Abstract
During the COMParative Activity of Carbapenems Testing (COMPACT) surveillance study, 448 Pseudomonas aeruginosa clinical isolates were obtained from 16 Spanish hospitals. Nonsusceptibility (EUCAST breakpoints) to imipenem (35%), meropenem (33%), and/or doripenem (33%) was observed with 175 isolates (39%). Simultaneous resistance to these three drugs was observed with 126 of the 175 isolates (72%). Except for colistin, high resistance rates were observed among noncarbapenem antibiotics. Clonal relatedness was investigated by pulsed-field gel electrophoresis (PFGE) with SpeI, discriminating 68 patterns. Multilocus sequence typing (MLST) was performed on 84 isolates representing different PFGE types and all participating hospitals. Thirty-nine sequence types (STs) could be distinguished, and of these, ST175 (48 isolates, 10 hospitals), ST646 (16 isolates, 4 hospitals), ST532 (13 isolates, 3 hospitals), and ST111 (13 isolates, 7 hospitals) were the most frequently encountered. Minimum-spanning tree analysis confirmed a wide dissemination of different clones among participant hospitals, particularly ST175. PFGE pattern comparison within the four most frequent STs revealed that ST175 isolates were relatively uniform, while ST646, ST532, and ST111 isolates were highly diverse, with almost every isolate belonging to a unique pulsotype, even when originating from the same center. The population of carbapenem-nonsusceptible P. aeruginosa isolates from 16 hospitals is highly diverse, with one ST (ST175) representing a highly conserved clone disseminated in 10 of the 16 participant hospitals. This ST175 clone should be added to the list of P. aeruginosa clones at high risk for epidemic spread, such as the Liverpool, Manchester, and Melbourne clones previously found in cystic fibrosis patients and ST235 in the nosocomial setting.
INTRODUCTION
Antibiotic surveillance studies are necessary not only for the better understanding of bacterial epidemiology, but also for the design of control strategies for preventing bacterial resistance and establishing therapeutic guidelines. In this context, the COMParative Activity of Carbapenems Testing (COMPACT) multicenter study was performed in 16 European countries, including Spain (23). The main objectives of this surveillance study were to test the activity of currently used carbapenems against Pseudomonas aeruginosa clinical isolates and to detect carbapenem nonsusceptible isolates (12).
In this multicenter study, Spanish authors concluded that percentages of carbapenem resistance among P. aeruginosa clinical isolates in their country (range of 21.4% to 32.6%) are very similar to those previously reported in other European countries. Doripenem was the most active drug against these resistant isolates (12). The presence of carbapenem-nonsusceptible P. aeruginosa isolates has been described worldwide, particularly in relation to outbreaks (1, 5, 6, 17, 18, 25, 28, 30, 31, 37). Most of these studies focused on clinical isolates, and genetic relatedness was established using molecular typing tools based on macrorestriction-fragment analysis or PCR of repetitive DNA sequences on the chromosome. These methods are well suited to study local outbreaks over a short time period. To study the genetic relatedness of isolates recovered from multicenters over an extended period of time and to indicate that isolates are members of particular clonal lineages, other typing methods that do not index rapidly evolving variation, like multilocus sequence typing (MLST) analysis, are preferred.
MLST analysis, initially proposed as a highly reproducible and portable epidemiological typing method to unambiguously identify clones within populations of pathogenic microorganisms, also allows better understanding of the population structure of bacterial species (21). For P. aeruginosa, the MLST scheme was developed by Curran et al. (8), with 1,059 different STs currently annotated (http://pubmlst.org/paeruginosa/). This scheme has demonstrated that P. aeruginosa has a nonclonal population structure with highly successful epidemic clones. Furthermore, MLST and multiple-locus variable-number tandem-repeat analysis (MLVA) revealed that P. aeruginosa clones colonizing cystic fibrosis patients are genotypically distinct from other clinical clones and thus seem to be well adapted to the lungs of these patients (33, 34).
The objectives of the present study were to analyze the population structure and the antimicrobial susceptibility of carbapenem-nonsusceptible P. aeruginosa isolates recovered from 2008 through 2009 in 16 Spanish hospitals during the COMPACT multicenter surveillance study.
MATERIALS AND METHODS
Bacterial isolates.
From an initial contemporary collection of 448 P. aeruginosa clinical isolates obtained from 16 Spanish hospitals during the COMPACT study (13), we selected the carbapenem-nonsusceptible isolates (175 isolates, 39.3%) for further studies. Of these, 35% were nonsusceptible to imipenem (MIC, >4 μg/ml), 33% nonsusceptible to meropenem (MIC, >2 μg/ml), and 33% nonsusceptible to doripenem (MIC, >1 μg/ml), using EUCAST breakpoints.
