Abstract
Participation criteria for clinical trials in pulmonary tuberculosis commonly include confirmation of sputum positive for mycobacteria and an indication of drug susceptibility before treatment is initiated. We investigated the suitability of two novel sputum-based nucleic acid amplification methods for patient selection in a recent early bactericidal activity study. Spontaneously expectorated sputum samples of 140 consecutive pulmonary tuberculosis patients were examined with direct fluorescence microscopy, Genotype MTBDRplus assay (MTBDR), Xpert MTB/RIF assay (Xpert), and liquid mycobacterial culture. The methods detected mycobacteria or mycobacterial DNA in 96.8%, 90.5%, 92.9%, and 92.1% of samples, respectively. MTBDR, Xpert, and liquid culture were 100% concordant for detection of resistance to rifampin. Sensitivity and specificity of MTBDR for detection of isoniazid resistance were 83.3% and 100%, respectively. For quantification of mycobacterial sputum load, we found a correlation between Xpert DNA amplification cycle thresholds, time to positivity, and microscopy smear grade. The best correlation was found between Xpert and time to positivity (r = 0.54), which were both correlated with smear microscopy with r values equal to −0.40 and −0.48, respectively. We conclude that MTBDR and Xpert are suitable screening tools for determining rifampin resistance in sputum microscopy smear-positive patients before participation in tuberculosis trials. Xpert should be further explored as a surrogate measurement for sputum mycobacterial load.
INTRODUCTION
Africa is seriously burdened by tuberculosis (TB), with 2.8 million cases and 385,000 deaths reported in 2008 (22). This epidemic is fuelled by a high rate of coinfection with HIV, especially in sub-Saharan Africa (6). The need for new antimycobacterial agents is obvious, as health care systems are observing high and rising rates of patients with multidrug-resistant (MDR) and extensively drug-resistant (XDR) TB (21). Early bactericidal activity (EBA) studies in smear-positive, treatment-naïve TB patients have become the recognized first step to evaluate novel antituberculosis agents (8).
Novel nucleic acid amplification tests (NAATs) that provide both diagnosis of TB and an indication of drug resistance from minimally processed sputum samples have recently become available. Handling of these NAATs has been greatly simplified, and they hold great promise for a wider rollout to facilitate both clinical research and TB management in high-burden settings. One representative is the GenoType MTBDRplus assay (MTBDR; Hain Lifescience GmbH, Nehren, Germany), for which mycobacterial DNA is amplified and reversely hybridized to detect Mycobacterium tuberculosis complex and common mutations of the rpoB gene causing resistance against rifampin (RIF) and of the katG and inhA genes leading to isoniazid (INH) resistance (12). Results are usually available within 48 h. Another example is the Xpert MTB/RIF assay (Xpert; Cepheid; Sunnyvale, CA), employing a fully automated real-time PCR mixture contained in a disposable cartridge (13). Xpert detects M. tuberculosis as well as RIF resistance-conferring mutations directly from sputum in an assay providing results within 120 min (1, 3, 4, 9, 19). Xpert does not test for INH resistance, but it offers a quantitative measurement that has shown a linear correlation to the CFU count in spiked sputum (3).
Clinical trials of antituberculosis medication are generally conducted on newly diagnosed, sputum smear-positive patients who are recruited from primary health care facilities. A catalogue of criteria must be fulfilled for trial participation. These include submission of a fresh sputum sample to the designated study laboratory for verification of the presence of M. tuberculosis, a sufficient sputum bacterial load, and an early indication of drug susceptibility. This screening process must be concluded in the shortest time possible to not unnecessarily prolong the patient's state of infectiousness by delaying the start of treatment. This study evaluated if and how the novel NAATs can be integrated into the process of patient selection for clinical trials of antituberculosis medication.
MATERIALS AND METHODS
Patients and samples.
EBA study TMC207-CL001 (sponsor, Global Alliance for TB Drug Development, Pretoria, South Africa; ClinicalTrials.gov no. NCT01215110) was carried out at two centers in Cape Town, South Africa, where a TB incidence of >1,000 cases per 100,000 inhabitants per year has been reported for parts of the city (17). Briefly, subjects aged 18 to 65 years were invited for screening if they were recently diagnosed with smear-positive first-time TB at a local TB clinic, untreated, free of severe comorbidities, and willing to be hospitalized. Bacteriological selection criteria at the study laboratory included examination of a separate sputum sample for confirmation of sufficient sputum bacterial load (microscopy grade, ≥1+ on at least one direct smear) and susceptibility to INH and RIF indicated by MTBDR. All microbiological testing for study purposes was carried out in the Department of Biomedical Sciences at Stellenbosch University. The relevant ethics and regulatory boards approved the study, which required patients to provide written informed consent.
