Abstract
Two mechanisms account for AmpC activity in Escherichia coli, namely, mutations in the ampC promoter and attenuator regions resulting in ampC overexpression and acquisition of plasmid-carried ampC genes. In this study, we analyzed 51 clinical E. coli isolates with reduced susceptibility to amoxicillin-clavulanic acid, piperacillin-tazobactam, or extended-spectrum cephalosporins for the presence of AmpC production. Three phenotypic AmpC confirmation assays (cefoxitin-cloxacillin disk diffusion test, cefoxitin-EDTA disk diffusion test, and AmpC Etest) were compared for the detection of AmpC activity. All 51 isolates were characterized genetically by mutational analysis of the chromosomal ampC promoter/attenuator region and by PCR detection of plasmid-carried ampC genes. Altogether, 21/51 (41%) E. coli isolates were considered true AmpC producers. AmpC activity due to chromosomal ampC promoter/attenuator mutations was found in 12/21 strains, and plasmid-carried ampC genes were detected in 8/21 isolates. One strain contained both ampC promoter mutations and a plasmid-carried ampC gene. All three phenotypic tests were able to detect the majority (>90%) of AmpC-positive strains correctly. Cefoxitin resistance was found to be a discriminative parameter, detecting 20/21 AmpC-producing strains. Susceptibility to extended-spectrum cephalosporins, e.g., ceftriaxone, ceftazidime, and cefotaxime, was found in 9 of the 21 AmpC-positive strains. Considering the elevated zone diameter breakpoints of the 2010 CLSI guidelines, 2/21 AmpC-positive strains were categorized as susceptible to extended-spectrum cephalosporins.
INTRODUCTION
The prevalence of multidrug-resistant Gram-negative bacteria has increased continuously over the past few years, and bacterial strains producing AmpC beta-lactamases and/or extended-spectrum beta-lactamases (ESBLs) are of particular concern. AmpC beta-lactamases can confer resistance to aminopenicillins, cephalosporins, oxyimino-cephalosporins (e.g., ceftriaxone, cefotaxime, and ceftazidime), cephamycins (e.g., cefoxitin and cefotetan), and monobactams (16). Cloxacillin and 3-aminophenylboronic acid inhibit AmpC beta-lactamases (2, 16, 36), while AmpC beta-lactamase activity is not affected by the ESBL inhibitor clavulanic acid. In Gram-negative bacteria, AmpC beta-lactamase production is chromosome or plasmid mediated. Chromosomal ampC genes are expressed constitutively at a low level. Some Enterobacteriaceae, such as Enterobacter spp., Citrobacter spp., and Serratia spp., carry an inducible ampC gene. In these cases, the gene is strongly induced by β-lactams, such as cefoxitin and imipenem, with expression mediated by the regulator AmpR. Mutations in the repressor gene ampD may lead to overproduction of AmpC beta-lactamases (16). The regulation of chromosomal ampC expression in Escherichia coli differs considerably from that in other Enterobacteriaceae. E. coli lacks ampR, and thus ampC expression is not inducible (15). In E. coli, ampC is expressed constitutively at a low level (17). Various mutations in the ampC promoter/attenuator region of E. coli have been identified that result in constitutive overexpression (7, 8, 13, 22, 24, 34, 39, 40). In addition to chromosomal ampC, E. coli may contain plasmids carrying ampC (pAmpC), transferred via horizontal gene transfer and derived from the chromosomal ampC genes of other Enterobacteriaceae spp. (16). Plasmid-based ampC genes are expressed constitutively in most cases. However, some plasmid-carried ampC genes, such as the DHA-1 gene, are inducible by β-lactams, with expression regulated similarly to that of inducible chromosomal ampC genes. All plasmid-carried ampC genes are considered to be of significant clinical relevance (23, 27). AmpC overproduction in addition to porin mutations of the outer membrane can reduce susceptibility to carbapenems, in particular in plasmid-mediated AmpC producers (19, 26).
AmpC producers may appear susceptible to extended-spectrum cephalosporins when initially tested (27, 37, 38, 40), and standardized procedures for the detection and identification of AmpC beta-lactamase-producing strains have not been established thus far. However, proper recognition of AmpC-overproducing E. coli strains is important for clinical management, as administration of beta-lactam antibiotics frequently results in therapeutic failure. For example, a recent study described the isolation of AmpC-overproducing E. coli strains from patients who did not respond to oxyimino-cephalosporin therapy (34). Another study analyzed the clinical outcomes of patients with bloodstream infection caused by plasmid-mediated AmpC-producing Klebsiella pneumoniae and showed high rates of treatment failure when cephalosporins were administered (27).
Different phenotypic AmpC confirmation tests have been reported in the literature (16). A recently described disk diffusion test is based on comparison of the inhibition zone diameters around a cefoxitin disk and a cefoxitin disk supplemented with the inhibitor cloxacillin. The test was shown to have a sensitivity and a specificity of 95% for the detection of plasmidic AmpC in 127 strains of E. coli, Klebsiella spp., and Proteus spp. (36). Another AmpC confirmation test is based on antagonism phenomena, using a cefoxitin-susceptible indicator strain. This test was evaluated for the detection of plasmid AmpC production in species lacking chromosomal ampC (4). Reportedly, the test had a sensitivity of 100% and a specificity of 98% with 140 isolates of Klebsiella spp., Proteus mirabilis, and Salmonella sp. (4). In this study, we aimed to evaluate and compare the diagnostic performances of the two disk diffusion tests and a commercially available assay (Etest; AB bioMérieux, Sweden) as a confirmation test for the detection of AmpC activity in clinical E. coli isolates with suspicion of AmpC production. Molecular analyses were used to assess the specificity of the phenotypic assays and to characterize the genetic basis for AmpC (over)production in these strains.
MATERIALS AND METHODS
Clinical isolates.
Fifty-one E. coli clinical strains with reduced susceptibility to amoxicillin-clavulanic acid, piperacillin-tazobactam, or oxyimino-cephalosporins (ceftazidime, cefotaxime, or ceftriaxone) were collected at the Institute of Medical Microbiology, Zurich, Switzerland, over a period of 2 years, from July 2006 until July 2008. The strains were isolated from urines (n = 12), blood cultures (n = 12), respiratory specimens (n = 8), perianal swabs (n = 4), wound swabs (n = 4), inguinal swabs (n = 3), abscesses (n = 2), tissue (n = 2), a vaginal swab (n = 1), a gastric aspirate (n = 1), cerebrospinal fluid (CSF) (n = 1), and a sample of unknown origin (n = 1).
Antibiotic susceptibility testing.
Antibiotic susceptibility testing was performed using susceptibility test disks (Becton Dickinson, Germany), and interpretation was done according to 2009 and 2010 CLSI guidelines (9, 10). For cefotetan susceptibility testing, the AmpC Etest strip (AB bioMérieux, Sweden) was used as described below. Susceptibility testing was performed on Mueller-Hinton agar (bioMérieux, France), using overnight cultures at a 0.5 McFarland standard followed by incubation at 35°C for 16 to 18 h.
Phenotypic AmpC and ESBL activity testing.
The AmpC Etest (AB bioMérieux, Sweden) for cefotetan susceptibility was performed according to the manufacturer's instructions. The AmpC Etest consists of a strip containing cefotetan on one end and cefotetan-cloxacillin on the other end. Ratios of the MICs of cefotetan and cefotetan-cloxacillin of ≥8 are considered positive for AmpC beta-lactamase production.