Susceptibility testing.
Susceptibility to imipenem, doripenem, and meropenem was determined by both standard microdilution (7) and Etest methods. Susceptibility to noncarbapenem antibiotics, including piperacillin-tazobactam, ceftazidime, cefepime, gentamicin, tobramycin, amikacin, netilmicin, ciprofloxacin, levofloxacin, fosfomycin, and colistin, were determined by microdilution using the Wider semiautomatic system (Francisco Soria Melguizo, Madrid, Spain) (4). As previously stated, EUCAST interpretative criteria were used (version 1.3; http://www.eucast.org).
Molecular typing.
In total, 175 carbapenem-nonsusceptible isolates were initially typed by pulsed-field gel electrophoresis (PFGE) using a modified protocol. Briefly, from an overnight culture of 5 ml of LB broth (Difco, Detroit, MI) of each isolate, an aliquot of 1 ml was centrifuged, and the pellet was mixed with 200 μl of SE (75 mM NaCl, 25 mM EDTA Na2 [filter sterilized]) solution, pH 7.5, in the same Eppendorf tube. Optical density was adjusted between 0.6 and 2.5 using the Nanodrop system (Thermo Scientific, Wilmington, DE) at 590 nm. Subsequently, 200 μl of 2% agarose was added to the Eppendorf tube, gently mixed, and deposited in appropriate plug molds. Overnight protein lysis was carried out at 56°C in 2 ml of lysis solution (EDTA, 0.5 M [pH 9], 10% Sarkosyl, and 25 mg/ml of proteinase K). Plugs were then washed twice in 2 ml of Tris-EDTA (TE) for 30 min. A third part of each plug was digested with 15 U of SpeI (Roche Diagnostics, Indianapolis, IN) at 37°C for at least 2 h. Electrophoresis was carried out in a 1.2% agarose gel in 0.5× Tris-borate-EDTA (TBE) at 14°C with the following settings: 6 v/cm2, 5 to 40 s, and 22 h. To define the different genetic lineages, a dendrogram was constructed using the Phoretix 5.0 software (Nonlinear Dynamics Ltd., United Kingdom) based on Dice's coefficient. The visual Tenover criteria were also applied (32).
Two or more isolates were considered to be clonally related when Dice's coefficient values were higher than 0.8. In order to avoid duplicate isolates, special care was taken with those strains recovered in the same hospital, and only one isolate per pulsotype and institution was further selected for MLST. Nevertheless, when similar PFGE patterns were found in different hospitals, one isolate per center was included in order to ascertain dissemination of clones between hospitals. The MLST technique was conducted according to the guidelines at http://pubmlst.org/paeruginosa/, which were developed by Keith Jolley (14), and new alleles were sent to curator Eleanor Pinnock. The minimum-spanning tree (MST) figure was constructed using the Bionumerics 5.1 program (Applied Maths, St.-Martens-Latem, Belgium).
Bacterial DNA was obtained using the QiaAmp kit (Qiagen, Hilden, Germany), and PCR amplicons were purified with ExoSAP-IT (GE Healthcare, Little Chalfont, United Kingdom). Nucleotide sequences were compared with those previously described in the MLST database to assign the corresponding sequence type (ST).
Statistic analysis.
Based on MLST data, the genetic diversity (D) of nonsusceptible P. aeruginosa was calculated for each individual hospital using Ridom Epicompare software (http://www.ridom.de/download.shtml).
RESULTS
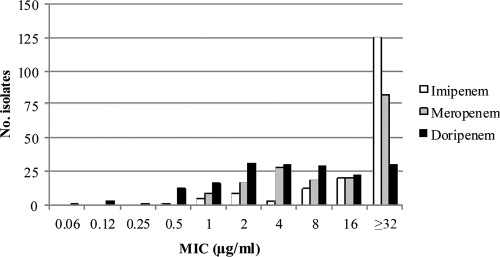
MIC distributions for imipenem, meropenem, and doripenem of the 175 carbapenem-nonsusceptible P. aeruginosa isolates recovered from the COMPACT surveillance study in 16 Spanish hospitals are shown in Fig. 1. Using the EUCAST clinical breakpoints, only 18 isolates (10.3%) remained susceptible to imipenem, 26 isolates (14.8%) to meropenem, and 31 isolates (17.7%) to doripenem. A high proportion of intermediate-resistant isolates was detected for doripenem (63 isolates, 36%) and meropenem (43 isolates, 24.6%) in comparison with imipenem (12 isolates, 6.9%). Multiresistance (including both resistant and intermediate isolates) against the three carbapenems was observed with 126 isolates (Table 1). Susceptibility testing results for the noncarbapenem antibiotics are shown in Table 2 and revealed high resistance rates for all antibiotics, except for colistin.