Sample collection and processing.
Sputum samples were spontaneously expectorated during a visit to the research ward. Samples were kept refrigerated at 2 to 8°C during transport and allowed to warm to room temperature for processing. The entire sample was homogenized by magnetic stirring for 30 min, and 30 μl of each sample was used to prepare a direct smear for detection of acid-fast bacilli (AFB) by fluorescence microscopy (20). These direct smears were scored according to WHO/IUATLD guidelines (16) as AFB negative, scanty, 1+, 2+, or 3+.
The homogenized sample was decontaminated with N-acetyl-l-cysteine–sodium hydroxide (BBL MycoPrep; Becton Dickinson, Sparks, MD) for 20 min, neutralized with sterile phosphate-buffered saline (PBS; pH 6.8), added to a final volume of 45 ml, and centrifuged at 3,000 × g for 15 min in a refrigerated centrifuge at 4°C. The supernatant was discarded, and the remaining pellet was resuspended in 1.5 ml of PBS. Pellets were processed further for MTBDR, liquid culture, and Xpert in that sequence and priority. A fresh sputum sample was requested if a patient had submitted a low-volume or poor-quality sample or when a <1+ positive direct smear was found. However, some patients were excluded from further participation on clinical grounds before they could submit a fresh sample to complete the laboratory testing.
Mycobacterial culture.
One mycobacterial growth indicator tube (MGIT; Becton Dickinson) was prepared for each resuspended sample pellet by adding 0.8 ml oleic acid-albumin-dextrose-catalase with PANTA (Becton Dickinson), and 0.5 ml of the sample was inoculated into the tube, which was incubated at 37°C in an automated Bactec MGIT 960 apparatus (Becton Dickinson) for a maximum of 42 days and monitored continuously. MGIT tubes flagging positive were removed from the machine, and the time to positivity (TTP) was recorded. The presence of AFB was confirmed by Ziehl-Neelsen microscopy, and the possibility of contamination was excluded by placing a drop of positive culture onto a blood agar plate and incubating it for 48 h at 37°C.
MTBDR.
A 0.5-ml aliquot of the resuspended pellet sample was heat inactivated (95°C for 30 min) and utilized for the detection of M. tuberculosis complex and possible gene mutations for resistance. This was conducted with the GenoType MTBDRplus96 assay (Hain Lifescience GmbH) following the instructions of the manufacturer (12). The results determined the presence of M. tuberculosis complex and of mutations coding for resistance to RIF and INH.
Xpert.
The remaining sputum pellet was further diluted with 2 ml PBS for testing with the Xpert MTB/RIF assay (Cepheid). As per manufacturer instructions, 1 ml of the resuspended sample was mixed with 2 ml Xpert sample reagent, inverted several times, and incubated for 15 min at room temperature to inactivate live bacteria. This mixture was transferred into an Xpert cartridge and placed into the GeneXpert instrument. Cycle thresholds (CTs) of 5 rpoB gene probes automatically reported the presence of M. tuberculosis (GeneXpert Dx software, version 2.1) (4, 13). The CT of probe B was used as a quantitative measurement as previously described elsewhere (3). Reported RIF resistance was confirmed manually by calculation of a change in CT (ΔCT) between the highest and the lowest signal of the 5 probes.
Susceptibility testing.
Culture-based drug susceptibility testing (DST) with four antibiotics (RIF, INH, streptomycin, and ethambutol) was carried out with the Bactec MGIT 960 SIRE kit (Becton Dickinson), according to the manufacturer's instructions.
Definitions and statistical analysis.
Dichotomous readings were compared by chi-square tests (microscopy, AFB positive or negative; MTBDR and Xpert, M. tuberculosis positive or negative; MGIT, culture positive or negative) and quantitative readings (smear grades, no AFB seen, scanty, 1+, 2+, or 3+; Xpert, CT of probe B; MGIT, TTP) with Spearman or Pearson correlation. The performance characteristics of the molecular tests for detecting resistance were measured against each other, and the MGIT DST was used as the “gold standard.”
