Cobas AmpliPrep/Cobas TaqMan HBV test, the Roche Cobas TaqMan HBV test with HighPure system, and the Qiagen artus HBV TM ASR. Limit of detection (LOD), linear range, reproducibility, and agreement were determined using a serially diluted plasma sample from a single chronically infected subject. Each assay was tested by at least three laboratories. The LOD of the RealTime and two TaqMan assays was approximately 1.0 log10 IU/ml; for artus HBV (which used the lowest volume of extracted DNA), it was approximately 1.5 log10 IU/ml. The linear range spanned 1.0 to at least 7.0 log10 IU/ml for all assays. Median values were consistently lowest for artus HBV and highest for Cobas AmpliPrep/Cobas TaqMan HBV. Assays incorporating automated nucleic acid extraction were the most reproducible; however, the overall variability was minor since the standard deviations for the means of all tested concentrations were ≤0.32 log10 IU/ml for all assays. False-positive results were observed with all assays; the highest rates occurred with tests using manual nucleic acid extraction. The performance characteristics of these assays suggest that they are useful for management and therapeutic monitoring of chronic HBV infection.
INTRODUCTION
Hepatitis B virus (HBV) has infected an estimated 400 million persons worldwide; cirrhosis and hepatocellular carcinoma, the major sequelae of chronic hepatitis B, result in over a half million deaths annually (4). HBV viremia is a critical risk factor for progression of chronic HBV infection (1); accordingly, quantification of HBV DNA in blood has become a critical tool in the assessment and management of chronic infection. In addition to serologic tests for HBV and measurement of serum transaminases, HBV viral load testing is used to determine the phase of chronic HBV infection (8) and is particularly useful in distinguishing active from inactive disease in individuals with no detectable HBeAg. A number of antiviral drugs have been introduced recently for the treatment of chronic HBV infection, and viremia is an important component in the decision to initiate treatment and in monitoring therapeutic response (5, 7).
Viremia in chronic HBV infection varies from very low or undetectable to >108 copies/ml. Effective quantitative assays must therefore measure a wide range of viral DNA concentrations. Commercially available quantitative assays utilize a variety of different detection methods, including signal amplification (Versant HBV bDNA; Siemens Healthcare Diagnostics), conventional PCR (Amplicor HBV Monitor test; Roche Diagnostics), and real-time PCR (Cobas AmpliPrep/Cobas TaqMan HBV test [Roche Diagnostics] and RealTime HBV assay [Abbott Molecular]). Of these methods, only real-time PCR is able to cover the wide dynamic range required for quantification of the virus in all stages of infection.
Reported studies of real-time assays have mainly focused on the performance of individual tests compared to signal amplification tests rather than the comparative performance of multiple real-time PCR tests (2, 3, 6, 9). The present study assessed the limit of detection, linear range, reproducibility, and agreement among four commercially available real-time PCR HBV viral load tests: the Abbott RealTime HBV IUO, the Roche Cobas AmpliPrep/Cobas TaqMan HBV test, the Roche Cobas TaqMan HBV test with HighPure system, and the Qiagen artus HBV TM ASR. The Abbott m2000sp and Roche AmpliPrep protocols were performed using automated extraction methods, while the extractions for the Roche HighPure and Qiagen ASR were performed manually.
MATERIALS AND METHODS
Quantification panels.
A 65-specimen panel was created to be used by participating laboratories to assess assay performance. The panel was created in a centralized laboratory (the Division of AIDS Viral Quality Assurance Laboratory, Rush University Medical Center, Chicago, IL) by serially diluting a unit of plasma obtained from a patient donor infected with HBV. The diluent was human plasmapheresis plasma containing the anticoagulant sodium citrate (SeraCare, Milford, MA) and found to be negative for HBsAg by serologic testing; this also served as negative sample material. HBV-infected donor blood was obtained under an institutional review board-approved protocol. The plasma was assigned a nominal concentration of 8.58 log10 IU/ml of genotype A virus as determined by Versant HBV DNA 3.0 (Siemens, Hoffman Estates, IL). Four replicates each of three different dilutions (1:100, 1:1,000, and 1:10,000) were tested to determine IU/ml.