Fig. 1.
MIC distribution of 175 clinical P. aeruginosa isolates for each carbapenem studied that were not susceptible to at least imipenem (MIC, >4 μg/ml), meropenem (MIC, >2 μg/ml), or doripenem (MIC, > 1 μg/ml).
Table 1.
Percentage of isolates displaying nonsusceptibility to one or more carbapenems
| Carbapenem(s)a | No. (%) of isolates displaying nonsusceptibility |
|---|---|
| Imipenem | 27 (15.4) |
| Meropenem | 5 (2.8) |
| Doripenem | 1 (0.5) |
| Imipenem and meropenem | 10 (5.7) |
| Meropenem and doripenem | 6 (3.4) |
| Imipenem, meropenem, and doripenem | 126 (72.0) |
Imipenem MIC, >4 μg/ml; meropenem MIC, >2 μg/ml; doripenem MIC, >1 μg/ml.
Table 2.
Antibiotic susceptibility results for noncarbapenem antibiotics using EUCAST criteria
| Antibiotic | No. of isolates inhibited at the following concn (μg/ml)a |
MIC (μg/ml)b |
% Sc | % Rc | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.12 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | 256 | 50% | 90% | |||
| Ceftazidime | [22 | 26 | 22 | 22 | 30] | 53 | 8 | 32 | 42.2 | 57.8 | ||||||
| Cefepime | [4 | 16 | 20 | 34 | 49] | 52 | 16 | 32 | 52 | 78 | ||||||
| Gentamicin | [23 | 32 | 33] | 87 | 8 | 16 | 31.4 | 68.6 | ||||||||
| Tobramycin | [82 | 8 | 4] | 81 | 4 | 16 | 51.4 | 48.6 | ||||||||
| Amikacin | [72 | 1 | 58 | 22] | 22 | 16 | 64 | 41.7 | 58.3 | |||||||
| Netilmicin | [23 | 0 | 34] | 118 | 16 | 16 | 13.1 | 86.9 | ||||||||
| Ciprofloxacin | [39 | 8 | 7 | 6] | 115 | 4 | 4 | 26.8 | 73.2 | |||||||
| Levofloxacin | [3 | 2 | 5 | 30 | 9 | 9] | 117 | 4 | 4 | 22.8 | 87.2 | |||||
| Fosfomycin | [30 | 46] | 99 | 128 | 128 | 17.1 | 82.9 | |||||||||
| Colistin | [169 | 5] | 1 | 2 | 2 | 99.4 | 0.6 | |||||||||
| Piperacillin-tazobactam | [85d | 17e | 28f] | 45g | 32/4 | ≥64/4 | 48.5 | 51.5 | ||||||||
Left and right brackets indicate the lowest and highest concentrations tested, respectively.
50% and 90%, MICs at which 50 and 90% of the isolates, respectively, were inhibited.
Percentage of susceptible (S) and resistant (R) isolates according to EUCAST criteria.
Piperacillin/tazobactam concentrations (μg/ml) were ≤16/4, respectively.
Piperacillin/tazobactam concentrations (μg/ml) were 32/4, respectively.
Piperacillin/tazobactam concentrations (μg/ml) were 64/4, respectively.
Piperacillin/tazobactam concentrations (μg/ml) were >64/4, respectively.
Genetic diversity among the 175 carbapenem-nonsusceptible isolates was initially explored by PFGE-SpeI by constructing a dendrogram based on Dice's coefficient using the Phoretix 5.0 database software (data not shown). In total, 68 different pulsotypes were obtained.
