Abstract
PCR ribotyping was modified to allow direct detection of Clostridium difficile from stool samples. Direct PCR ribotyping was possible in 86 out of 99 C. difficile-positive stool samples, and in 84 cases (84.8%), the ribotype determined directly from the stool sample was identical to the ribotype of the strain isolated from the same stool sample.
TEXT
Clostridium difficile infections represent a significant burden on the health care system. Although many infections are sporadic, nosocomial transmission is still important and outbreaks are a constant threat in the hospital environment. The ability to detect such outbreaks quickly is critical to infection control.
Several typing techniques, all of them based on having a pure culture of the organism, have been described for C. difficile (3). Pulsed-field gel electrophoresis (PFGE) and PCR ribotyping are the methods of choice in North America and in Europe, respectively. Three variations of PCR ribotyping have been described, two of them differing in the primers used, and while two use traditional agarose gel-based analysis (1, 5), the third uses capillary gel electrophoresis-based analysis of the results (2). Here, we describe a modification of PCR ribotyping that can be used for detection of C. difficile ribotypes directly in stool samples.
Direct PCR ribotyping from stool samples.
A total of 105 stool samples submitted to the Institute of Public Health Maribor (MB laboratory) for routine C. difficile testing and 84 samples from the Institute of Public Health Murska Sobota (MS laboratory) were tested. Samples from the MB laboratory were identified as C. difficile positive according to positive culture on CLO selective plates (bioMérieux) after ethanol shock. Samples from the MS laboratory were tested using the Cepheid Xpert C. difficile assay. From these samples, C. difficile was isolated as described above.
For direct PCR ribotyping from stool samples, we designed new primers located partially within the C. difficile 16S–23S rRNA intergenic spacer region (ISR) and partially within the 16S (forward primer) and 23S (reverse primer) rRNA genes. New primers were defined on the basis of DNA sequences of the ISRs of 29 different PCR ribotypes (data not shown). These primers resulted in increased specificity for C. difficile and could be used for direct typing from the stool sample.
The primer sequences were 5′ GCTGGATCACCTCCTTTCTAAG (forward primer) and 5′ TGACCAGTTAAAAAGGTTTGATAGATT (reverse primer). A QIAamp DNA stool minikit (Qiagen, Germany) was used to extract total DNA from stool samples, and 5 μl was used as template DNA for PCR. The ISRs were amplified in a final volume of 50 μl containing 5 μl of DNA, 50 pmol of each primer, 1.5 mM MgCl2, 0.1 mg/ml bovine serum albumin (BSA), and 1.25 U of Taq DNA polymerase (Roche Diagnostics, Germany). The amplification conditions were as follows: an initial denaturation step of 5 min at 95°C, followed by 35 cycles of 1 min at 95°C for denaturation, 1 min at 57°C for annealing, and 1 min at 72°C for elongation, plus 10 min at 72°C for final elongation. Amplification products were concentrated to a final volume of 25 μl by heating at 75°C for 45 min before electrophoresis in 3% agarose gel (Bio-Rad, United States) for 5 h at 2.5 V/cm. BioNumerics software (Applied Maths, Belgium) was used to analyze the banding patterns. PCR ribotypes for which the reference strains were available were designated by standard Cardiff nomenclature (001, 002,…), while others are designated by internal nomenclature (SLO and a 3-digit code).
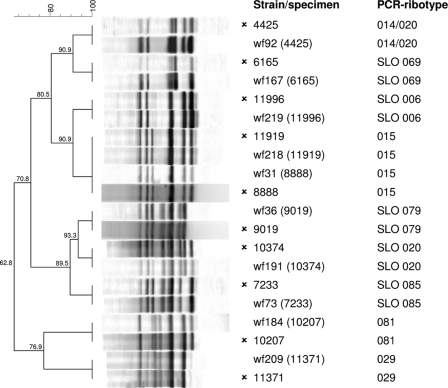
Altogether, 189 samples were tested; among them, 99 were C. difficile positive and 90 were C. difficile negative. By using the new primers, direct PCR ribotyping from stool samples was possible in 86 out of 99 C. difficile-positive samples. In 84 out of 86 cases, the PCR ribotype determined directly from the stool sample was identical to the PCR ribotype of the strain isolated from the same stool sample (sensitivity, 84.8%; 84 out of 99 positive samples) (Fig. 1). The two exceptions had very similar but not identical banding patterns (one additional band in both cases). The remaining 13 C. difficile-positive samples either were negative (n = 11) or represented fragments that were too weak to be analyzed (n = 2) with direct ribotyping. However, strains isolated from these samples reacted with the modified primers, indicating that lack of amplification was not due to the primer specificity. Of 9 false-negative samples that were also tested with Bidet primers (as described below), 5 were negative and the other 4 samples reacted with the primers but the banding patterns were not comparable with the PCR ribotype of the strain isolated from same stool sample. This suggests that low concentrations of C. difficile DNA could be a cause of negative direct typing with the modified primers.
