Abstract
Limited data are available on the performance of different automated extraction platforms and commercially available quantitative real-time PCR (QRT-PCR) methods for the quantitation of cytomegalovirus (CMV) DNA in plasma. We compared the performance characteristics of the Abbott mSample preparation system DNA kit on the m24 SP instrument (Abbott), the High Pure viral nucleic acid kit on the COBAS AmpliPrep system (Roche), and the EZ1 Virus 2.0 kit on the BioRobot EZ1 extraction platform (Qiagen) coupled with the Abbott CMV PCR kit, the LightCycler CMV Quant kit (Roche), and the Q-CMV complete kit (Nanogen), for both plasma specimens from allogeneic stem cell transplant (Allo-SCT) recipients (n = 42) and the OptiQuant CMV DNA panel (AcroMetrix). The EZ1 system displayed the highest extraction efficiency over a wide range of CMV plasma DNA loads, followed by the m24 and the AmpliPrep methods. The Nanogen PCR assay yielded higher mean CMV plasma DNA values than the Abbott and the Roche PCR assays, regardless of the platform used for DNA extraction. Overall, the effects of the extraction method and the QRT-PCR used on CMV plasma DNA load measurements were less pronounced for specimens with high CMV DNA content (>10,000 copies/ml). The performance characteristics of the extraction methods and QRT-PCR assays evaluated herein for clinical samples were extensible at cell-based standards from AcroMetrix. In conclusion, different automated systems are not equally efficient for CMV DNA extraction from plasma specimens, and the plasma CMV DNA loads measured by commercially available QRT-PCRs can differ significantly. The above findings should be taken into consideration for the establishment of cutoff values for the initiation or cessation of preemptive antiviral therapies and for the interpretation of data from clinical studies in the Allo-SCT setting.
INTRODUCTION
Quantitative real-time PCR (QRT-PCR) assays are being increasingly used for the surveillance of active cytomegalovirus (CMV) infection in allogeneic stem cell transplant (Allo-SCT) recipients (23). Several CMV QRT-PCR assays targeting different CMV genes and using different chemistries are commercially available (23). The analytical performance and clinical usefulness of these assays have been assessed, mostly in comparison with the pp65 antigenemia test or quantitative endpoint PCR assays (1, 2, 6, 9, 10-13, 15, 17, 19, 21, 22, 24-26). Limited data are available on how these QRT-PCR tests compare to each other for the quantitation of CMV plasma DNAemia (5, 11, 12, 18, 24). This information may allow, at least to some extent, direct comparisons of CMV DNA loads measured at different centers.
Nucleic acid extraction is a critical step in QRT-PCR testing, and it has been shown to be a major source of assay variability in viral DNA quantitation (7). Automated nucleic acid extraction systems are less time-consuming, less prone to analytical errors, and, overall, more efficient than manual methods. A few studies have directly compared the extraction efficiency of different automated systems for the quantitation of CMV DNA in plasma by QRT-PCR assays (5, 14, 16). In this study, we evaluated the performance characteristics of three automated DNA extraction platforms coupled with three different QRT-PCR assays for CMV DNA quantitation in plasma obtained from Allo-SCT recipients.
MATERIALS AND METHODS
Clinical specimens and reference standards.
A total of 42 plasma specimens were included in the study. These samples were obtained from 23 patients (11 males and 12 females; mean age, 48 years; range, 19 to 65 years) who had undergone related/HLA-matched (n = 10), related/HLA-mismatched (n = 3), unrelated/HLA-matched (n = 7), or unrelated/HLA-mismatched (n = 3) peripheral blood Allo-SCT at our institution between March 2009 and October 2010. Paired CMV serostatuses of donors (D) and recipients (R) were D+/R+ in 16 cases and D−/R+ in the remaining 7 cases. The underlying disease was of hematological origin in all patients. All plasma specimens were obtained within the first 120 days following Allo-SCT. The aliquots of plasma used in the current study were frozen at −20°C shortly after collection and had not been thawed prior to use for the analyses described herein. The specimens were grouped into three categories according to the CMV DNA loads measured using the Abbott real-time PCR assay (coupled with the Abbott mSample preparation system DNA kit on the m24 SP instrument), the method currently used at our institution (see below): (i) 10 samples with low CMV DNA contents (<1,000 copies/ml; mean, 394 copies/ml; range, 25 to 734 copies/ml); (ii) 22 samples with intermediate CMV DNA loads (>1,000 and <10,000 copies/ml; mean, 5,126 copies/ml; range, 1,088 to 9,824 copies/ml), and (iii) 10 samples with high CMV DNA loads (>10,000 copies/ml; mean, 23,766; range, 10,961 to 48,406 copies/ml). Plasma specimens were tested once by each nucleic acid extraction method/QRT-PCR assay combination. A unique aliquot of each plasma specimen was used for all the analyses reported herein.