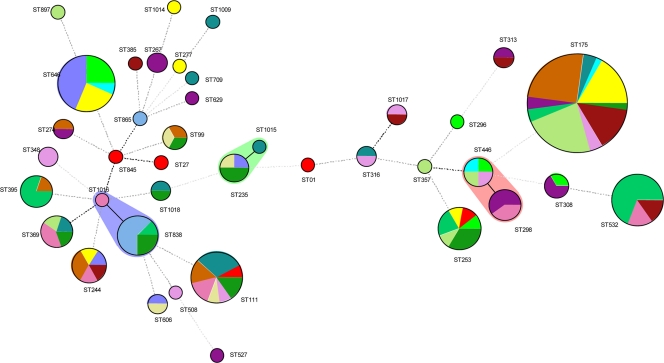
MLST was performed on 84 isolates selected according to their pulsotypes and/or their hospital origin and revealed 39 STs. MLST results are summarized in Table 3 and Fig. 2. The most prevalent ST was ST175, representing 48 isolates from 10 hospitals, followed by ST646 with 16 isolates from 4 hospitals, and ST532 and ST111 with 13 isolates each from 3 and 7 hospitals, respectively. Other prevalent STs were ST838, with 8 isolates from 3 hospitals; ST244, with 6 isolates from 5 hospitals; and ST298, ST369, and ST395, with 5 isolates each from 2, 4, and 2 hospitals, respectively. Sixteen STs were unique (one isolate/one hospital), while 8 STs were represented by two isolates. ST1014, ST1015, ST1016, ST1017, and ST1018 were described for the first time in our collection and were annotated in the MLST database. Genetic distance among the different STs was analyzed by making use of an MST (Fig. 2). This MST displays the genotypic heterogeneity of this collection of carbapenem-nonsusceptible P. aeruginosa by revealing three small clusters of two STs (ST98 to ST446; ST838 to ST1016; and ST235 to ST1015), while the other 33 STs were singletons. This indicates that carbapenem nonsusceptibility is not confined to a single genetic lineage or subpopulation.
Table 3.
Genetic diversity among the 16 hospitals included in the study
| Hospital no. | No. of isolates | No. of PFGE patterns | No. of STs | D (95% CI) | ST no. (no. of isolates) |
|---|---|---|---|---|---|
| 148 | 8 | 8 | 5 | 0.786 (0.521-1.00) | 208 (1), 253 (1), 296 (1), 446 (1), 646 (4) |
| 149 | 5 | 5 | 5 | 1 | 27 (1), 111 (1), 253 (1), 701 (1), 845 (1) |
| 150 | 10 | 6 | 4 | 0.533 (0.186-0.881) | 235 (1), 244 (1), 606 (1), 646 (7) |
| 152 | 16 | 9 | 6 | 0.717 (0.532-0.901) | 175 (8), 244 (1), 253 (1), 277 (1), 646 (4), 1014 (1) |
| 153 | 3 | 3 | 3 | 1 | 175 (1), 446 (1), 646 (1) |
| 154 | 12 | 10 | 8 | 0.927 (0.833-1.00) | 111 (4), 175 (2), 316 (1), 369 (1), 709 (1), 1009 (1), 1015 (1), 1018 (1) |
| 155 | 19 | 8 | 6 | 0.602 (0.359-0.846) | 111 (2), 175 (12), 244 (2), 274 (1), 395 (1), 699 (1) |
| 156 | 13 | 11 | 8 | 0.923 (0.861-0.985) | 175 (2), 267 (2), 274 (1), 298 (3), 308 (2), 313 (1), 527 (1), 629 (1) |
| 157 | 18 | 8 | 5 | 0.712 (0.544-0.881) | 175 (2), 253 (2), 395 (4), 532 (9), 838 (1) |
| 158 | 16 | 7 | 6 | 0.542 (0.252-0.831) | 175 (11), 253 (1), 357 (1), 369 (1), 446 (1), 897 (1) |
| 159 | 6 | 3 | 2 | 0.333 (0.0-0.739) | 838 (5), 865 (1) |
| 161 | 10 | 7 | 6 | 0.911 (0.861-0.962) | 111 (2), 244 (1), 298 (2), 369 (2), 532 (2), 1016 (1) |
| 162 | 4 | 4 | 4 | 1 (1.00-1.00) | 111 (1), 235 (1), 606 (1), 699 (1) |
| 165 | 9 | 7 | 7 | 0.944 (0.871-1.00) | 111 (1), 175 (2), 316 (1), 348 (2), 446 (1), 508 (1), 1017 (1) |
| 167 | 13 | 6 | 6 | 0.718 (0.475-0.961) | 175 (7), 244 (1), 313 (1), 385 (1), 532 (2), 1017 (1) |
| 168 | 13 | 10 | 8 | 0.923 (0.861-0.985) | 111 (2), 175 (1), 235 (2), 253 (3), 369 (1), 699 (1), 838 (2), 1018 (1) |
Fig. 2.
Minimum-spanning tree of 175 P. aeruginosa clinical isolates. Each color represents a single hospital (n = 16). Wide lines represent single-locus variants, dotted lines represent multilocus variants, and the size of the circles represents the number of isolates found with the respective ST.