The cefoxitin-cloxacillin disk diffusion test was performed as described by Tan et al. (36). The test is based on the inhibitory effect of cloxacillin on AmpC. In brief, 30-μg cefoxitin disks (Becton Dickinson, Germany) were supplemented with 200 μg cloxacillin. The test strain was inoculated on Mueller-Hinton agar. The diameters of the cefoxitin inhibition zones were compared with and without cloxacillin; if the difference in inhibition was ≥4 mm, the strain was considered positive for AmpC production.
The cefoxitin-EDTA disk test was performed as described by Black et al. (4). In brief, a lawn of the cefoxitin-susceptible E. coli strain ATCC 25922 was inoculated on a Mueller-Hinton agar plate. A 30-μg cefoxitin disk (Becton Dickinson, Germany) was placed on the bacterial lawn and flanked by two disks (A and B), each containing 20 μl of a 1:1 mixture of saline and 100× Tris-EDTA solution. Colonies of the test strain were applied to disk A, and colonies of the cefoxitin-susceptible E. coli strain ATCC 25922 (as a negative control) were applied to disk B. Flattening or indentation of the growth inhibition zone of the cefoxitin disk at the side of disk A containing the test strain indicated the release of AmpC beta-lactamase.
To analyze the induction of plasmid-encoded DHA AmpC, a disk approximation assay was used, with imipenem as an inducer and ceftazidime, cefoxitin, ceftriaxone, and piperacillin-tazobactam as substrate antibiotics (12).
For phenotypic detection of ESBL activity according to CLSI guidelines, a DDS test using ceftazidime and cefotaxime (30 μg) disks, with and without clavulanic acid (10 μg) (Liofilchem, Roseto degli Abruzzi, Italy), was used. The bacterial test strains were inoculated onto Mueller-Hinton agar at a 0.5 McFarland standard, followed by incubation at 35°C for 16 to 18 h. Diameters of inhibition zones were measured with a standard caliper. A difference in inhibition zones of ≥5 mm for at least one extended-spectrum cephalosporin-clavulanic acid combination versus the corresponding extended-spectrum cephalosporin alone was considered indicative of ESBL production.
Beta-lactamase hydrolysis assays.
For phenotypic detection of beta-lactamase activity, the chromogenic substrate nitrocefin (Calbiochem, San Diego, CA) was used (35). 3-Aminophenylboronic acid (Sigma-Aldrich Chemie, GmbH, Zug, Switzerland) was used as a specific AmpC inhibitor (2), and clavulanic acid (Sigma-Aldrich Chemie, GmbH, Zug, Switzerland) was used as an inhibitor of Ambler class A beta-lactamases (e.g., ESBL and TEM-1 beta-lactamases) (5). A bacterial suspension at a 0.5 McFarland standard in 0.45% NaCl was prepared from overnight cultures. E. coli strain DH5α was used as a negative-control strain. Reaction mixtures consisted of 50 μl bacterial cell suspension, 25 μl nitrocefin (0.5 mg/ml in 10 mM phosphate buffer, pH 6.8), 25 μl 3-aminophenylboronic acid (3.6 mg/ml in 10 mM phosphate buffer, pH 6.8), and/or 25 μl potassium clavulanate (2.2 mg/ml in 10 mM phosphate buffer, pH 6.8). In cases where 3-aminophenylboronic acid and/or clavulanic acid was not added, the end volume of 125 μl was reached by adding 10 mM phosphate buffer, pH 6.8. Reaction mixtures were incubated in microtiter plates at 37°C. The nitrocefin hydrolysis product was detected by quantifying the optical density at 492 nm (OD492) after 8 h, using a titer plate spectrophotometer (Biochrom Asys Expert Plus microplate reader; Biochrom Ltd., Cambridge, United Kingdom).
ampC promoter/attenuator sequencing.
DNAs were extracted from colonies grown on agar medium by using an InstaGene matrix (Bio-Rad, Switzerland) following the manufacturer's instructions. For ampC promoter/attenuator mutation analysis, a 271-bp fragment was amplified using primers AB1 (5′-GATCGTTCTGCCGCTGTG-3′) and ampC2 (5′-GGGCAGCAAATGTGGAGCAA-3′) (11). PCR amplicons were purified with a QIAquick PCR purification kit (Qiagen, Switzerland) followed by cycle sequencing using a BigDye reagent kit (Applied Biosystems, Switzerland). Sequence analysis was performed on an ABI Prism model 3100 DNA sequencer (Applied Biosystems, Switzerland) following standard protocols. Sequences were analyzed and edited using Lasergene 7 MegAlign software (DNASTAR Inc.). The ampC promoter/attenuator sequences were compared to the ampC wild-type sequence of E. coli strain ATCC 25922.
Molecular detection of plasmid-carried ampC beta-lactamase genes.
A multiplex PCR was used for the detection of plasmid-carried ampC beta-lactamase genes (29). This assay is able to detect the six plasmid-carried ampC gene families. Resulting PCR amplicons were sequenced with the amplification primers following the protocol described above. The sequences were compared to reference sequences in the NCBI GenBank (http://www.ncbi.nlm.nih.gov/).
Detection of ESBL and KPC genes.
For detection of TEM and SHV beta-lactamase genes, a multiplex PCR was performed as described previously (21). Sequences were analyzed and edited using Lasergene 7 MegAlign software (DNASTAR Inc.). The TEM beta-lactamase sequences were compared to the wild-type E. coli TEM-1 sequence (GenBank accession no. AF427133.1) by using the publicly available database at http://www.lahey.org/studies. For the detection of CTX-M beta-lactamase genes, a multiplex PCR was performed as described by Pitout et al. (31). For detection of Klebsiella carbapenemase (KPC) genes, a PCR was used as described previously (33).
Interpretation.
E. coli strains positive for AmpC activity in at least one phenotypic test (AmpC Etest, AmpC cefoxitin-EDTA disk test, or AmpC cefoxitin-cloxacillin disk test) and validated by genetic analysis (presence of plasmid-carried ampC genes or ampC promoter/attenuator mutations associated with chromosomal ampC overexpression) were considered to be true AmpC producers. Strains with discrepant test results were analyzed in further detail for beta-lactamase production, using nitrocefin hydrolysis assays, phenotypic ESBL assays, molecular assays for ESBL and KPC detection, and sequence analysis of detected TEM and SHV genes.
RESULTS
Phenotypic screening for AmpC production.
For 18/51 (35%) E. coli isolates, all three phenotypic tests gave a positive result for AmpC production (Table 1). The MIC ratios obtained with the AmpC Etest ranged from 8 to 64. In the AmpC cefoxitin-cloxacillin disk test, the differences in zone diameters ranged from 4 mm to 14 mm (Table 1). For 28/51 (55%) E. coli strains, negative results were obtained in all three phenotypic assays (Table 2). For these strains, the MIC ratios obtained with the AmpC Etest ranged from 1 to 4, and the differences in zone diameters measured in the cefoxitin-cloxacillin disk test were 0 mm (n = 22), 1 mm (n = 4), 2 mm (n = 1), and 3 mm (n = 1) (Table 2). Molecular testing confirmed the results of the concordant phenotypic testing (18/18 positive results and 28/28 negative results).
Table 1.