The OptiQuant CMV DNA quantification panel (AcroMetrix Corp., Benicia, CA), which contains normal human plasma spiked with four concentrations of CMV strain Ad169, was used to compare the performance characteristics of the extraction methods and the QRT-PCR assays evaluated herein. The standards were tested in two different runs at 100, 1,000, 5,000, and 10,000 copies/ml. Dilutions were prepared from the panel using the NAT dilution matrix (AcroMetrix).
Nucleic acid extraction.
The following automated nucleic acid extraction methods were evaluated: the Abbott mSample preparation system DNA kit on the m24 SP instrument (Abbott Diagnostics, IL), the High Pure viral nucleic acid kit on the COBAS AmpliPrep system (Roche Diagnostics, Mannheim, Germany), and the EZ1 Virus 2.0 kit (Qiagen, Valencia, CA) on the BioRobot EZ1 extraction platform (Qiagen, Valencia, CA), following the instructions of the respective manufacturers. The DNA extractions were performed using 500 μl of plasma for the m24 SP and COBAS AmpliPrep systems with an elution volume of 70 μl in both methods and using 400 μl for the BioRobot EZ-1 (maximum volume) with an elution volume of 60 μl.
QRT-PCR assays.
The following QRT-PCR assays were evaluated. The Abbott CMV PCR kit (produced by Qiagen GmbH, Hilden, Germany, for Abbott Diagnostics, Des Plaines, IL), which amplifies a 105-bp region of the UL122 (IE-1) gene, was evaluated using the m2000RT system (Abbott Molecular, IL). The limit of detection of the assay is approximately 25 copies/ml (9). The LightCycler CMV Quant kit (Roche Diagnostics GmbH, Mannheim, Germany), which targets the UL54 DNA polymerase gene of CMV, was evaluated using the LightCycler 2.0 instrument (Roche). The limit of detection of this assay is approximately 250 to 300 copies/ml for plasma specimens (5). The Q-CMV complete kit (Cepheid, Turin, Italy; manufactured by Nanogen Advanced Diagnostics), which targets the exon 4 region of the CMV major immediate early gene (UL123), was evaluated using the LightCycler 2.0 instrument (Roche). According to the manufacturer, the limit of detection of this assay is approximately 700 copies/ml. The mean intra-assay variability (percent coefficient of variation) of the above-described QRT-PCR assays for noncellular specimens is <0.2 log10 over the linear range of the assays (according to the manufacturers). All assays were performed following the instructions of the respective manufacturers. The three QRT-PCR assays were approved via the CE-labeling system.
Statistical methods.
The data were log10 transformed prior to analysis. Differences between mean CMV DNA loads measured using the three assays (coupled with the three automated extraction methods) were analyzed using Pearson's chi-square test. Any quantitative correlations between the CMV DNA loads obtained using the different extraction methods and the QRT-PCR assays were detected using Pearson's correlation tests. The method of Bland and Altman (3) was used to assess the agreement between viral loads measured using the different analytical systems. Statistical calculations were performed with the aid of the SPSS 17.0 program (SPSS Inc., Chicago, IL); P values of ≤0.05 were considered statistically significant.
RESULTS
Comparison of extraction efficiencies of the automated methods.