The genetic diversity (D) of carbapenem-nonsusceptible P. aeruginosa in the different hospitals ranged between 0.333 (95% confidence interval [CI], 0.00 to 0.739) for hospital 159 (6 isolates) and 1 for hospitals 149 (4 isolates), 153 (3 isolates) and 162 (4 isolates). A value of 1, found for these three hospitals, means that each isolate had a different ST. This situation might indicate independent emergence or acquisition of nonsusceptible P. aeruginosa from different sources and not via cross-transmission and possibly reflects effective infection control measures. On the other hand, these three hospitals corresponded to those with a low number of isolates (3 or 4 isolates), whereas hospitals with more isolates in general presented lower values of D. When taking into account only hospitals from which ≥10 isolates were typed, hospital 158 had a significantly lower D than hospitals 154, 156, 161, and 168, suggesting more-frequent events of cross-transmission in hospital 158 compared with other hospitals. In hospital 158, 11 out of 16 (68.7%) isolates belonged to ST175.
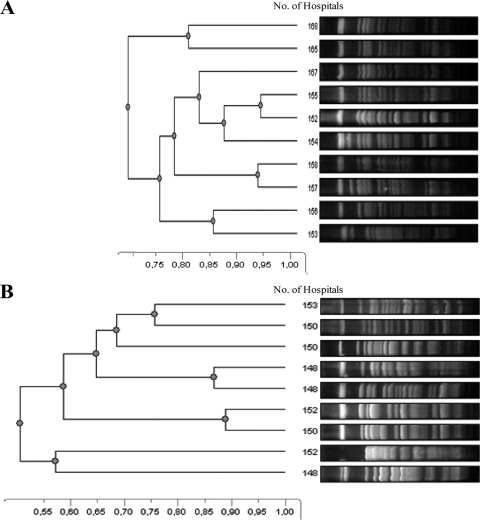
To investigate microevolution within each ST in the different hospitals, PFGE patterns from isolates with the same ST were compared (Fig. 3). This revealed that all isolates with ST175 had a uniform PFGE band pattern (Fig. 3A), while for isolates with ST646 (Fig. 3B) and ST532 and ST111 isolates (data not shown for these STs), each pulsotype was almost unique, even when isolates originated from the same center.
Fig. 3.
PFGE-SpeI of the different pulsotypes detected for ST175 (A) and ST646 (B) in isolates from different hospitals.
Despite the fact that ST175 is genetically highly homogenous, based on PFGE, isolates displayed a heterogeneous antibiotic resistance profile, ranging from resistance to three antibiotics to resistance to nine antibiotics. Highly different antibiograms, ranging from susceptibility to ceftazidime, cefepime, ciprofloxacin, levofloxacin, amikacin, tobramycin, and/or fosfomycin to isolates resistant to these antibiotics, were even observed among isolates recovered from the same hospital. In the case of ST175, 91.5% of the isolates (43 out of 47) were resistant to fluoroquinolones, 97.9% (46 out of 47) to gentamicin, 91.4% (43 out of 47) to tobramycin, and 17% to amikacin (8 out of 47). Moreover, a high proportion of isolates was resistant to ceftazidime (63.8%) and cefepime (70.2%). This high diversity in the resistant phenotype was also observed with isolates with prevalent STs, such as ST646, ST532, and ST111. In general, different phenotypes were associated with the same ST and similar profiles with different STs in the same and different centers.
DISCUSSION
Nosocomial outbreaks caused by P. aeruginosa have previously been described for several Spanish hospitals, and some of these outbreaks included carbapenem-resistant isolates (2, 15, 24, 25, 27, 29). As part of the COMPACT multicenter study, in which the rates of susceptibility to carbapenems of common Gram-negative bacilli causing serious infections in hospitalized patients were compared, 448 P. aeruginosa clinical isolates were recovered. These isolates displayed a higher intrinsic activity of doripenem than of meropenem and imipenem (resistance rates of 21.4%, 22.7%, and 32.6%, respectively) (12). The present study was focused on the analysis of the population structure and diversity of the carbapenem-nonsusceptible (n = 175) P. aeruginosa clinical isolates recovered from 16 institutions during the COMPACT study.