RESULTS
Patients and samples.
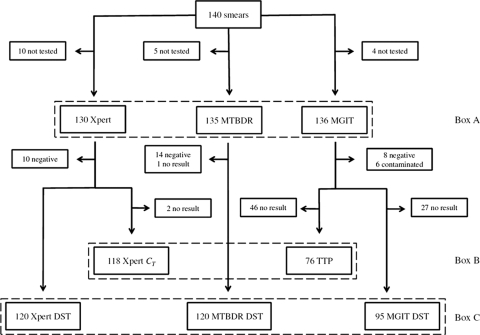
One hundred forty-three patients were screened for participation between 14 August and 27 November 2009. Three patients were excluded before a sputum sample was submitted, and 110, 28, and 2 patients submitted 1, 2, and 3 samples, respectively. In 120 patients (85.7%), all results were reported from a single sample. Patients whose results were derived from more than one sample were excluded from a comparative analysis of values that were not from the same sputum sample. A diagram of the sample flow is shown in Fig. 1.
Fig. 1.
One hundred forty patients provided a sputum sample for a direct smear. Detection of mycobacteria with all tests available could be directly compared between 126 patients (Box A). A quantitative comparison between Xpert and TTP was possible on 74 patients with a valid TTP result (Box B). Drug resistance detection (Box C) could be compared between NAATs in 118 patients, and the NAATs could be compared to MGIT-based DST in 95 patients.
Presence of Mycobacterium tuberculosis.
The ability to detect M. tuberculosis by NAAT and culture could be directly compared for 126 patients (Fig. 1). Six contaminated MGIT cultures were microscopy positive for AFB and were counted as positive for this analysis. Smear microscopy had the highest proportion of positive readings (96.8%), which was expected because smears were performed directly on homogenized sputa and not from the decontaminated pellets as for the other assays. The second most positive detection method was Xpert, closely followed by liquid culture and MTBDR (Table 1). Relating to the EBA study selection procedure, the sensitivities of MTBDR and Xpert for detection of a smear grade of ≥1+ were 97.3% and 100%, respectively, with a specificity of 64.3% for each. Xpert and MTBDR scored the same 5 scanty smears as positive, but Xpert recognized all ≥1+ smears as positive, while MTBDR missed 3 of the ≥1+ smears. This difference is not significant (P = 0.351, Fisher exact test). The main outcome endpoint in many clinical trials is sputum culture conversion from positive to negative during treatment. Xpert best predicted a positive culture (sensitivity, 95%), followed by MTBDR (92.4%) and direct smear ≥1+ (90.8%).
Table 1.
Detection of AFB or mycobacterial DNA in sputum (all n = 126)
| Smear status | No. (%) of patients |
||||||
|---|---|---|---|---|---|---|---|
| Total | MTBDR |
Xpert MTB/RIF |
MGIT |
||||
| Positive | Negative | Positive | Negative | Positive | Negative | ||
| All positive | 122 (96.8) | 114 (90.5) | 8 (6.3) | 117 (92.9) | 5 (4.0) | 116 (92.1) | 6 (4.8) |
| 3+ | 46 (36.5) | 46 (36.5) | 0 | 46 (36.5) | 0 | 44 (34.9) | 2 (1.6) |
| 2+ | 52 (41.3) | 49 (38.8) | 3 (2.4) | 52 (41.3) | 0 | 50 (39.7) | 2 (1.6) |
| 1+ | 14 (11.1) | 14 (11.1) | 0 | 14 (11.1) | 0 | 14 (11.1) | 0 |
| Scanty | 10 (7.9) | 5 (4.0) | 5 (4.0) | 5 (4.0) | 5 (4.0) | 8 (6.3) | 2 (1.6) |
| Negative | 4 (3.2) | 0 | 4 (3.2) | 0 | 4 (3.2) | 3 (2.4) | 1 (0.8) |
Quantitative analyses.
Semiquantitative microscopy smear grades were compared with continuous readings of MGIT TTP (n = 74, parallel positive readings) and Xpert CTs (n = 117, parallel positive readings) in Table 2. Only a limited number of TTP readings were available owing to a temporary fault in an incubator that affected incubation conditions and data storage and made the affected TTP readings unreliable. There was no significant difference between smear grade and Xpert CT values with and without a parallel TTP value, which excludes bias for the correlation analysis.
Table 2.