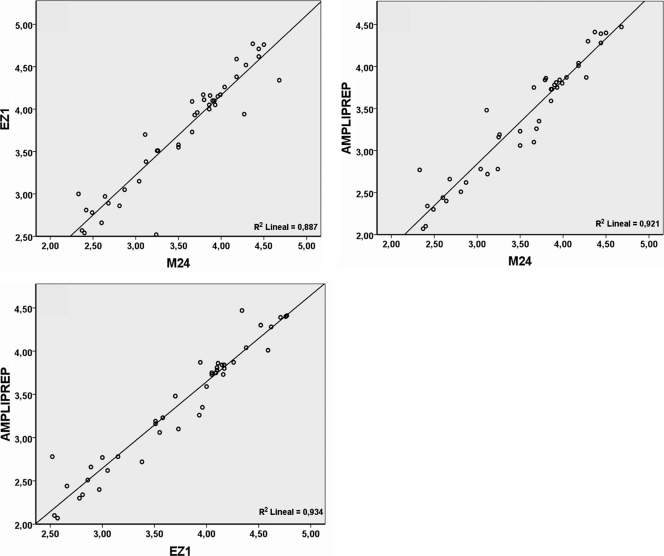
The CMV DNA loads in the nucleic acid extracts from the plasma specimens obtained using the three automated extraction methods were significantly correlated, irrespective of the QRT-PCR assay employed for quantitation (P = <0.001 for all correlations). The data obtained using the Abbott QRT-PCR kit test are shown in Fig. 1. The extraction efficiency of the EZ1 platform was superior to that of the other two systems (Table 1). The CMV DNA loads measured using the three QRT-PCR assays following extraction with the EZ1 BioRobot were consistently higher than those obtained after extraction by the m24 SP system. The differences in the CMV DNA loads measured following extraction by these platforms varied depending on the QRT-PCR assay used and the CMV DNA content of the specimen. For specimens with <1,000 copies/ml, the differences in the mean CMV DNA loads ranged from 0.10 log10 copies/ml (P = 0.280) in the Nanogen PCR assay to 0.25 log10 copies/ml (P = 0.07) in the Abbott PCR assay. For specimens with a higher CMV DNA content (>10,000 CMV DNA copies/ml), the differences were less marked and ranged from 0.15 log10 copies/ml (P = 0.157) in the Abbott PCR assay to 0.01 log10 copies/ml (P = 0.853) in the Nanogen PCR assay. However, the differences were more striking when the extraction efficiency of the EZ1 BioRobot was compared to that of the AmpliPrep system. In effect, for specimens with <1,000 copies/ml, the mean differences ranged from 0.66 log10 copies/ml (P = <0.001) in the Abbott PCR assay to 0.27 log10 copies/ml (P = 0.005) in the Nanogen PCR assay. For specimens with intermediate CMV DNA loads, the differences ranged from 0.41 log10 copies/ml (P = <0.001) in the Roche PCR assay to 0.33 log10 copies/ml (P = <0.001) in the Nanogen PCR. The differences were also significant for specimens with >10,000 copies/ml (P = 0.019 in the Abbott PCR assay; P = 0.014 in the Roche PCR assay; P = 0.005 in the Nanogen PCR assay). Finally, the m24 SP system displayed greater extraction efficiency than the AmpliPrep method over the entire range of CMV DNA concentrations, irrespective of the QRT-PCR employed. Nevertheless, the differences reached statistical significance only when the specimens were analyzed by the Nanogen PCR (P = 0.026 for specimens with <1,000 copies/ml, P = 0.007 for specimens with 1,000 to 10,000 copies/ml, and P = 0.006 for samples with >10,000 copies/ml). As shown in Table 2, the above differences in the efficiency of CMV DNA extraction were also observed when the OptiQuant proficiency panel was tested (at different CMV DNA concentrations, ranging from 2.0 to 4.0 log10 CMV DNA copies/ml).
Fig. 1.
Correlation and linear regression analysis of cytomegalovirus (CMV) DNA load values (copies/ml) obtained for all positive specimens by the Abbott CMV PCR kit following DNA extraction using the Abbott mSample preparation system DNA kit on the m24 SP instrument (M24), the High Pure viral nucleic acid kit on the COBAS AmpliPrep system (AMPLIPREP), and the EZ1 Virus 2.0 kit on the BioRobot EZ1 extraction platform (EZ).
Table 1.