As expected, high diversity was observed by both PFGE and MLST. MLST revealed that of the 39 STs, only 6 were evolutionally linked into three small clonal complexes of two STs each, while the other 33 STs were part of distinct evolutionary lineages, indicating that carbapenem resistance was not confined to a single P. aeruginosa lineage. Despite the high level of genetic diversity, some STs were recovered from different hospitals, suggesting intra- and interhospital dissemination of carbapenem-nonsusceptible P. aeruginosa. The most prevalent lineages were ST175, ST646, ST111, and ST532. In addition to its countrywide distribution, ST175 appears to be capable of causing outbreaks, as occurred in two hospitals in which this ST was identified in 12 of 19 isolates and in 11 of 16 isolates (Table 3).
ST175 has previously been recognized as a contaminant of the hospital environment and colonizer of respiratory secretions in cystic fibrosis patients (19). Moreover, the ST175 clone seems to have spread extensively in Central Europe. It has been associated with multiresistant isolates, particularly with those producing the VIM-2 metallo-β-lactamase (MBL), and can be considered a high-risk clone (10, 20, 22). It is of note that within ST175 isolates, six isolates were VIM-2 cluster producers, each from different centers (data not shown; see reference 26 for further information). The other high-prevalence clone, ST646, has also been found among invasive isolates in Central Europe associated with a multiresistance phenotype (23). The most successful clones, with respect to worldwide dissemination, are ST111, ST235, and ST357 (9, 11, 18). Of these, ST111 was also frequently found in our study, while the other two STs (ST235 and ST357) were represented by only a small number of isolates (9, 11, 18). Nevertheless, the ST235 clone was recently reported in Spain, linked either to MBL production (16) or to a large outbreak by a multidrug-resistant strain producing the extended-spectrum beta-lactamases (ESBLs) GES-1 and GES-5 (35).
A more detailed investigation of the genetic microevolution of different clones using PFGE confirmed the highly clonal nature of ST175. In contrast, PFGE patterns of ST646, ST532, and ST111 were much more diverse, suggesting microevolution among isolates with these STs. This might be due to mutations or the acquisition of exogenous DNA, possibly coding for antimicrobial resistance. In fact, no clear association between pulsotype and antibiogram was obtained. This finding is supported by previous results described for ST111, which is associated with the production of VIM-2 β-lactamase, in which different PFGE and resistance profiles were obtained (9).
Susceptibility testing showed that carbapenem resistance is frequently associated with resistance to non-β-lactam antibiotics, including fluoroquinolones (73.2% to 87.2%) and aminoglycosides (48.6% to 86.9%). This fact was observed in other surveillance studies and could be due to the frequent linkage of different resistance mechanisms, including OprD inactivation and AmpC and efflux pump overexpression. It could also be due to topoisomerase mutations and acquisition of resistant determinants, such as aminoglycoside-modifying enzymes (3, 13, 22). The fact that only 57.8% and 78% of the carbapenem-nonsusceptible P. aeruginosa isolates were resistant to ceftazidime and cefepime, respectively, denotes the presence of β-lactam resistance mechanisms that variably affect different β-lactam antibiotics. Interestingly, and despite the multiresistant nature of this collection, only one isolate belonging to the ST348 clone was resistant to colistin.
In summary, we found that carbapenem-nonsusceptible P. aeruginosa isolates recovered during the COMPACT multicenter surveillance study displayed a multiresistance phenotype, including resistance against both β-lactam and non-β-lactam antibiotics. The population structure determined for the first time in a well-defined collection of carbapenem-nonsusceptible P. aeruginosa isolates is characterized by polyclonality in which most STs appeared not to be evolutionally linked. Some clones, particularly ST175, displaying multiresistance were found to be well dispersed among different hospitals and should be added to the list of the so-called high-risk clones, such as the Liverpool, Manchester, and Melbourne clones from cystic fibrosis patients or ST235 in the nosocomial setting (36).
ACKNOWLEDGMENTS
R.D.C. has a contract from the Instituto de Salud Carlos III-FIS (CB05/137). This work was supported with a grant from Janssen-Cilag (Madrid, Spain), Ministerio de Ciencia e Innovación of Spain and Instituto de Salud Carlos III, through the Spanish Network for the Research in Infectious Diseases (REIPI C03/14 and RD06/0008) and the European Project TROCAR (HEALTH-F3-2008-223031).
This study used the Pseudomonas aeruginosa MLST website (http://pubmlst.org/paeruginosa/) developed and managed by Keith Jolley and curated by Eleanor Pinnock.
Footnotes
Published ahead of print on 22 June 2011.