HBV genotype was determined by direct sequencing of the sequence of the polymerase gene (nucleotides 1 to 1615 and 2528 to 3221 of the circular genome) by BLAST analysis and alignment with sequences in GenBank. The panel consisted of 5 to 7 replicates of 10 concentrations ranging from 1.0 to 7.7 log10 IU/ml each in a volume of 750 μl (Table 1). Panel members were tested in a blinded manner by participating laboratories.
Table 1.
HBV panel composition
| No. of samples | Nominal concn |
|
|---|---|---|
| IU/ml | Log10 IU/ml | |
| 7 | 0 | 0 |
| 7 | 10 | 1.0 |
| 7 | 30 | 1.5 |
| 7 | 100 | 2.0 |
| 7 | 300 | 2.5 |
| 5 | 1,000 | 3.0 |
| 5 | 10,000 | 4.0 |
| 5 | 100,000 | 5.0 |
| 5 | 1,000,000 | 6.0 |
| 5 | 10,000,000 | 7.0 |
| 5 | 50,000,000 | 7.7 |
Viral load assays.
Four HBV viral load assays were evaluated, including RealTime HBV IUO with m2000sp automated sample preparation (Abbott Molecular, Des Plaines, IL), TaqMan48 HBV ASR with AmpliPrep automated sample preparation (Roche Diagnostics, Indianapolis, IN), TaqMan HBV RUO with HighPure system manual sample preparation (Roche), and artus HBV TM ASR with QIAamp MinElute Virus Spin kit manual sample preparation (Qiagen, Germantown, MD). The plasma extraction volume was 500 μl for the AmpliPrep TaqMan, HighPure TaqMan, and RealTime assays and 200 μl for the Qiagen artus HBV TM assay. An internal control (IC) for the RealTime and artus HBV assays and a quantitation standard (QS) for the TaqMan assays were processed simultaneously with the samples during the extraction. The elution volumes from the extractions were 75 μl for the two TaqMan assays, 70 μl for the RealTime assay, and 60 μl for the artus HBV assay. The real-time PCR amplification mixtures for the two TaqMan assays and the RealTime assay contained 50 μl of extracted sample plus 50 μl of amplification master mix supplied with the manufacturer's reagents (total volume of 100 μl) for each assay, while the artus HBV amplification used 20 μl of sample extract plus 30 μl of master mix for a total amplification volume of 50 μl. Amplification and quantification were performed per package insert if available; otherwise, protocols were developed in collaboration with manufacturers' research and development expertise.
Study design.
The panels were used to determine the limit of detection, linear range, reproducibility, and agreement for each assay. The two Roche assays and the Qiagen assay were each performed by four different laboratories. Only three laboratories were able to perform the Abbott assay; therefore, one laboratory tested two complete panels so that the same number of replicates was tested for all assays.
Statistical analysis.
HBV concentrations were log10 transformed for analysis. Results that were reported as “not extracted” in the AmpliPrep TaqMan assay or as invalid in any assay were excluded from the analysis. For each assay, results from all laboratories were combined when calculating means and medians. Standard deviations accounted for variation between laboratories within a given assay platform. Medians were not generated for a given nominal concentration if fewer than 50% of the samples produced a quantitative estimate. Means and standard deviations were not generated for a given nominal concentration if a valid quantitative result was not obtained for all samples tested.
RESULTS
The detection rates of the different tests are summarized in Table 2. All tests were highly sensitive, with the RealTime, AmpliPrep TaqMan, and High Pure TaqMan assays qualitatively detecting all 27 or 28 replicates of the 1.0-log10-IU/ml sample. The artus HBV test detected 24/28 1.0-log10-IU/ml replicates and all 28 replicates of the 1.5-log10-IU/ml sample. Although false-positive results occurred with all of the assays, they were seen more frequently in the assays that used manual extraction methods (4/28 for the HighPure TaqMan test and 5/28 for the artus HBV assay, compared to 1/28 for the RealTime and 2/28 for the AmpliPrep TaqMan assays). False-positive results were not confined to any one laboratory; 50% of the false-positive results were from samples that were adjacent to a sample with a very high viral load (>10 million IU/ml).