Quantitative comparison of smear grade, MGIT TTP, and Xpert CT
| Microscopy grade | TTP (h) |
CT |
||
|---|---|---|---|---|
| Mean (±SD) | No. of patients | Mean (±SD) | No. of patients | |
| 3+ | 121.0 (41.17) | 26 | 19.48 (3.24) | 44 |
| 2+ | 128.1 (26.45) | 31 | 21.13 (2.80) | 54 |
| 1+ | 146.7 (26.58) | 10 | 23.01 (2.49) | 14 |
| Scanty | 298.0 (167.9) | 7 | 22.94 (2.68) | 5 |
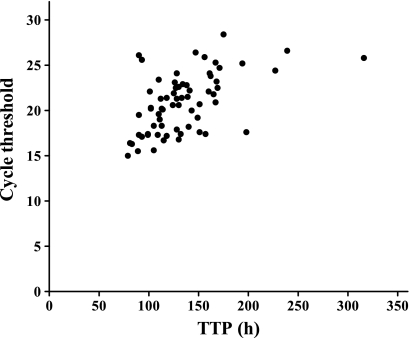
Table 2 shows a trend of increasing TTP and increasing CT with lower smear grades. A moderate negative correlation was found between smear grades and Xpert (r = −0.4017) and smear grade and TTP (r = −0.4783) (for both, Spearman P < 0.0001). Figure 2 shows the two continuous variables MGIT TTP and Xpert CT (n = 66, parallel readings), which correlated better with each other than either of them did with smear grade (r = 0.5394, Pearson P < 0.0001).
Fig. 2.
Linear correlation between Xpert cycle thresholds and MGIT TTP. The correlation is moderate but significant (P < 0.0001), with r equal to 0.5394 (Pearson).
Detection of resistance.
Three cases of RIF resistance and 6 cases of INH resistance were identified on culture-based MGIT DST (INH and RIF resistance, n = 2; RIF monoresistance, n = 1; INH monoresistance, n = 4). RIF susceptibility results for verification against MGIT DST were available in 93 and 91 cases for Xpert and MTBDR, respectively. MTBDR was 100% concordant with MGIT DST. RIF resistance determination with Xpert depends on the indication of a mutation by absent or delayed CT tracings. The Xpert system available at the time of the study indicated 10 potentially resistant samples. Manual verification revealed 3 certain cases of RIF resistance and 7 cases with borderline readings. Upon release of an updated Xpert cartridge version and software (version 4.0, July 2010), these uncertain samples were retested. The instrument detected M. tuberculosis in all samples and confirmed the original manual readings. Thus, 3 cases were RIF resistant on Xpert (100% accuracy). In a further 24 cases, we found concordant RIF susceptibility on Xpert and MTBDR without confirmatory MGIT DST. INH resistance was reported only by MTBDR. Compared to MGIT DST, MTBDR missed one case of INH resistance, resulting in a sensitivity of 83.3% and a specificity of 100% for INH resistance detection.
DISCUSSION
This study covered a series of 140 consecutive patients with smear-positive TB who were screened for participation in an EBA study. The novel molecular diagnostic tests Genotype MTBDRplus and Xpert MTB/RIF were more than 95% sensitive for an AFB smear grade of ≥1+ and accurately predicted RIF resistance (100%) confirmed with liquid culture. The quantitative measurement of Xpert was correlated with both TTP of liquid culture and smear positivity grade.
MTBDR was used per protocol to exclude patients with INH or RIF resistance from participation in the present study. The test confirmed its high accuracy in recognizing M. tuberculosis (90.5% of positive direct smears, 92.4% of positive liquid cultures) and drug resistance on sputum (100%), with results generally being available within 24 h (2, 15). The ability to objectively exclude patients from participation in a clinical trial constitutes major progress over soft criteria, such as previous exposure to antituberculosis drugs or to individuals with drug-resistant TB. Resistant patients can now be referred for adequate treatment without delay, and the risk of exposure of research staff and other participants to resistant TB is greatly reduced. Such a diagnostic test also allows immediate identification not only of clinical trial patients but also of all patients with drug-resistant pulmonary TB for the timely initiation of appropriate therapy.