Characterization of the 21 E. coli strains positive for AmpC activitya
| Strain | AmpC Etest |
AmpC cefoxitin-cloxacillin disk test |
AmpC cefoxitin-EDTA disk test result | ampC plasmid PCR resultd (class) | ampC chromosomal sequence analysis resulte (promoter variant) | Final AmpC resultf | Antibiotic resistance disk test result (zone of inhibition [mm])g |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cefotetan susceptibility (MIC [μg/ml]) | MIC ratiob | AmpC result | Inhibition zone diam difference (mm)c | AmpC result | CAZ | CTX | CRO | FEP | AMC | TZP | FOX | |||||
| 1 | S (12) | 24 | Positive | 8 | Positive | Positive | Negative | Positive (1) | Positive | S (18) | S (23) | S (23) | S (29) | R (7) | S (21) | R (7) |
| 2 | S (4) | 8 | Positive | 8 | Positive | Positive | Negative | Positive (1) | Positive | S (21) | I (22) | S (21) | S (27) | R (10) | S (23) | R (12) |
| 3 | I (32) | 43 | Positive | 8 | Positive | Positive | Negative | Positive (1) | Positive | R (12) | I (19) | S (21) | S (28) | R (7) | I (20) | R (7) |
| 4 | I (32) | 64 | Positive | 11 | Positive | Positive | Negative | Positive (1) | Positive | S (21) | S (24) | S (26) | S (30) | R (7) | I (20) | R (13) |
| 5 | I (32) | 21 | Positive | 5 | Positive | Positive | Negative | Positive (1) | Positive | S (20) | I (19) | S (21) | S (26) | I (16) | S (22) | R (7) |
| 6 | S (12) | 24 | Positive | 5 | Positive | Positive | Negative | Positive (1) | Positive | S (18) | S (25) | S (26) | S (28) | R (7) | I (18) | I (15) |
| 7 | S (4) | 8 | Positive | 8 | Positive | Positive | Negative | Positive (3) | Positive | S (28) | S (27) | S (28) | S (32) | R (11) | S (26) | I (17) |
| 8 | S (24) | 48 | Positive | 5 | Positive | Positive | Negative | Positive (2) | Positive | S (19) | S (23) | S (23) | S (30) | R (7) | I (19) | S (18) |
| 9 | S (16) | 32 | Positive | 10 | Positive | Positive | Negative | Positive (5) | Positive | S (20) | S (23) | S (25) | S (32) | R (10) | S (23) | R (13) |
| 10 | S (12) | 24 | Positive | 6 | Positive | Positive | Negative | Positive (6) | Positive | S (19) | S (25) | S (28) | S (29) | R (12) | S (21) | R (14) |
| 11 | R (>32) | 8 | Positive | 5 | Positive | Positive | Positive (CIT) | Negative (12) | Positive | R (7) | R (7) | R (7) | S (24) | R (7) | I (18) | R (7) |
| 12 | R (>32) | 8 | Positive | 4 | Positive | Positive | Positive (CIT) | Negative (12) | Positive | R (7) | R (7) | R (7) | S (20) | R (7) | R (14) | R (7) |
| 13 | I (32) | 8 | Positive | 9 | Positive | Positive | Positive (CIT) | Negative (12) | Positive | R (7) | R (10) | R (7) | S (25) | R (7) | I (20) | R (7) |
| 14 | I (32) | 64 | Positive | 13 | Positive | Positive | Positive (CIT) | Negative (15) | Positive | I (15) | I (15) | I (16) | S (29) | R (7) | S (21) | R (7) |
| 15 | I (32) | 64 | Positive | 14 | Positive | Positive | Positive (CIT) | Negative (15) | Positive | I (16) | I (17) | I (16) | S (25) | R (10) | S (25) | R (7) |
| 16 | I (32) | 64 | Positive | 10 | Positive | Positive | Positive (CIT) | Negative (11) | Positive | R (7) | R (14) | R (12) | S (28) | R (10) | S (21) | R (7) |
| 17 | S (8) | 16 | Positive | 10 | Positive | Positive | Positive (CIT) | Negative (11) | Positive | I (17) | I (20) | I (18) | S (26) | R (11) | S (24) | R (9) |
| 18 | I (32) | 43 | Positive | 10 | Positive | Positive | Positive (CIT) | Positive (5) | Positive | R (7) | I (18) | I (15) | S (28) | R (7) | S (22) | R (7) |
| 19 | I (32) | 32 | Positive | 0 | Negative | Positive | Negative | Positive (3) | Positive | S (23) | S (25) | S (26) | S (31) | R (10) | S (24) | R (14) |
| 20 | S (0.5) | 1 | Negative | 6 | Positive | Positive | Negative | Positive (4) | Positive | S (26) | S (28) | S (37) | S (28) | R (12) | S (25) | I (17) |
| 21 | S (0.75) | 2 | Negative | 4 | Positive | Negative | Positive (DHA) | Negative (14) | Positive | R (11) | R (13) | R (7) | S (18) | R (9) | S (23) | R (7) |
Strains were analyzed for AmpC production by use of three phenotypic confirmation assays. Genetic analysis of AmpC production was done by multiplex PCR for plasmid ampC detection and by sequence analysis of the chromosomal ampC promoter/attenuator region. In addition, antimicrobial susceptibility data were generated for each strain. Data for strains with discrepant results in the three phenotypic AmpC confirmation assays are shaded. Positive results and results indicating resistance and intermediate susceptibility are shown in bold. Abbreviations: S, susceptible; R, resistant; I, intermediate; AMC, amoxicillin-clavulanic acid; CAZ, ceftazidime; CRO, ceftriaxone; CTT, cefotetan; CTX, cefotaxime; FEP, cefepime; FOX, cefoxitin; TZP, piperacillin-tazobactam.
Ratio of cefotetan to cefotetan-cloxacillin MICs, determined by the AmpC Etest (ratios of ≥8 are considered AmpC-positive results).
Differences between diameters of cefoxitin inhibition zones with and without cloxacillin (increases of ≥4 mm for the cefoxitin-cloxacillin disk are considered AmpC-positive results).
Plasmid-mediated ampC was detected by multiplex PCR (29). CIT, plasmid-carried ampC originating from Citrobacter freundii; DHA, plasmid-carried ampC originating from Morganella morganii.
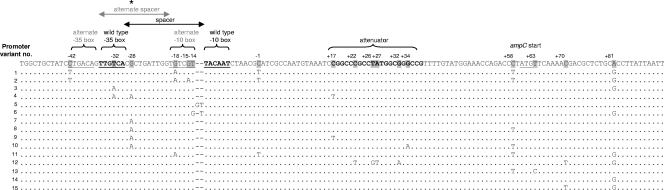
For ampC promoter/attenuator sequence variants, see Fig. 1.
A final assignment for AmpC activity, combining phenotypic and genetic results, was made according to the definition specified in Materials and Methods. A strain was scored positive when at least one phenotypic test was positive and validated by genetic analysis (plasmid-carried ampC or ampC promoter mutations associated with ampC overexpression).
The zone diameter breakpoints of the 2009 CLSI guidelines were applied (9).
Table 2.