REFERENCES
- 1. Baumgart A. M., Molinari M. A., Silveira A. C. 2010. Prevalence of carbapenem resistant Pseudomonas aeruginosa and Acinetobacter baumannii in high complexity hospital. Braz. J. Infect. Dis. 14: 433–436 [DOI] [PubMed] [Google Scholar]
- 2. Bou R., et al. 2006. Nosocomial outbreak of Pseudomonas aeruginosa infections related to a flexible bronchoscope. J. Hosp. Infect. 64: 129–135 [DOI] [PubMed] [Google Scholar]
- 3. Cabot G., et al. 2011. Overexpression of AmpC and efflux pumps in Pseudomonas aeruginosa isolate from bloodstream infections: prevalence and impact on resistance in a Spanish multicenter study. Antimicrob. Agents Chemother. 55: 1906–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cantón R., et al. 2000. Evaluation of the Wider system, a new computer-assisted image-processing device for bacterial identification and susceptibility testing. J. Clin. Microbiol. 38: 1339–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carvalho A. P., et al. 2006. Characterization of an epidemic carbapenem-resistant Pseudomonas aeruginosa producing SPM-1 metallo-β-lactamase in a hospital located in Rio de Janeiro, Brazil. Microb. Drug Resist. 12: 103–108 [DOI] [PubMed] [Google Scholar]
- 6. Cezário R. C., et al. 2009. Nosocomial outbreak by imipenem-resistant metallo-β-lactamase-producing Pseudomonas aeruginosa in an adult intensive care unit in a Brazilian teaching hospital. Enferm. Infecc. Microbiol. Clin. 27: 269–274 [DOI] [PubMed] [Google Scholar]
- 7. Clinical Laboratory Standards Institute 2010. Performance standards for antimicrobial susceptibility testing; 20th informational supplement. M100-S20. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 8. Curran B., Jonas D., Grundmann H., Pitt T., Dowson C. G. 2004. Development of a multilocus sequence typing scheme for the opportunistic pathogen Pseudomonas aeruginosa. J. Clin. Microbiol. 42: 5644–5649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Edalucci E., et al. 2008. Acquisition of different carbapenem resistance mechanisms by an epidemic clonal lineage of Pseudomonas aeruginosa. Clin. Microbiol. Infect. 14: 88–90 [DOI] [PubMed] [Google Scholar]
- 10. Elias J., et al. 2010. Nosocomial outbreak of VIM-2 metallo-β-lactamase producing Pseudomonas aeruginosa associated with retrograde urography. Clin. Microbiol. Infect. 16: 1494–1500 [DOI] [PubMed] [Google Scholar]
- 11. Empel J., et al. 2007. Outbreak of Pseudomonas aeruginosa infections with PER-1 extended-spectrum β-lactamase in Warsaw, Poland: further evidence for an international clonal complex. J. Clin. Microbiol. 45: 2829–2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gimeno C., Cantón R., García A., Gobernado M., Grupo Español de Estudio de Doripenem 2010. Comparative activity of doripenem, meropenem, and imipenem in recent clinical isolates obtained during the COMPACT-Spain epidemiological surveillance study. Rev. Esp. Quimioter. 23: 144–152 [PubMed] [Google Scholar]
- 13. Hocquet D., et al. 2007. Pseudomonas aeruginosa may accumulate drug resistance mechanisms without losing its ability to cause bloodstream infections. Antimicrob. Agents Chemother. 51: 3531–3536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jolley K. A., Chan M. S., Maiden M. C. 2004. mlstdbNet—distributed multi-locus sequence typing (MLST) databases. BMC Bioinformatics 5: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Juan C., et al. 2005. Contribution of clonal dissemination and selection of mutants during therapy to Pseudomonas aeruginosa antimicrobial resistance in an intensive care unit setting. Clin. Microbiol. Infect. 11: 887–892 [DOI] [PubMed] [Google Scholar]
- 16. Juan C., et al. 2010. Metallo-beta-lactamase-producing Pseudomonas putida as a reservoir of multidrug resistance elements that can be transferred to successful Pseudomonas aeruginosa clones. J. Antimicrob. Chemother. 65: 474–478 [DOI] [PubMed] [Google Scholar]
- 17. Kikuchi T., et al. 2007. Contaminated oral intubation equipment associated with an outbreak of carbapenem-resistant Pseudomonas in an intensive care unit. J. Hosp. Infect. 65: 54–57 [DOI] [PubMed] [Google Scholar]