Table 2.
Detection rate of the HBV viral load assays
| Assay | No. detected/no. of valid results at each nominal concn (log10 IU/ml) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1.0 | 1.5 | 2.0 | 2.5 | 3.0 | 4.0 | 5.0 | 6.0 | 7.0 | 7.70 | |
| RealTime HBV | 1/28 | 28/28 | 28/28 | 28/28 | 28/28 | 20/20 | 20/20 | 20/20 | 20/20 | 20/20 | 20/20 |
| AmpliPrep TaqMan HBVa | 2/28 | 27/27 | 28/28 | 28/28 | 28/28 | 19/19 | 18/18 | 20/20 | 20/20 | 20/20 | 20/20 |
| HighPure TaqMan HBVb | 4/28 | 28/28 | 28/28 | 28/28 | 28/28 | 19/19 | 20/20 | 20/20 | 20/20 | 18/18 | 20/20 |
| artus HBV TM ASR | 5/28 | 24/28 | 28/28 | 28/28 | 28/28 | 20/20 | 20/20 | 20/20 | 20/20 | 20/20 | 20/20 |
Four samples invalid or not extracted.
Three samples invalid or not extracted.
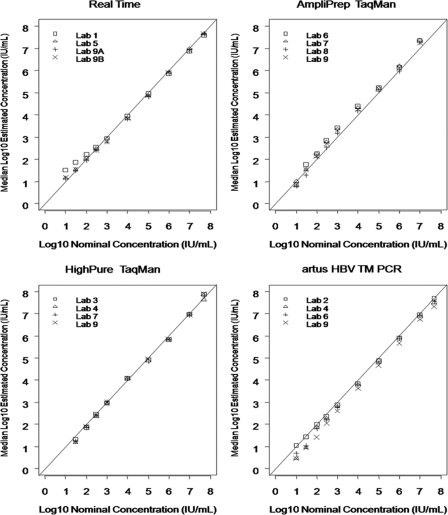
The median viral load values and intraquartile ranges for the four viral load assays are shown in Table 3. Overall, there was good agreement in viral load values between the assays. For samples with a nominal concentration ranging from 1.5 log10 IU/ml to 7.0 log10 IU/ml, the differences in the median viral load values across the four assays were between 0.29 log10 IU/ml and 0.56 log10 IU/ml. The samples with nominal concentrations of 1.0 log10 IU/ml and 7.7 log10 IU/ml were not analyzed because values were not available for all four assays. The standard deviations of the mean viral load values are shown in Table 4. In most instances, lower standard deviations were observed at concentrations greater than 2.0 log10 IU/ml. All assays were linear to 7.7 log10 IU/ml with the exception of the AmpliPrep TaqMan assay, as all 7.7-log10-IU/ml values were above the upper linear range of the assay (8.04 log10 IU/ml) and were reported as such by the TaqMan instrument (Fig. 1).
Table 3.