Characterization of the 30 E. coli strains negative for AmpC activitya
| Strain | AmpC Etest |
AmpC cefoxitin-cloxacillin disk test |
AmpC cefoxitin-EDTA disk test result | Plasmid ampC PCR resultd | ampC chromosomal sequence analysis resulte (promoter variant) | Final AmpC resultf | Antibiotic resistance disk test result (zone of inhibition [mm])g |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cefotetan susceptibility (MIC [μg/ml]) | MIC ratiob | AmpC result | Inhibition zone diam difference (mm)c | AmpC result | CAZ | CTX | CRO | FEP | AMC | TZP | FOX | |||||
| 22 | S (2) | 1 | Negative | 0 | Negative | Positive | Negative | Negative (10) | Negative | R (14) | R (7) | R (7) | I (15) | R (13) | S (23) | I (16) |
| 23 | S (3) | 2 | Negative | 0 | Negative | Positive | Negative | Negative (8) | Negative | S (30) | S (26) | S (25) | S (29) | I (16) | S (22) | R (12) |
| 24 | S (0.5) | 1 | Negative | 0 | Negative | Negative | Negative | Negative (12) | Negative | S (28) | S (25) | S (29) | S (28) | I (16) | S (22) | S (25) |
| 25 | S (0.75) | 1 | Negative | 0 | Negative | Negative | Negative | Negative (15) | Negative | S (23) | S (28) | S (23) | S (24) | R (10) | R (9) | S (24) |
| 26 | S (0.5) | 1 | Negative | 0 | Negative | Negative | Negative | Negative (15) | Negative | S (20) | S (28) | S (27) | S (28) | R (10) | I (18) | S (20) |
| 27 | S (2) | 1 | Negative | 0 | Negative | Negative | Negative | Negative (15) | Negative | S (22) | S (29) | S (26) | S (27) | R (7) | R (7) | S (20) |
| 28 | S (1) | 1 | Negative | 3 | Negative | Negative | Negative | Negative (15) | Negative | S (22) | S (25) | S (26) | S (25) | I (17) | S (23) | S (27) |
| 29 | S (0.5) | 1 | Negative | 0 | Negative | Negative | Negative | Negative (15) | Negative | S (29) | S (28) | S (28) | S (33) | I (14) | S (21) | S (24) |
| 30 | S (0.5) | 1 | Negative | 0 | Negative | Negative | Negative | Negative (11) | Negative | S (26) | S (28) | S (26) | S (26) | R (10) | I (18) | S (23) |
| 31 | S (0.5) | 1 | Negative | 0 | Negative | Negative | Negative | Negative (11) | Negative | S (28) | S (25) | S (25) | S (24) | R (7) | R (14) | S (25) |
| 32 | S (0.5) | 1 | Negative | 0 | Negative | Negative | Negative | Negative (11) | Negative | S (26) | S (32) | S (28) | S (30) | R (12) | R (14) | S (30) |
| 33 | S (0.5) | 1 | Negative | 0 | Negative | Negative | Negative | Negative (10) | Negative | S (27) | S (23) | S (25) | S (26) | R (10) | R (15) | S (21) |
| 34 | S (0.5) | 1 | Negative | 0 | Negative | Negative | Negative | Negative (10) | Negative | S (28) | S (25) | S (28) | S (28) | I (15) | S (23) | S (21) |
| 35 | S (0.5) | 1 | Negative | 0 | Negative | Negative | Negative | Negative (8) | Negative | S (26) | R (7) | R (7) | S (23) | S (19) | R (7) | S (24) |
| 36 | S (0.5) | 1 | Negative | 0 | Negative | Negative | Negative | Negative (7) | Negative | S (20) | R (7) | R (7) | I (17) | R (10) | R (14) | S (23) |
| 37 | S (0.5) | 1 | Negative | 0 | Negative | Negative | Negative | Negative (7) | Negative | S (25) | I (19) | I (14) | S (25) | I (15) | I (20) | S (24) |
| 38 | I (32) | 1 | Negative | 0 | Negative | Negative | Negative | Negative (7) | Negative | R (7) | R (7) | R (7) | R (7) | R (7) | R (7) | S (18) |
| 39 | S (0.5) | 1 | Negative | 0 | Negative | Negative | Negative | Negative (7) | Negative | S (24) | I (19) | R (7) | S (25) | R (11) | I (18) | S (23) |
| 40 | S (0.75) | 1 | Negative | 0 | Negative | Negative | Negative | Negative (7) | Negative | S (26) | I (20) | R (7) | S (26) | R (13) | S (22) | S (25) |
| 41 | S (0.5) | 1 | Negative | 0 | Negative | Negative | Negative | Negative (7) | Negative | S (25) | R (7) | R (7) | R (13) | I (14) | R (7) | S (28) |
| 42 | S (0.5) | 1 | Negative | 0 | Negative | Negative | Negative | Negative (14) | Negative | S (25) | S (25) | S (28) | S (25) | I (15) | S (23) | S (22) |
| 43 | S (0.5) | 1 | Negative | 0 | Negative | Negative | Negative | Negative (14) | Negative | S (27) | S (28) | S (28) | S (26) | I (14) | S (23) | S (25) |
| 44 | S (0.5) | 1 | Negative | 1 | Negative | Negative | Negative | Negative (15) | Negative | S (29) | S (33) | S (26) | S (31) | R (13) | S (22) | S (28) |
| 45 | S (0.5) | 1 | Negative | 1 | Negative | Negative | Negative | Negative (14) | Negative | S (26) | S (28) | S (27) | S (28) | I (17) | S (21) | S (21) |
| 46 | S (1.5) | 1 | Negative | 1 | Negative | Negative | Negative | Negative (9) | Negative | S (21) | S (27) | S (25) | S (25) | R (10) | R (10) | S (19) |
| 47 | S (0.5) | 1 | Negative | 2 | Negative | Negative | Negative | Negative (13) | Negative | S (26) | I (15) | I (15) | S (22) | I (17) | S (25) | S (30) |
| 48 | S (0.75) | 2 | Negative | 0 | Negative | Negative | Negative | Negative (7) | Negative | S (26) | I (18) | I (16) | S (24) | I (16) | I (18) | S (25) |
| 49 | S (0.75) | 2 | Negative | 0 | Negative | Negative | Negative | Negative (7) | Negative | S (25) | I (20) | S (25) | I (15) | R (7) | R (10) | S (24) |
| 50 | S (0.75) | 2 | Negative | 0 | Negative | Negative | Negative | Negative (14) | Negative | S (25) | S (28) | S (26) | S (25) | I (17) | I (20) | S (18) |
| 51 | S (8) | 4 | Negative | 1 | Negative | Negative | Negative | Negative (14) | Negative | S (26) | S (27) | S (26) | S (25) | R (13) | I (18) | S (21) |
Strains were analyzed for AmpC production by use of three phenotypic AmpC assays. Genetic analysis of AmpC production was done by multiplex PCR for plasmid ampC detection and by sequence analysis of the chromosomal ampC promoter/attenuator region. In addition, antimicrobial susceptibility data were generated for each strain. Data for strains with discrepant results in the three phenotypic AmpC confirmation assays are shaded. Positive results and results indicating resistance and intermediate susceptibility are shown in bold. Interpretation of susceptibility results was done according to the 2009 CLSI guidelines. For abbreviations and explanations of the footnotes, see the footnotes in Table 1.
Discrepant test results were obtained for 5/51 (10%) isolates, i.e., strains 19, 20, 21, 22, and 23. The analysis of the discrepant test results and their resolution are given in detail below.
Strain 19 was positive in the AmpC Etest (MIC ratio = 32), negative in the AmpC cefoxitin-cloxacillin disk diffusion test (difference in zone diameters, 0 mm), and positive in the AmpC cefoxitin-EDTA disk diffusion test. In this strain, the ampC promoter/attenuator sequence showed mutations associated with upregulation of chromosomal ampC gene expression (sequence variant 3) (Table 3). Strain 20 was negative in the AmpC Etest (MIC ratio = 1) and positive in both AmpC disk diffusion tests; the difference in zone diameters for the AmpC cefoxitin-cloxacillin disk diffusion test was 6 mm. Genetic analysis showed promoter/attenuator mutations associated with chromosomal ampC overexpression (variant 4) (Table 3) (7, 17, 40).