- 18. Koh T. H., et al. 2010. Multilocus sequence types of carbapenem-resistant Pseudomonas aeruginosa in Singapore carrying metallo-β-lactamase genes, including the novel bla(IMP-26) gene. J. Clin. Microbiol. 48: 2563–2564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lanini S., et al. 2011. Molecular epidemiology of a Pseudomonas aeruginosa hospital outbreak driven by a contaminated disinfectant-soap dispenser. PLoS One 16: e17064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Libisch B., Balogh B., Füzi M. 2009. Identification of two multidrug-resistant Pseudomonas aeruginosa clonal lineages with a countrywide distribution in Hungary. Curr. Microbiol. 58: 111–116 [DOI] [PubMed] [Google Scholar]
- 21. Maiden M. C., et al. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. U. S. A. 95: 3140–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nemec A., Krizova L., Maixnerova M., Musilek M. 2010. Multidrug-resistant epidemic clones among bloodstream isolates of Pseudomonas aeruginosa in the Czech Republic. Res. Microbiol. 161: 234–242 [DOI] [PubMed] [Google Scholar]
- 23. Nordmann P., et al. 2011. Comparative activity of carbapenem testing: the COMPACT study. J. Antimicrob. Chemother. 66: 1070–1078 [DOI] [PubMed] [Google Scholar]
- 24. Peña C., et al. 2003. An outbreak of carbapenem-resistant Pseudomonas aeruginosa in a urology ward. Clin. Microbiol. Infect. 9: 938–943 [DOI] [PubMed] [Google Scholar]
- 25. Peña C., et al. 2009. Nosocomial outbreak of a non-cefepime-susceptible ceftazidime-susceptible Pseudomonas aeruginosa strain overexpressing MexXY-OprM and producing an integron-borne PSE-1 beta-lactamase. J. Clin. Microbiol. 47: 2381–2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Riera E., et al. 2011. Pseudomonas aeruginosa carbapenem resistance mechanisms in Spain: impact on the activity of imipenem, meropenem and doripenem. J. Antimicrob. Chemother. [Epub ahead of print.] doi:10.1093/jac/dkr232 [DOI] [PubMed] [Google Scholar]
- 27. Rosselló J., et al. 1992. Investigation of an outbreak of nosocomial infection due to a multiply drug-resistant strain of Pseudomonas aeruginosa. J. Hosp. Infect. 20: 87–96 [DOI] [PubMed] [Google Scholar]
- 28. Ryoo N. H., et al. 2009. Outbreak by meropenem-resistant Pseudomonas aeruginosa producing IMP-6 metallo-β-lactamase in a Korean hospital. Diagn. Microbiol. Infect. Dis. 63: 115–117 [DOI] [PubMed] [Google Scholar]
- 29. Sánchez-Carrillo C., et al. 2009. Contaminated feeding bottles: the source of an outbreak of Pseudomonas aeruginosa infections in a neonatal intensive care unit. Am. J. Infect. Control 37: 150–154 [DOI] [PubMed] [Google Scholar]
- 30. Satoh R., et al. 2008. An outbreak and isolation of drug-resistant Pseudomonas aeruginosa at Niigata University Hospital, Japan. J. Infect. Chemother. 14: 325–329 [DOI] [PubMed] [Google Scholar]
- 31. Scheffer M. C., Gales A. C., Barth A. L., do Carmo Filho J. R., Dalla-Costa L. M. 2010. Carbapenem-resistant Pseudomonas aeruginosa: clonal spread in southern Brazil and in the state of Goiás. Braz. J. Infect. Dis. 14: 508–509 [PubMed] [Google Scholar]
- 32. Tenover F. C., et al. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33: 2233–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. van Mansfeld R., et al. 2009. Pseudomonas aeruginosa genotype prevalence in Dutch cystic fibrosis patients and age dependency of colonization by various P. aeruginosa sequence types. J. Clin. Microbiol. 47: 4096–4101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. van Mansfeld R., et al. 2010. The population genetics of Pseudomonas aeruginosa isolates from different patient populations exhibits high-level host specificity. PLoS One 5: e13482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Viedma E., et al. 2009. Nosocomial spread of colistin-only-sensitive sequence type 235 Pseudomonas aeruginosa isolates producing the extended-spectrum beta-lactamases GES-1 and GES-5 in Spain. Antimicrob. Agents Chemother. 53: 4930–4933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Woodford N., Turton J. F., Livermore D. M. 9 February 2011. Multiresistant Gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol. Rev. doi:10.1111/j.1574-6976.2011.00268.x [DOI] [PubMed] [Google Scholar]
- 37. Zilberberg M. D., Chen J., Mody S. H., Ramsey A. M., Shorr A. F. 2010. Imipenem resistance of Pseudomonas in pneumonia: a systematic literature review. BMC Pulm. Med. 10: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]