Fig. 1.
Comparison of PCR ribotyping profiles obtained directly from stool samples and the ribotyping profiles of the strains isolated from the samples (isolates are marked with the symbol  ).
).
In 24 of the 90 C. difficile-negative samples, direct ribotyping with modified primers generated 1 to 4 nonspecific fragments which were clearly distinct from the common C. difficile ribotyping profiles and could easily be interpreted as C. difficile negative.
Samples positive with direct ribotyping (n = 84) were distributed into 25 PCR ribotypes; the most common were 027 (n = 32; due to an outbreak), 014/020 (n = 7), 070 (n = 4), 023 (n = 4), 002 (n = 4), SLO 011 (n = 3), SLO 006 (n = 3), and 003 (n = 3). Thirteen samples positive by culture for C. difficile but negative or weak on direct ribotyping contained 10 different ribotypes (014/020, 027, 001, 023, 106, 010, SLO 076, SLO 064, SLO 011, and SLO 036). When analyzed by direct ribotyping, none of the C. difficile-positive samples showed band profiles that would indicate the presence of two different ribotypes that is occasionally noticed (4, 6).
Conventional PCR ribotyping performed on total stool DNA.
For comparison, Bidet primers were used for standard PCR ribotyping of strains and also for direct ribotyping from 97 of the 189 stool samples. Amplification conditions were as described in Bidet et al. (1). While a ribotype profile was obtained from 37 of 51 (72.5%) C. difficile-positive samples, most of the C. difficile-negative samples (31 out of 46; 67.4%) reacted with this primer pair but with distinctively different fragment profiles. In a majority of cases (33 out of 37; 89.2%), profiles obtained from C. difficile-positive samples could not be assigned to known ribotypes due to the nonspecific bands (data not shown).
Summary.
Direct PCR ribotyping gives the information on the presence and type of C. difficile within hours, in contrast to standard culture-dependent methods where typing results can be obtained only after 3 days or more (48 h for culture and 1 day for PCR ribotyping). Direct PCR ribotyping on DNA isolated from stool samples is convenient, rapid, and useful for the detection of specific types of C. difficile in fecal samples.
Acknowledgments
S.J. was supported by grant 1000-08-310144 from the Slovenian Research Agency. Part of this work was supported by ERA NET Pathogenomic grant CDIFFGEN, contract number 3211-09-000141.
Footnotes
Published ahead of print on 1 June 2011.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. Bidet P., Barbut F., Lalande V., Burghoffer B., Petit J. C. 1999. Development of a new PCR-ribotyping method for Clostridium difficile based on ribosomal RNA gene sequencing. FEMS Microbiol. Lett. 175:261–266 [DOI] [PubMed] [Google Scholar]
- 2. Indra A., et al. 2008. Characterization of Clostridium difficile isolates using capillary gel electrophoresis-based PCR ribotyping. J. Med. Microbiol. 57:1377–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Janezic S., Rupnik M. 2010. Molecular typing methods for Clostridium difficile: pulsed-field gel electrophoresis and PCR ribotyping. Methods Mol. Biol. 646:55–65 [DOI] [PubMed] [Google Scholar]
- 4. Permoser M., et al. 2010. Clostridium difficile infection—monoclonal or polyclonal?, abstract P13, p. 69.In Rupnik M., Janezic S., Zemljic M. (ed.), Abstract book, Zavod za zdravstveno varstvo Maribor, Maribor, Slovenia. [Google Scholar]
- 5. Stubbs S. L., Brazier J. S., O'Neill G. L., Duerden B. I. 1999. PCR targeted to the 16S-23S rRNA gene intergenic spacer region of Clostridium difficile and construction of a library consisting of 116 different PCR ribotypes. J. Clin. Microbiol. 37:461–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. van den Berg R. J., et al. 2005. Coexistence of multiple PCR-ribotype strains of Clostridium difficile in faecal samples limits epidemiological studies. J. Med. Microbiol. 54:173–179 [DOI] [PubMed] [Google Scholar]