Table 3.
Genetic characterization of 51 E. coli isolates by ampC promoter region sequence analysis and multiplex PCR for detection of plasmid-mediated ampC
| Promoter sequence varianta | E. coli strain no. | No. of strains (n = 51) | No. (%) of AmpC-positive strains | No. of strains carrying plasmid-mediated ampC | Position(s) of mutation(s) in ampC promoter/attenuator regionb,c | Localization and function of mutationsc |
|---|---|---|---|---|---|---|
| 1 | 1, 2, 3, 4, 5, 6 | 6 | 6 (100) | −42, −18, (−1), (+58), +81 | Alternate displaced promoter (−35 box and −10 box) and mutations in the AmpC coding region | |
| 2 | 8 | 1 | 1 (100) | −42, −18, −15, (−1), (+58), +81 | Alternate displaced promoter (−35 box and −10 box) and mutations in the AmpC coding region | |
| 3 | 7, 19 | 2 | 2 (100) | −32, +81 | Promoter mutation and mutation in the AmpC coding region | |
| 4 | 20 | 1 | 1 (100) | −32, −28, +17 | Promoter mutation, mutations in the spacer region and in the AmpC coding region | |
| 5 | 9, 18 | 2 | 2 (100) | 1 | INS (−13.1), INS (−13.2) | Increased distance between −35 and −10 boxes |
| 6 | 10 | 1 | 1 (100) | −14, INS (−13.1), +81 | Increased distance between −35 and −10 boxes, promoter mutation, and mutation in the ampC coding region | |
| 7 | 36, 37, 38, 39, 40, 41, 48, 49 | 8 | None | −28 | Mutation in the spacer region | |
| 8 | 23, 35 | 2 | None | −28, (+58) | Mutation in the spacer region | |
| 9 | 46 | 1 | None | −28, +17 | Mutation in the spacer region and mutation in the attenuator | |
| 10 | 22, 33, 34 | 3 | None | −28, +34, (+58) | Mutation in the spacer, attenuator mutation, and mutation in the ampC coding region | |
| 11 | 16, 17, 30, 31, 32 | 5 | 2 (40) | 2 | −18, (−1), (+58), +81 | Alternate displaced promoter (−10 box only) and mutation in the ampC coding region |
| 12 | 11, 12, 13, 24 | 4 | 3 (75) | 3 | +22, +26, +27, +32, +70, +81 | Attenuator mutations and mutations in the ampC coding region |
| 13 | 47 | 1 | None | (+58), +63 | Mutation in the ampC coding region | |
| 14 | 21, 42, 43, 45, 50, 51 | 6 | 1 (17) | 1 | +81 | Mutation in the ampC coding region |
| 15 | 14, 15, 25, 26, 27, 28, 29, 44 | 8 | 2 (25) | 2 | +70, +81 | Mutations in the ampC coding region |
All isolates with promoter sequence variants 1, 2, 3, 4, 5, and 6 are considered positive for AmpC activity due to chromosomal overexpression of ampC. Promoter variants 11, 12, 14, and 15 are found in phenotypically AmpC-positive and AmpC-negative strains; the AmpC-positive strains all harbor plasmid-encoded AmpC. Promoter sequence variants 7, 8, 9, 10, and 13 are not associated with increased phenotypic AmpC activity.
For detailed sequence analysis, see Fig. 1.
Mutations and mechanisms resulting in overexpression leading to ampC upregulation are typed in bold and shaded. INS, insertion of nucleotides. Mutations outside functional promoter elements are displayed in parentheses (4).
Strain 21 was negative in the AmpC Etest (MIC ratio = 1.5), negative in the AmpC cefoxitin-EDTA disk diffusion test, and positive in the AmpC cefoxitin-cloxacillin disk diffusion test (diameter difference, 4 mm). The ampC promoter and attenuator region of strain 21 resembles that of the wild-type E. coli K-12 strain. The multiplex PCR for plasmid-mediated ampC genes was positive for the DHA gene. Induction of the DHA gene in strain 21 was revealed by a disk approximation assay (12) using imipenem as the inducer and ceftazidime, cefoxitin, ceftriaxone, and piperacillin-tazobactam as substrate antibiotics. Nitrocefin hydrolysis assays showed that the beta-lactamase activity of strains 19, 20, and 21 was inhibited by the AmpC inhibitor 3-aminophenylboronic acid (Table 4). Phenotypic assays for ESBL detection (DDS assays) revealed that strains 19 and 20 were ESBL negative and strain 21 was ESBL positive, which was confirmed by the identification of a CTXM-1 gene in strain 21. A non-ESBL TEM-1 beta-lactamase was detected in strains 19 and 21. A corresponding inhibitory effect of the TEM-1 inhibitor clavulanic acid on hydrolysis of nitrocefin was detected in strains 19 and 21, whereas strain 20 did not show such an inhibitory effect. A KPC PCR was negative for strains 19, 20, and 21 (for a summary of the results, see Table 4). Based on our interpretation criteria and the additional beta-lactamase analyses, strains 19, 20, and 21 were considered true AmpC producers (Table 1).
Table 4.
Characterization of β-lactamase activity in E. coli strains (n = 5) considered falsely positive or falsely negative in phenotypic screening for AmpC overproductiond
| E. coli straina | Nitrocefin hydrolysis (OD492)b |
ESBLc |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Without inhibitor | With aminophenylboronic acid | Inhibition | With clavulanic acid | Inhibition | With boronic acid and clavulanic acid | Inhibition | Phenotype | Genetic result | |
| 19 | 0.309 ± 0.006 | 0.106 ± 0.008 | + | 0.080 ± 0.010 | + | 0.041 ± 0.004 | + | − | (TEM-1) |
| 20 | 0.151 ± 0.014 | 0.006 ± 0.005 | + | 0.120 ± 0.008 | − | 0.015 ± 0.006 | + | − | − |
| 21 | 0.344 ± 0.020 | 0.225 ± 0.002 | + | 0.083 ± 0.0 | + | 0.051 ± 0.006 | + | + | (TEM-1)/CTXM-1 |
| 22 | 0.777 ± 0.007 | 0.847 ± 0.015 | − | 0.332 ± 0.006 | + | 0.395 ± 0.009 | + | + | CTXM-1 |
| 23 | 0.366 ± 0.005 | 0.314 ± 0.005 | − | 0.074 ± 0.001 | + | 0.066 ± 0.007 | + | − | (TEM-1) |
Strain numbers correspond to those in Tables 1 and 2.
Nitrocefin hydrolysis was tested in the absence and presence of boronic acid (as a specific AmpC inhibitor) and/or clavulanic acid (as a specific ESBL and TEM-1 inhibitor). Presented values are averages and standard deviations for duplicate reactions.
The presence of ESBLs was detected phenotypically by the DDS test as described in Materials and Methods. PCR detection of TEM, SHV, and CTX-M genes was done as described previously (21, 31). Note that TEM-1 is not an ESBL but is able to hydrolyze nitrocefin. Therefore, the detection of TEM-1 is shown in parentheses.
PCR detection of KPC genes was performed as described previously (33), and all strains tested negative.