Median viral load values and intraquartile ranges
| Log10 nominal concn (IU/ml) | Median (Q1, Q3) |
|||
|---|---|---|---|---|
| RealTime HBV | AmpliPrep TaqMan HBV | HighPure TaqMan HBV | artus HBV TM ASR | |
| 1.0 | 1.18 (1.10, 1.34) | 0.85 (0.78, 1.00) | —a | 0.65 (0.39, 0.90) |
| 1.5 | 1.57 (1.52, 1.73) | 1.54 (1.40, 1.74) | 1.26 (1.11, 1.33) | 1.11 (0.90, 1.34) |
| 2.0 | 2.02 (1.96, 2.13) | 2.20 (2.11, 2.24) | 1.89 (1.82, 1.95) | 1.83 (1.55, 1.98) |
| 2.5 | 2.44 (2.40, 2.49) | 2.73 (2.67, 2.78) | 2.42 (2.38, 2.45) | 2.25 (2.11, 2.35) |
| 3.0 | 2.89 (2.80, 2.93) | 3.34 (3.23, 3.40) | 3.00 (2.96, 3.03) | 2.78 (2.69, 2.82) |
| 4.0 | 3.85 (3.84, 3.90) | 4.30 (4.24, 4.38) | 4.08 (4.03, 4.11) | 3.79 (3.64, 3.85) |
| 5.0 | 4.87 (4.83, 4.92) | 5.18 (5.12, 5.22) | 4.92 (4.85, 4.95) | 4.78 (4.69, 4.88) |
| 6.0 | 5.91 (5.86, 5.94) | 6.14 (6.10, 6.17) | 5.85 (5.84, 5.87) | 5.86 (5.71, 5.92) |
| 7.0 | 6.93 (6.89, 6.96) | 7.32 (7.26, 7.37) | 6.98 (6.94, 7.02) | 6.90 (6.77, 6.96) |
| 7.7 | 7.66 (7.61, 7.69) | —b | 7.86 (7.80, 7.91) | 7.52 (7.35, 7.59) |
Nineteen of 28 samples reported as “detected, <6 IU/ml.”
All 20 samples reported as >8.04 log10 IU/ml (110,000,000 copies/ml).
Table 4.
Standard deviations of the mean viral load values
| Log10 nominal concn (IU/ml) | RealTime HBV |
AmpliPrep TaqMan HBV |
HighPure TaqMan HBV |
artus HBV TM ASR |
||||
|---|---|---|---|---|---|---|---|---|
| n | SD | n | SD | n | SD | n | SD | |
| 1.00 | ||||||||
| 1.50 | 28 | 0.19 | 28 | 0.22 | 27 | 0.14 | 28 | 0.32 |
| 2.00 | 28 | 0.13 | 28 | 0.11 | 28 | 0.25 | 28 | 0.27 |
| 2.50 | 28 | 0.08 | 28 | 0.13 | 28 | 0.12 | 28 | 0.16 |
| 3.00 | 20 | 0.09 | 19 | 0.11 | 19 | 0.10 | 20 | 0.13 |
| 4.00 | 20 | 0.10 | 18 | 0.10 | 20 | 0.10 | 20 | 0.13 |
| 5.00 | 20 | 0.06 | 20 | 0.10 | 20 | 0.07 | 20 | 0.11 |
| 6.00 | 20 | 0.05 | 20 | 0.09 | 20 | 0.06 | 20 | 0.27 |
| 7.00 | 20 | 0.07 | 20 | 0.11 | 18 | 0.13 | 20 | 0.13 |
| 7.70 | 20 | 0.07 | —a | —b | 20 | 0.16 | ||
All 20 samples reported as >8.04 log10 IU/ml.
Three samples reported as >8.04 log10 IU/ml.
Fig. 1.
Linear range of the viral load assays.
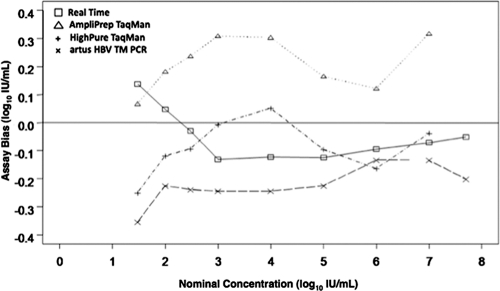
The assay bias, expressed as log10 IU/ml viral load values minus the nominal concentration, is shown in Fig. 2. The RealTime assay showed very consistent bias for samples greater than or equal to 3.0 log10 IU/ml. The AmpliPrep TaqMan assay consistently gave the highest viral load values, and there was variable bias throughout the linear range of the assay. A similar pattern of bias was seen with the HighPure TaqMan assay, although the values were lower than those seen with the AmpliPrep TaqMan assay. The artus HBV assay showed consistent bias for samples above 2.0 log10 IU/ml and overall showed the lowest viral load values.