Cytomegalovirus (CMV) DNA load values in plasma obtained with three different QRT-PCR assays coupled to three distinct automated extraction systems
| QRT-PCR assay and extraction system | CMV DNA load, log10 copies/ml, for groupa: |
||
|---|---|---|---|
| <3 (n = 10) | 3–4 (n = 22) | >4 (n = 10) | |
| Abbottb | |||
| m24 | 2.55 (0.19) | 3.61 (0.31) | 4.33 (0.18) |
| AmpliPrep | 2.14 (0.77) | 3.43 (0.38) | 4.20 (0.23) |
| EZ1 | 2.80 (0.17) | 3.80 (0.41) | 4.48 (0.26) |
| Rochec | |||
| m24 | ND | 3.79 (0.37) | 4.38 (0.23) |
| AmpliPrep | ND | 3.63 (0.31) | 4.17 (0.23) |
| EZ1 | ND | 4.04 (0.31) | 4.47 (0.25) |
| Nanogend | |||
| m24 | 3.22 (0.15) | 4.03 (0.26) | 4.81 (0.20) |
| AmpliPrep | 3.05 (0.11) | 3.80 (0.26) | 4.47 (0.28) |
| EZ1 | 3.32 (0.18) | 4.13 (0.30) | 4.82 (0.21) |
Specimens were subgrouped into three categories according to CMV DNA loads measured by using the Abbott real-time PCR assay (coupled with the Abbott mSample preparation system DNA kit on the m24 SP instrument). Data are reported as mean values (standard deviation) of CMV DNA loads quantitated in specimens testing positive by a given QRT-PCR assay. ND, not detected.
One of 10 specimens extracted with the AmpliPrep method tested negative by the Abbott QRT-PCR.
Only one out of 10 specimens with <3 log10 copies/ml extracted with the m24 SP and AmpliPrep systems and two extracted with the EZ1 system tested positive by the Roche QRT-PCR. Two specimens with CMV DNA loads between 3 and 4 log10 copies/ml extracted with the m24 SP, AmpliPrep, and EZ1 methods tested negative by the Roche QRT-PCR.
Three specimens (<3 log10 copies/ml) extracted with the AmpliPrep system and one (<3 log10 copies/ml) extracted with the EZ1 system tested negative by the Nanogen QRT-PCR.
Table 2.
Cytomegalovirus (CMV) DNA values obtained with three QRT-PCR assays coupled with three automated extraction methods with the OptiQuant proficiency panel
| QRT-PCR assay and extraction system | CMV DNA load in OptiQuant standards (log10 copies/ml) per dilutiona |
|||
|---|---|---|---|---|
| 2.0 | 3.0 | 3.7 | 4.0 | |
| Abbott | ||||
| m24 | 1.58 | 2.61 | 3.46 | 3.93 |
| AmpliPrep | 1.57 | 2.28 | 3.23 | 3.83 |
| EZ1 | 1.63 | 3.11 | 3.78 | 4.01 |
| Roche | ||||
| m24 | ND | 2.56 | 3.48 | 3.92 |
| AmpliPrep | ND | 2.41 | 3.32 | 3.80 |
| EZ1 | ND | 3.11 | 3.87 | 4.04 |
| Nanogen | ||||
| m24 | ND | ND | 3.60 | 3.94 |
| AmpliPrep | ND | ND | 3.41 | 3.92 |
| EZ1 | ND | 3.28 | 3.94 | 4.05 |
Dilutions of the OptiQuant standards were prepared from the panel using the NAT dilution matrix (AcroMetrix). Data are reported as mean results of two experiments.
Comparison of the performance of QRT-PCR assays.
The Roche PCR assay gave positive results for 31/42 samples following extraction either with the AmpliPrep or with the m24 system and for 32 specimens after extraction with the EZ1 BioRobot. The Nanogen PCR assay yielded positive results for 39, 41, and 42 specimens following extraction with the AmpliPrep, the EZ1 BioRobot, and the m24 platforms, respectively. Finally, the Abbott PCR assay gave positive results for all specimens extracted with the EZ1 BioRobot and for 41/42 of specimens extracted with the AmpliPrep platform.
The Nanogen PCR assay yielded significantly higher CMV DNA loads than the Abbott and the Roche PCR assays, regardless of the platform used for nucleic acid extraction or the CMV DNA content of the specimen (P = <0.042 for all comparisons between means) (Table 1). In turn, the CMV DNA loads measured by the Abbott PCR and the Roche PCR did not differ significantly (P = >0.102 for all comparisons between mean values), regardless of the extraction method used and the concentration of the CMV DNA specimen (Table 1). When DNA extraction was performed using the most efficient extraction system (the EZ1 BioRobot) we found that (i) the Nanogen and the Abbott QRT-PCR assays gave CMV DNA loads differing by less than 0.5 log10 DNA copies/ml in 33/42 samples (78.5%) (Fig. 2 A), the Nanogen and the Roche QRT-PCR assays in 27/32 specimens (84.3%) (Fig. 2B), and the Roche and the Abbott PCR assays in 31/32 specimens (96.8%) and (ii) the highest differences in CMV DNA loads measured by the PCR assays were found for specimens with low CMV DNA content (<1,000 copies/ml). In keeping with the above data, the Nanogen PCR assay yielded higher CMV DNA loads than the Abbott and Roche PCR assays when the OptiQuant panel was tested, whereas the Roche and Abbott PCR assays gave equivalent results, irrespective of the extraction platform used (Table 2). When samples were extracted using the EZ1 BioRobot, all QRT-PCR assays gave CMV DNA loads above the expected values.