Two strains (22 and 23) were positive in the AmpC cefoxitin-EDTA disk diffusion test and negative in the AmpC cefoxitin-cloxacillin disk diffusion test and the AmpC Etest. Both strains were negative for plasmid-carried ampC genes, and genetic analysis of the ampC promoter/attenuator region did not reveal mutations typically associated with chromosomal ampC upregulation. Additional beta-lactamase analysis of strains 22 and 23 showed that nitrocefin hydrolysis was not inhibited by the AmpC inhibitor 3-aminophenylboronic acid (Table 4). Phenotypic ESBL testing (DDS assay) revealed that strain 22 was ESBL positive and strain 23 was ESBL negative, which was confirmed by the identification of a CTXM-1 gene in strain 22. A TEM-1 beta-lactamase was detected in strain 23. A corresponding inhibitory effect of clavulanic acid on hydrolysis of nitrocefin was detected in both strains. KPC PCR was negative for strains 22 and 23 (Table 4). Based on our interpretation criteria and the additional beta-lactamase analyses, strains 22 and 23 were considered AmpC negative (Table 2).
In total, 21/51 (41%) E. coli strains investigated in this study were considered AmpC producers, and 30/51 (59%) strains were negative for AmpC production (Tables 1 and 2). The AmpC Etest detected 19/21 (90.5%) positive strains and showed no false-positive results. The AmpC cefoxitin-cloxacillin disk test was correctly positive for 20/21 (95.2%) AmpC-positive strains and did not give false-positive results. The AmpC cefoxitin-EDTA disk test was correctly positive for 20/21 (95.2%) strains and gave two false-positive results.
ampC promoter/attenuator mutations and plasmid-encoded AmpC beta-lactamases.
In the 51 E. coli strains, 15 different ampC promoter/attenuator sequence variants were detected (Fig. 1). For the 21 AmpC-positive strains, 10 different promoter/attenuator sequence variants (1, 2, 3, 4, 5, 6, 11, 12, 14, and 15) were found (Table 3). Promoter/attenuator sequence variants 1, 2, 3, 4, 5, and 6 were found in 11 strains with a positive AmpC production phenotype and a negative result for plasmid-carried ampC. Sequence variant 5 comprised two AmpC-positive strains, one positive for plasmid-carried ampC genes and one negative for plasmid-carried ampC. Mutations detected in sequence variants 1, 2, 3, 4, 5, and 6 included (i) mutations that created an alternate displaced promoter (variants 1 and 2), (ii) mutations in the wild-type promoter/attenuator (variants 3 and 4), and (iii) mutations that increased the spacer length between the −35 and −10 boxes (variants 5 and 6) by insertion of 1 or 2 base pairs. The mutations found in these sequence variants are associated with an increase of ampC expression (11). Sequence variants 11, 12, 14, and 15 were found in phenotypically AmpC-positive strains which were positive for the presence of plasmid-carried ampC genes. Mutations in these sequence variants were located in the attenuator region or coding region for AmpC or resulted in an alternate displaced −10 box. None of these changes has been reported to be associated with significant chromosomal AmpC overproduction (see below). Variant 14 resembled the wild-type E. coli K-12 ampC promoter/attenuator (Table 3). In total, 13/21 AmpC-positive strains harbored changes in the promoter/attenuator region typically associated with chromosomal AmpC overproduction (32).
Fig. 1.
Alignment of the chromosomal ampC promoter, attenuator, and 5′-end regions. For the 51 E. coli isolates, 15 different sequence variants were identified. *, chromosomal ampC sequence variant classifications and descriptions of functional elements are used as reported in the work of Tracz et al. (40).
Plasmid-carried ampC genes were detected in 9 of the 21 strains. In one strain, both chromosome- and plasmid-mediated mechanisms responsible for AmpC production were found, e.g., a 2-bp insertion in the spacer of the ampC chromosomal promoter/attenuator region and a plasmid-carried ampC gene (Table 3). The plasmid-carried ampC genes found in the isolates belonged to the CIT family (n = 8) and the DHA family (n = 1). Sequences of the PCR products showed 100% homology to the blaCMY-2 gene for the CIT family isolates and 100% homology to the blaDHA-1 gene for the DHA family isolate (data not shown).
Susceptibility to amoxicillin-clavulanic acid, piperacillin-tazobactam, cefoxitin, and extended-spectrum cephalosporins.
All 21 AmpC-positive strains showed reduced susceptibility to amoxicillin-clavulanic acid. Twenty of 21 strains tested were resistant, and 1 strain was intermediate. In contrast, only 1/21 strains was resistant to piperacillin-tazobactam, 6/21 isolates showed an intermediate level, and 14/21 strains were susceptible.
For extended-spectrum cephalosporins, the following test results were obtained for the 21 AmpC-positive strains, applying the 2009 CLSI guideline (9) zone diameter breakpoints: for ceftazidime, 11 strains were susceptible, 3 strains were intermediate, and 7 strains were resistant; for cefotaxime, 9 strains were susceptible, 7 strains were intermediate, and 5 strains were resistant; and for ceftriaxone, 12 strains were susceptible, 4 strains were intermediate, and 5 strains were resistant. All AmpC-positive strains were susceptible to cefepime (Table 1). Resistance patterns for AmpC-negative strains are summarized in Table 2. In 2010, the CLSI zone diameter breakpoints for ceftazidime, cefotaxime, and ceftriaxone were elevated (10). Applying these breakpoints resulted in the following interpretation of susceptibility testing for AmpC-positive strains: for ceftazidime, 5 strains were susceptible, 6 strains were intermediate, and 10 strains were resistant; for cefotaxime, 2 strains were susceptible, 7 strains were intermediate, and 12 strains were resistant; and for ceftriaxone, 9 strains were susceptible, 3 strains were intermediate, and 9 strains were resistant (see Table S1A in the supplemental material). Results for the AmpC-negative strains are summarized in Table S1B in the supplemental material.
By disk diffusion susceptibility testing and according to 2009 CLSI guidelines, 17/21 (81%) AmpC-producing strains were resistant to cefoxitin, 3/21 strains were intermediate, and 1/21 strains was susceptible (inhibition zone diameter, 18 mm) (Table 1). A total of 28/30 AmpC-negative strains (93%) were susceptible to cefoxitin, for 1/30 strains an intermediate result was obtained, and 1/30 strains was resistant (Table 2).
DISCUSSION
Detection of AmpC beta-lactamases in E. coli poses a challenge to microbiological laboratories. For practical reasons, it is not feasible to routinely test all E. coli isolates for AmpC production in detail. In our study, we selected 51 E. coli clinical isolates collected during a 2-year period for putative AmpC production based on reduced susceptibility to amoxicillin-clavulanic acid, piperacillin-tazobactam, or oxyimino-cephalosporins (ceftriaxone, ceftazidime, and cefotaxime).