Fig. 2.
Assay bias of the four real-time PCR assays for HBV quantification. Bias is defined as measured viral load (log10 IU/ml) minus the nominal concentration of the sample (log10 IU/ml).
DISCUSSION
This comparison of four real-time PCR assays demonstrated that they have very similar performance characteristics, although some differences were noted. For example, the two TaqMan and the RealTime assays had a lower limit of detection of 1.0 log10 IU/ml, compared to 1.5 log10 IU/ml for the artus HBV assay. This minor difference is likely due to plasma input volume differences between artus HBV (200 μl) and the other assays (500 μl). In addition, a lower percentage of the extracted DNA was added to the master mix with the artus HBV assay compared to the other three assays (33% versus ∼70%). Despite this analytical difference, all assays had detection limits within the necessary range for clinical decision making.
Another difference that was observed pertained to false-positive rates. Assays that relied on manual extraction (HighPure TaqMan and artus HBV) had higher false-positive rates than did those that employed automated extraction (RealTime and AmpliPrep TaqMan assays). The occurrence of false-positive results with automated extraction platforms is an important observation that may reflect the performance expected in the clinical laboratory, where samples with viral load values in excess of 8.0 log10 IU/ml may be tested alongside samples containing no virus. These data suggest that regardless of the extraction method employed, careful attention to good laboratory practices will be needed to avoid false-positive results due to the extraordinarily high viral loads that occur in chronic HBV infections.
All four assays demonstrated a broad linear range of approximately 7 log10 IU/ml; we were unable to obtain a large-volume sample with a higher viral load to better define the upper limits of linearity of the assays. Quantification of the 7.7-log10-IU/ml sample resulted in concentrations that exceeded the upper limit of the AmpliPrep TaqMan assay but not those of the other three assays. According to the AmpliPrep TaqMan package insert, it would have been acceptable to dilute these high-concentration samples up to 1:100 in order to report out a value up to 10.23 log10 IU/ml; however, specimen dilution was not part of the study protocol and, therefore, it was not performed. This approach is appropriate for all of the assays as long as the dilution process is validated by the laboratory.
The reproducibility of the assays was similar to that seen with other real-time PCR tests. For viral load values in the middle of the linear range where the standard deviations are 0.05 to 0.10 log10 IU/ml, a change in viral load that is greater than 3-fold would be interpreted to be a significant difference. For viral load values less than 2.0 log10 IU/ml, where the standard deviation is higher, 5-fold changes would be significant. Overall, the artus HBV assay was the least precise of the four assays evaluated; this may in part reflect the manual extraction method.
The differences in the median viral load values obtained with the four assays ranged from 0.29 log10 IU/ml to 0.56 log10 IU/ml. The smallest difference was seen for the 6.0-log10-IU/ml sample, and the largest difference was seen for the 3.0-log10-IU/ml sample. The most consistent bias (difference between nominal concentration and observed concentration) was seen with the artus HBV assay for values of >2.0 log10 IU/ml and with the RealTime assay for values of ≥3.0 log10 IU/ml. Both TaqMan assays showed an inconsistent bias throughout the linear range of the assays. One limitation of this study is that only a genotype A sample was studied, so it is not possible to determine if any of these assays has a genotype bias.
The availability and regulatory status of the investigated assays have changed since the conclusion of experimentation. The artus HBV assay is available outside the United States as a Conformité Européenne (CE)-marked product. The RealTime and TaqMan reagents have been approved for use by the U.S. Food and Drug Administration (U.S. FDA) and are CE marked. Plasma volumes identical to those used in this study are used in the U.S. FDA-approved assays. The RealTime assay is additionally approved for use with 200 μl of plasma. Comparative performance of the assay with this reduced volume was not investigated in this study.