Fig. 2.
Bland-Altman representation of CMV DNA loads (copies/ml) measured using the Abbott CMV PCR kit in the m2000RT (ABBOTT), the LightCycler CMV Quant kit in the Light Cycler 2.0 instrument (ROCHE), and the Q-CMV complete kit in the LightCycler 2.0 instrument (NANOGEN), following DNA extraction by the EZ1 Virus 2.0 kit on the BioRobot EZ1 extraction platform.
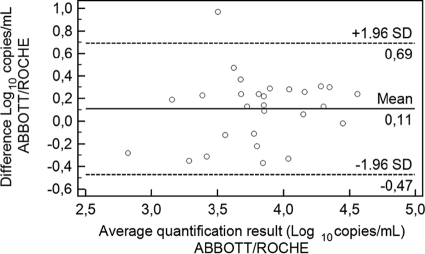
Comparison of CMV DNA loads measured using the Abbott and Roche reagents and extraction platforms.
The CMV DNA loads obtained using the Abbott and Roche reagents (QRT-PCRs coupled with the respective extraction methods) were directly compared for specimens that tested positive in both systems (n = 32). CMV DNA loads differing by less than 0.5 log10 DNA copies/ml were obtained by both methods in all but one sample (Fig. 3). In fact, the mean CMV DNA load values obtained using both systems were not significantly different for either the specimens with intermediate CMV DNA loads (P = 0.547) or samples with high CMV DNA content (P = 0.106) (Table 1). For specimens with 1,000 to 10,000 copies/ml, the CMV loads measured using the Roche assay were slightly higher than those obtained using the Abbott PCR assay (mean, 1.1-fold; range, 0.3- to 2.1-fold). Conversely, for specimens with >10,000 copies/ml, the Abbott PCR assay gave higher CMV DNA values than the Roche PCR assay (mean, 1.49-fold; range, 0.77- to 2.96-fold).
Fig. 3.
Bland-Altman representation of CMV DNA loads (copies/ml) measured using the Abbott CMV PCR kit in the m2000RT (ABBOTT) after extraction by the Abbott mSample preparation system DNA kit on the m24 SP instrument and those measured by the LightCycler CMV Quant kit in the Light Cycler 2.0 instrument (ROCHE), following extraction by the High Pure viral nucleic acid kit on the COBAS AmpliPrep system.
DISCUSSION
Limited data are available on how commercially available QRT-PCR assays compare to each other for the measurement of CMV DNA loads in plasma (5, 11, 12, 18, 24). Likewise, there are scarce data published on the performance of automated extraction systems for CMV plasma DNA quantitation (5, 11, 14-16, 20). The above information could, to some extent, allow the direct comparison and interpretation of CMV DNA loads obtained using different methods. In the current study, a comparative evaluation of three widely used automated systems for DNA extraction coupled with three commercially available QRT-PCR assays for the quantitation of cytomegalovirus DNA in plasma was performed for both clinical specimens from Allo-SCT recipients and the OptiQuant proficiency panel. Specimens were grouped into three categories according to the CMV DNA loads measured using Abbott reagents and the m24 SP platform (Abbott). We found consistent differences in the nucleic acid extraction efficiencies of the different automated systems. Although CMV DNA loads in the nucleic acid extracts from the plasma specimens obtained by using different methods correlated significantly, irrespective of the QRT-PCR assay employed for quantitation, the extraction efficiency of the EZ1 platform was superior to those of the Abbott m24 SP and AmpliPrep systems, both for clinical specimens (despite using a lower starting volume of plasma) and for the OptiQuant standards. In turn, the m24 SP system displayed greater extraction efficiency than the AmpliPrep system over the entire range of CMV DNA loads and irrespective of the QRT-PCR assay employed. Nevertheless, the differences between the latter two methods appeared to be subtle and reached statistical significance only when the specimens were analyzed using the Nanogen PCR assay. The impact of automated extraction methods on CMV DNA quantitation has also been noted in other studies. Caliendo et al. (5) found substantial differences in mean viral loads measured in plasma samples by the artus CMV LC/TM/RG PCR Kit (Qiagen, Valencia, CA)—an assay equivalent to the Abbott PCR kit used in the present study—depending upon the automated extraction method used (MagNA pure from Roche versus NucliSens easyMAG from bioMérieux). Miller et al. (16) reported that the QIAsymphony platform (Qiagen) displayed greater extraction efficiency than the BioRobot EZ1 for CMV DNA quantitation with the QRT-PCR ASR (analyte-specific reagents) from Roche (equivalent to the LightCycler CMV Quant kit used in the present study).