Xpert has recently been validated as a diagnostic test for pulmonary TB (4). It is technically easier to handle than MTBDR. Its quick turnaround time means that results are available on the same day, but it does not include resistance testing for INH. The significance of this shortcoming is debatable. In practice, the vast majority of patients with RIF resistance are found to also carry INH resistance (11). Moreover, INH monoresistance is usually not considered a reason for an immediate change of treatment, whereas patients with RIF monoresistance would be approached similarly to multidrug-resistant patients irrespective of INH sensitivity (5). In the present study, Xpert identified all patients with at least ≥1+ positive smears, all patients with RIF resistance, and 95% of patients with a positive liquid culture. Five samples scanty positive on direct smear were not detected by either NAAT. This may be explained by the decontamination procedure and subsequent loss of bacteria. Interestingly, Boehme et al. could not find a significant difference between direct sputum and decontaminated samples in the Xpert validation study on diagnostic sputum samples (4).
TTP of growth in liquid culture has increasingly been proposed to be a measurement of sputum bacterial load. Baseline TTP and its increase during treatment have been shown to correlate with TB outcomes, and TTP has served as a secondary endpoint in EBA studies (7, 10, 14). Of note, Xpert CTs were correlated to both TTP and sputum smear grades in the present study. This adds to a previous study showing a negative correlation between TTP and CFU count (18). The correlation of CT and TTP raises the question of whether the Xpert result could be explored for use as a marker of treatment response in TB trials. As a measurement of mycobacterial DNA, Xpert CTs do not relate to the viability of mycobacteria, but this is probably a minor handicap in EBA studies of 14 days' duration. In phase IIB or III trials, however, the ability to identify patients still expectorating viable bacteria is essential. Xpert would have to be compared to mycobacterial culture for this purpose.
What are the possible conclusions for the design of clinical trials? Xpert would have excluded the same RIF-resistant patients as MTBDR. A requirement of a positive Xpert result would have included the same patients with the addition of 5 with only scanty positive sputum. The current practice of including only patients with a ≥1+ positive smear is based on the experience that such patients still have measureable CFU counts after 14 days of treatment with the current standard drug combination. This has stood the test of time, but it is an arbitrary standard, considering that drugs with unknown activity are explored. The selection of TTP as a trial endpoint might allow inclusion of scanty positive patients owing to the higher sensitivity of liquid culture.
This study's strong point is its relatedness to a clinical situation where the need for timely delivery of results does not allow the luxury of repeating tests from retained material. Such a scenario will allow the tests to reveal their practical value. As a drawback, some patients' results were incomplete or put together from more than one sputum specimen, as we were unable to request better-quality sputum samples in patients no longer considered for participation in the clinical study. An untimely technical breakdown due to a hardware failure in the MGIT machine further reduced the data available for analysis, but we do not believe that this introduced bias into the study. From the perspective of a manager of a busy TB clinic, the results should be interpreted with caution. Only patients already diagnosed with TB on at least one smear entered screening for the EBA study. This means that no individuals only suspected to have TB were included, as it would have been the case in the evaluation of a diagnostic test. This limited the chance for false-positive results.
In conclusion, this study has shown Xpert and MTBDR to be equally useful screening tools for clinical TB trials. If protocols do not explicitly require INH susceptibility to be determined, Xpert has the edge due to its fast turnover time and easy technical handling. The Xpert's all-in-one capability not only to determine the presence of M. tuberculosis and drug resistance but also to quantify the mycobacterial sputum load should be further explored, including during drug exposure.
ACKNOWLEDGMENTS
We thank all patients who volunteered to participate in the trial. Mark Nicol and Widaad Zemanay helped with the Xpert resistance readings.
The Foundation of Innovative New Diagnostics (FIND) contributed materials to this study.
S.O.F., X.A.K., A.V., R.D., P.R.D., and A.H.D. have no conflict of interest to state.
Footnotes
Published ahead of print on 8 June 2011.