Though there is reasonable agreement in viral load values across the four assays, the intra-assay and between-assay variability are such that patients should be monitored with a single assay. False positives were observed on all platforms, and careful attention should be paid to avoid cross-contamination by samples that may contain extremely high concentrations of this virus.
In summary, all four assays are similarly sensitive and have a broad linear range, providing clinical utility for both diagnostic testing and therapeutic monitoring.
ACKNOWLEDGMENTS
Assay reagents for this study were generously supplied by the manufacturers. We thank Michael S. Forman, Jess Ingersoll, Salvatore Scianna, Heather B. Steinmetz, Debra Kohn, Stephanie Merritt, Najma Akbany, and Ray Mills for their expert technical assistance.
This work was supported in part by National Institutes of Health contract NO1-AI35172 to J.W.B., an Emory University Center for AIDS Research grant (P30 AI050409, A.M.C.), and the HIV Prevention Trials Network (HPTN) sponsored by the NIAID, National Institutes of Child Health and Human Development (NICH/HD), National Institute on Drug Abuse, National Institute of Mental Health, and Office of AIDS Research, of the NIH, DHHS (U01-AI-46745 and U01-AI-068613 to A.V.).
Disclosures of potential conflicts of interest include the following: A.M.C., scientific advisory boards of Roche Diagnostics and Abbott Molecular and research support from Roche Diagnostics and Qiagen; S.Y., scientific advisory board of Roche Diagnostics; A.V., scientific advisory boards of Roche Diagnostics, Abbott Molecular, and Qiagen and research support from Roche Diagnostics and Qiagen; L.S., speaking honorarium from Abbott Molecular.
Footnotes
Published ahead of print on 22 June 2011.
REFERENCES
- 1. Chen C. J., Yang H. I., Iloeje U. H. 2009. Hepatitis B virus DNA levels and outcomes in chronic hepatitis B. Hepatology 49:S72–S84 [DOI] [PubMed] [Google Scholar]
- 2. Chevaliez S., Bouvier-Alias M., Laperche S., Pawlotsky J. M. 2008. Performance of the Cobas AmpliPrep/Cobas TaqMan real-time PCR assay for hepatitis B virus DNA quantification. J. Clin. Microbiol. 46:1716–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ciotti M., Marcuccilli F., Guenci T., Prignano M. G., Perno C. F. 2008. Evaluation of the Abbott RealTime HBV DNA assay and comparison to the Cobas AmpliPrep/Cobas TaqMan 48 assay in monitoring patients with chronic cases of hepatitis B. J. Clin. Microbiol. 46:1517–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dienstag J. L. 2008. Hepatitis B virus infection. N. Engl. J. Med. 359:1486–1500 [DOI] [PubMed] [Google Scholar]
- 5. European Association for the Study of the Liver 2009. EASL clinical practice guidelines: management of chronic hepatitis B. J. Hepatol 50:227–242 [DOI] [PubMed] [Google Scholar]
- 6. Hochberger S., et al. 2006. Fully automated quantitation of hepatitis B virus (HBV) DNA in human plasma by the COBAS AmpliPrep/COBAS TaqMan system. J. Clin. Virol. 35:373–380 [DOI] [PubMed] [Google Scholar]
- 7. Lok A. S., McMahon B. J. 2007. Chronic hepatitis B. Hepatology 45:507–539 [DOI] [PubMed] [Google Scholar]
- 8. McMahon B. J. 2009. The natural history of chronic hepatitis B virus infection. Hepatology 49:S45–55 [DOI] [PubMed] [Google Scholar]
- 9. Ronsin C., Pillet A., Bali C., Denoyel G. A. 2006. Evaluation of the COBAS AmpliPrep-total nucleic acid isolation-COBAS TaqMan hepatitis B virus (HBV) quantitative test and comparison to the VERSANT HBV DNA 3.0 assay. J. Clin. Microbiol. 44:1390–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]