Several AmpC confirmation tests have recently been evaluated (4, 36) or become commercially available (AmpC Etest; AB bioMérieux). In this study, we compared the performances of three of these tests for accurate identification of AmpC-producing E. coli strains: the AmpC Etest (AB bioMérieux, Sweden), the AmpC cefoxitin-cloxacillin disk test (36), and the AmpC cefoxitin-EDTA disk test (4). AmpC-producing E. coli strains were validated by genetic analyses. In addition, strains with discrepant AmpC screening results were analyzed further for beta-lactamase production by nitrocefin hydrolysis assays, ESBL phenotypic testing, and genetic testing for the presence of SHV, TEM, CTX-M, and KPC beta-lactamases (Table 4). The additional test results confirmed the accuracy of our interpretation criteria for AmpC production. In total, 21 of the selected 51 E. coli isolates were identified as true AmpC-producing strains (for plasmidic ampC, n = 8; for overexpression of chromosomal ampC, n = 12; and for a combination of plasmidic ampC and overexpression of chromosomal ampC, n = 1). We found that the cefoxitin-cloxacillin disk test detected 20/21 AmpC-positive E. coli strains (Table 1) and gave 1 false-negative result. The cefoxitin-EDTA disk test (4) detected 20/21 AmpC-positive strains and gave 1 false-negative result and 2 false-positive results. A drawback of the cefoxitin-EDTA disk assay is that carbapenemases may give rise to false-positive results, because carbapenemases are able to inactivate cefoxitin (4), although KPC was not detected in the 2 false-positive strains. The AmpC Etest strip uses cefotetan for AmpC screening. While cefotetan resistance (MIC of >64 mg/liter) was not consistently present in the AmpC-positive strains (Table 1), the AmpC Etest was able to detect 19/21 positive strains, and 2 strains gave a false-negative test result.
The ampC promoter/attenuator mutations detected in the 51 E. coli isolates (Table 3) included 6 previously described variants associated with ampC overexpression (7, 17, 18, 24, 34, 40). Overall, 13/21 (61.9%) positive AmpC strains were associated with chromosomal ampC promoter mutations resulting in hyperproduction of AmpC, and 9/21 strains (42.9%) were AmpC positive due to the presence of plasmid-carried ampC genes. One strain had a 2-bp insertion in the ampC promoter spacer region (variant 5) and a plasmid-carried ampC gene; both mechanisms may have contributed to AmpC activity in this strain. The observed ratio of AmpC production due to chromosomal ampC upregulation versus plasmid-mediated AmpC is in accordance with the distribution observed in studies conducted in France, Spain, and Norway (6, 14, 20). We did not detect any strain that was positive in the genetic analysis and negative in all three phenotypic confirmation tests.
Because the cefoxitin- and cefotetan-based AmpC disk assays effectively identify AmpC producers, we decided to evaluate whether cefoxitin and cefotetan susceptibility testing can be used as a screening test for AmpC production. In the group of AmpC-positive strains, 20 of 21 (95%) isolates were resistant or intermediate in the cefoxitin disk test. One strain showed an inhibition zone of 18 mm, which is just within the susceptible range (Table 1). For the AmpC-negative strains, 26 of 30 (87%) strains were susceptible to cefoxitin, with inhibition zones of >18 mm. Two strains scored within the susceptible range, with inhibition zones of 18 mm, one strain was intermediate, and one strain was resistant to cefoxitin (Table 2). Applying a screening criterion of a cefoxitin inhibition zone of ≤18 mm, all AmpC-positive strains would have been detected, plus an additional 4 false-positive results. However, the use of cefoxitin as a screening marker is compromised by isolates producing plasmid-encoded AmpC beta-lactamases of the ACC family. ACC-1 itself is inhibited by cefoxitin, and thus strains carrying it may appear cefoxitin susceptible (1, 16, 32). ACC-1 was first isolated in Germany and in several other European countries (1, 25, 30, 32). Recently, the AmpC beta-lactamase ACC-4 was identified in E. coli, conferring increased MICs for oxyimino-cephalosporins, with low MICs for cefoxitin and cefepime (28). In our study, strains with a plasmid ACC beta-lactamase gene were not detected.
Analyzing cefotetan MICs for the AmpC Etest revealed that 2 of the 21 (10%) AmpC-positive strains were resistant to cefotetan. Ten of 21 (48%) AmpC-positive strains were susceptible to cefotetan, and intermediate results were obtained for 9 (42%) isolates (Table 1). On the basis of these results, we cannot recommend cefotetan susceptibility testing for initial AmpC screening.
We also evaluated whether reduced sensitivity to extended-spectrum cephalosporins can be used as a screening parameter for AmpC testing. Several studies showed that cephalosporin susceptibility screening of E. coli isolates with the initial purpose of ESBL identification resulted in selection for AmpC-producing strains (3, 23). Nine of the 21 (43%) AmpC-positive strains were susceptible to ceftazidime, cefotaxime, and ceftriaxone in vitro according to the CLSI 2009 guidelines. Another two strains were susceptible to ceftazidime and ceftriaxone, and one strain was susceptible to ceftriaxone only (Table 1). Applying the elevated 2010 CLSI zone diameter breakpoints, two strains were susceptible to ceftazidime, cefotaxime, and ceftriaxone, another two strains were susceptible to ceftazidime and ceftriaxone, one strain was susceptible to ceftazidime only, and five strains were susceptible to ceftriaxone only (see Table S1A in the supplemental material). On the basis of our results, we cannot recommend extended-spectrum cephalosporins as screening parameters for AmpC.
In summary, we demonstrate that after a first screening procedure, each of the three phenotypic AmpC tests used in this study was capable of confirming the majority of AmpC beta-lactamase-producing E. coli strains (>90%), including those producing plasmid-mediated AmpC beta-lactamases and chromosomal AmpC hyperproduction strains. Each of the three tests is an acceptable phenotypic confirmation tool when AmpC production in E. coli is suspected.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported in part by the University of Zurich.
We have no conflicts of interest to disclose.
Footnotes
Supplemental material for this article may be found at http://jcm.asm.org/.
Published ahead of print on 8 June 2011.