REFERENCES
- 1. Banada P. P., et al. 2010. Containment of bioaerosol infection risk by the Xpert MTB/RIF assay and its applicability to point-of-care settings. J. Clin. Microbiol. 48:3551–3557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barnard M., Albert H., Coetzee G., O'Brien R., Bosman M. E. 2008. Rapid molecular screening for multidrug-resistant tuberculosis in a high-volume public health laboratory in South Africa. Am. J. Respir. Crit. Care Med. 177:787–792 [DOI] [PubMed] [Google Scholar]
- 3. Blakemore R., et al. 2010. Evaluation of the analytical performance of the Xpert MTB/RIF assay. J. Clin. Microbiol. 48:2495–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boehme C. C., et al. 2010. Rapid molecular detection of tuberculosis and rifampin resistance. N. Engl. J. Med. 363:1005–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chiang C. Y., Schaaf H. S. 2010. Management of drug-resistant tuberculosis. Int. J. Tuberc. Lung Dis. 14:672–682 [PubMed] [Google Scholar]
- 6. Corbett E. L., et al. 2003. The growing burden of tuberculosis: global trends and interactions with the HIV epidemic. Arch. Intern. Med. 163:1009–1021 [DOI] [PubMed] [Google Scholar]
- 7. Diacon A. H., et al. 2010. Time to detection of the growth of Mycobacterium tuberculosis in MGIT 960 for determining the early bactericidal activity of antituberculosis agents. Eur. J. Clin. Microbiol. Infect. Dis. 29:1561–1565 [DOI] [PubMed] [Google Scholar]
- 8. Donald P. R., Diacon A. H. 2008. The early bactericidal activity of anti-tuberculosis drugs: a literature review. Tuberculosis. (Edinb.) 88(Suppl. 1):S75–S83 [DOI] [PubMed] [Google Scholar]
- 9. El Hajj H. H., Marras S. A., Tyagi S., Kramer F. R., Alland D. 2001. Detection of rifampin resistance in Mycobacterium tuberculosis in a single tube with molecular beacons. J. Clin. Microbiol. 39:4131–4137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Epstein M. D., et al. 1998. Time to detection of Mycobacterium tuberculosis in sputum culture correlates with outcome in patients receiving treatment for pulmonary tuberculosis. Chest 113:379–386 [DOI] [PubMed] [Google Scholar]
- 11. Gandhi N. R., et al. 2010. Multidrug-resistant and extensively drug-resistant tuberculosis: a threat to global control of tuberculosis. Lancet 375:1830–1843 [DOI] [PubMed] [Google Scholar]
- 12. Hain Lifescience GmbH 2009. GenoType MTBDRplus version 1.0 product manual. Hain Lifescience GmbH, Nehren, Germany [Google Scholar]
- 13. Helb D., et al. 2010. Rapid detection of Mycobacterium tuberculosis and rifampin resistance by use of on-demand, near-patient technology. J. Clin. Microbiol. 48:229–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hesseling A. C., et al. 2010. Baseline sputum time to detection predicts month two culture conversion and relapse in non-HIV-infected patients. Int. J. Tuberc. Lung Dis. 14:560–570 [PubMed] [Google Scholar]
- 15. Hillemann D., Rusch-Gerdes S., Richter E. 2006. Application of the Genotype MTBDR assay directly on sputum specimens. Int. J. Tuberc. Lung Dis. 10:1057–1059 [PubMed] [Google Scholar]
- 16. International Union against Tuberculosis and Lung Disease 2000. Technical guide: sputum examination for tuberculosis by direct microscopy in low income countries, 5th ed. International Union against Tuberculosis and Lung Disease, Paris, France [Google Scholar]
- 17. Kapp C. 2009. South Africa tries new approach to resistant tuberculosis. Lancet 374:441. [DOI] [PubMed] [Google Scholar]
- 18. Pheiffer C., et al. 2008. Time to detection of Mycobacterium tuberculosis in BACTEC systems as a viable alternative to colony counting. Int. J. Tuberc. Lung Dis. 12:792–798 [PubMed] [Google Scholar]
- 19. Piatek A. S., et al. 1998. Molecular beacon sequence analysis for detecting drug resistance in Mycobacterium tuberculosis. Nat. Biotechnol. 16:359–363 [DOI] [PubMed] [Google Scholar]
- 20. Vestal A. L. 2011. Procedures for the isolation and identification of mycobacteria (publication no. 77-8230). U.S. Department of Health and Human Services, Washington, DC [Google Scholar]
- 21. World Health Organization 2008. Anti-tuberculosis drug resistance in the world. The WHO/IUATLD Global Project on Anti-Tuberculosis Drug Resistance Surveillance. http://www.who.int/tb/publications/2008/drs_report4_26feb08.pdf World Health Organization, Geneva, Switzerland [Google Scholar]
- 22. World Health Organization 2010. Tuberculosis fact sheet no. 104. WHO Media Center. http://www.who.int/mediacentre/factsheets/fs104/en/print.html World Health Organization, Geneva, Switzerland [Google Scholar]