REFERENCES
- 1. Bauernfeind A., Schneider I., Jungwirth R., Sahly H., Ullmann U. 1999. A novel type of AmpC beta lactamase, ACC-1, produced by a Klebsiella pneumoniae strain causing nosocomial pneumonia. Antimicrob. Agents Chemother. 43:1924–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beesley T., et al. 1983. The inhibition of class C β-lactamases by boronic acids. Biochem. J. 209:229–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bell J. M., et al. 2007. Prevalence and significance of a negative extended-spectrum β-lactamase (ESBL) confirmation test result after a positive ESBL test result for isolates of Escherichia coli and Klebsiella pneumoniae: results from the SENTRY Asia-Pacific Surveillance Program. J. Clin. Microbiol. 45:1478–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Black J. A., Moland E. S., Thomson K. S. 2005. AmpC disk test for detection of plasmid-mediated AmpC beta-lactamases in Enterobacteriaceae lacking chromosomal AmpC beta-lactamases. J. Clin. Microbiol. 43:3110–3113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bradford P. A. 2001. Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 14:933–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Briñas L., et al. 2005. Mechanisms of resistance to expanded-spectrum cephalosporins in Escherichia coli isolates recovered in a Spanish hospital. J. Antimicrob. Chemother. 56:1107–1110 [DOI] [PubMed] [Google Scholar]
- 7. Caroff N., Espaze E., Bérard I., Richet H., Reynaud A. 1999. Mutations in the ampC promoter of Escherichia coli isolates resistant to oxyiminocephalosporins without extended spectrum beta-lactamase production. FEMS Microbiol. Lett. 173:459–465 [DOI] [PubMed] [Google Scholar]
- 8. Caroff N., Espaze E., Gautreau D., Richet H., Reynaud A. 2000. Analysis of the effects of −42 and −32 ampC promoter mutations in clinical isolates of Escherichia coli hyperproducing AmpC. J. Antimicrob. Chemother. 45:783–788 [DOI] [PubMed] [Google Scholar]
- 9. Clinical and Laboratory Standards Institute 2009. Performance standards for antimicrobial susceptibility testing; 19th informational supplement. M100–S19. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 10. Clinical and Laboratory Standards Institute 2010. Performance standards for antimicrobial susceptibility testing; 20th informational supplement. M100–S20. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 11. Corvec S., et al. 2007. Most Escherichia coli strains overproducing chromosomal AmpC beta-lactamase belong to phylogenetic group A. J. Antimicrob. Chemother. 60:872–876 [DOI] [PubMed] [Google Scholar]
- 12. Dunne W. M., Jr., Hardin D. J. 2005. Use of several inducer and substrate antibiotic combinations in a disk approximation assay format to screen for AmpC induction in patient isolates of Pseudomonas aeruginosa, Enterobacter spp., Citrobacter spp., and Serratia spp. J. Clin. Microbiol. 43:5945–5949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Forward K. R., et al. 2001. Molecular mechanisms of cefoxitin resistance in Escherichia coli from the Toronto area hospitals. Diagn. Microbiol. Infect. Dis. 41:57–63 [DOI] [PubMed] [Google Scholar]
- 14. Haldorsen B., et al. 2008. The AmpC phenotype in Norwegian clinical isolates of Escherichia coli is associated with an acquired ISEcp1-like ampC element or hyperproduction of the endogenous AmpC. J. Antimicrob. Chemother. 62:694–702 [DOI] [PubMed] [Google Scholar]
- 15. Honoré N., Nicolas M. H., Cole S. T. 1986. Inducible cephalosporinase production in clinical isolates of Enterobacter cloacae is controlled by a regulatory gene that has been deleted from Escherichia coli. EMBO J. 5:3709–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jacoby G. A. 2009. AmpC β-lactamases. Clin. Microbiol. Rev. 22:161–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jaurin B., Grundström T., Edlund T., Normark S. 1981. The E. coli β-lactamase attenuator mediates growth rate-dependent regulation. Nature 290:221–225 [DOI] [PubMed] [Google Scholar]
- 18. Jørgensen R. L., Nielsen J. B., Friis-Møller A., Fjeldsøe-Nielsen H., Schønning K. 2010. Prevalence and molecular characterization of clinical isolates of Escherichia coli expressing an AmpC phenotype. J. Antimicrob. Chemother. 65:460–464 [DOI] [PubMed] [Google Scholar]
- 19. Mammeri H., Guillon H., Eb F., Nordmann P. 2010. Phenotypic and biochemical comparison of the carbapenem-hydrolyzing activities of five plasmid-borne AmpC beta-lactamases. Antimicrob. Agents Chemother. 54:4556–4560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mammeri H., Eb F., Berkani A., Nordmann P. 2008. Molecular characterization of AmpC-producing Escherichia coli clinical isolates recovered in a French hospital. J. Antimicrob. Chemother. 61:498–503 [DOI] [PubMed] [Google Scholar]
- 21. Moland E. S., et al. 2006. Prevalence of newer beta-lactamases in gram-negative clinical isolates collected in the United States from 2001 to 2002. J. Clin. Microbiol. 44:3318–3324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mulvey M. R., et al. 2005. Molecular characterization of cefoxitin-resistant Escherichia coli from Canadian hospitals. Antimicrob. Agents Chemother. 49:358–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Munier G. K., et al. 2010. Positive extended-spectrum β-lactamase (ESBL) screening results may be due to AmpC β-lactamases more often than to ESBLs. J. Clin. Microbiol. 48:673–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nelson E. C., Elisha B. G. 1999. Molecular basis of AmpC hyperproduction in clinical isolates of Escherichia coli. Antimicrob. Agents Chemother. 43:957–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ohana S., et al. 2005. Spread of a Klebsiella pneumoniae strain producing a plasmid-mediated ACC-1 AmpC β-lactamase in a teaching hospital admitting disabled patients. Antimicrob. Agents Chemother. 49:2095–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Oteo J., et al. 2008. Emergence of imipenem resistance in clinical Escherichia coli during therapy. Int. J. Antimicrob. Agents 32:534–537 [DOI] [PubMed] [Google Scholar]
- 27. Pai H., et al. 2004. Epidemiology and clinical features of bloodstream infections caused by AmpC-type-beta-lactamase-producing Klebsiella pneumoniae. Antimicrob. Agents Chemother. 48:3720–3728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Papagiannitsis C. C., Tzouvelekis L. S., Tzelepi E., Miriagou V. 2007. Plasmid-encoded ACC-4, an extended-spectrum cephalosporinase variant from Escherichia coli. Antimicrob. Agents Chemother. 51:3763–3767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pérez-Pérez F. J., Hanson N. D. 2002. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J. Clin. Microbiol. 40:2153–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Philippon A., Arlet G., Jacoby G. A. 2002. Plasmid-determined AmpC type β-lactamases. Antimicrob. Agents Chemother. 46:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pitout J. D., Hanson N. D., Church D. L., Laupland K. B. 2004. Population-based laboratory surveillance for Escherichia coli-producing extended-spectrum beta-lactamases: importance of community isolates with blaCTX-M genes. Clin. Infect. Dis. 38:1736–1741 [DOI] [PubMed] [Google Scholar]
- 32. Ruppé E., Bidet P., Verdet C., Arlet G., Bingen E. 2006. First detection of the Ambler class C 1 AmpC beta-lactamase in Citrobacter freundii by a new, simple double-disk synergy test. J. Clin. Microbiol. 44:4204–4207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schechner V., et al. 2009. Evaluation of PCR-based testing for surveillance of KPC-producing carbapenem-resistant members of the Enterobacteriaceae family. J. Clin. Microbiol. 47:3261–3265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Siu L. K., Lu P. L., Chen J. Y., Lin F. M., Chang S. C. 2003. High-level expression of ampC β-lactamase due to insertion of nucleotides between −10 and −35 promoter sequences in Escherichia coli clinical isolates: cases not responsive to extended-spectrum-cephalosporin treatment. Antimicrob. Agents Chemother. 47:2138–2144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sutton L. D., Biedenbach D. J., Yenn A., Jones R. N. 1995. Development, characterization and initial evaluations of S1. A new chromogenic cephalosporin for β-lactamase detection. Diagn. Microbiol. Infect. Dis. 21:1–8 [DOI] [PubMed] [Google Scholar]
- 36. Tan T.Y., et al. 2009. Evaluation of screening methods to detect plasmid-mediated AmpC in Escherichia coli, Klebsiella pneumoniae, and Proteus mirabilis. Antimicrob. Agents Chemother. 53:146–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Thomson K. S. 2001. Controversies about extended-spectrum and AmpC beta-lactamases. Emerg. Infect. Dis. 7:333–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Thomson K. S. 2010. Extended-spectrum-β-lactamase, AmpC and carbapenemase issue. J. Clin. Microbiol. 48:1019–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tracz D. M., et al. 2005. Increase in ampC promoter strength due to mutations and deletion of the attenuator in a clinical isolate of cefoxitin-resistant Escherichia coli as determined by RT-PCR. J. Antimicrob. Chemother. 55:758–772 [DOI] [PubMed] [Google Scholar]
- 40. Tracz D. M., et al. 2007. ampC gene expression in promoter mutants of cefoxitin-resistant Escherichia coli clinical isolates. FEMS Microbiol. Lett. 270:265–271 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.