Whole blood is routinely used at many centers as the sample material for monitoring of active CMV infection in Allo-SCT recipients. Whole blood specimens yield higher CMV DNA loads than plasma samples and permit the earlier detection of active CMV infection in this clinical setting (23). In a prior study (14), differences in the performance of several automated extraction methods for CMV DNA quantitation in whole blood specimens using “in-house” QRT-PCR assays were reported. Overall, the magnitude of these differences was slightly higher than those observed in the current study for plasma specimens. This is a clinically relevant issue that must be thoroughly addressed in future studies. In the current study, the differences between the EZ1 and the other two automated extraction systems were particularly marked when specimens with low CMV DNA loads (specimens with <1,000 copies/ml) were analyzed, and their magnitude depended on the QRT-PCR used.
The analytical performance of commercially available QRT-PCR assays has mostly been compared with that of the pp65 antigenemia test or quantitative endpoint PCR assays (1, 2, 6, 9, 10-13, 15, 17, 19, 21, 22, 24-26). Nevertheless, a few studies directly compared the performance of these assays for the quantitation of CMV plasma DNAemia (5, 11, 12, 18, 24). In addition, to the best of our knowledge, data on the performance of the QRT-PCR from Nanogen have not been published. Our data indicated the following observations. (i) The QRT-PCR assay from Abbott was more sensitive than those from Roche and Nanogen. This was expected on the basis of previously published data (5, 9) and the specifications of the manufacturers. Nevertheless, our data suggested that the sensitivity of the Nanogen PCR assay may have been underestimated. (ii) The QRT-PCR assay from Nanogen yielded significantly higher CMV DNA loads than the other two PCR assays, regardless of the platform used for DNA extraction and the CMV DNA contents of the specimens. These differences were overall of greater magnitude than those that would be expected according to the intra-assay variability of the respective QRT-PCR assays, and they were greatest at the lowest CMV DNA concentration. Nevertheless, the CMV DNA loads measured with the Nanogen PCR assay and those quantitated by the Abbott and Roche PCR methods, following extraction with the most efficient system (EZ1 BioRobot), differed by less than 0.5 log10 copies/ml for 78.5% and 84.3% of samples, which, as suggested by Pang et al. (17, 18), represents the upper limit for divergence from the expected reference values for adequate interlaboratory comparisons of CMV DNA loads with clinical purposes. The CMV DNA loads measured by the Abbott PCR and the Roche PCR assays were not significantly different, irrespective of the extraction method and the CMV DNA content of specimens. In fact, the differences found were less than 0.5 log10 copies/ml for 96.6% of plasma samples. Interestingly, we found that the CMV DNA loads obtained using the Abbott QRT-PCR assay coupled with the m24 SP extraction system were slightly higher than those obtained by the Roche QRT-PCR assay coupled with the AmpliPrep extraction method for specimens with high CMV DNA concentrations (>10,000 copies/ml), but they were slightly lower for specimens with intermediate CMV DNA contents (>1,000 but <10,000 copies/ml). In this context, Hanson et al. (12) compared the Roche CMV UL54 analyte-specific reagent (equivalent to the Roche QRT-PCR assay used in the present study) and the Qiagen realArt CMV LightCycler PCR reagent (essentially the same product as the Abbott QRT-PCR assay used in the current study) and found that the CMV plasma DNA loads measured using the Qiagen assay tended to be lower than those measured with the Roche reagent when using clinical samples but not when employing the OptiQuant panel, in both cases after DNA extraction with the Roche MagnaPure automated platform. (iii) The performance characteristics of the QRT-PCR assays evaluated herein for clinical samples were extensible at cell-based standards from AcroMetrix, despite relevant differences in viral DNA conformation (highly fragmented CMV DNA in plasma specimens and large amounts of concatemers in cell-derived preparations) (4, 5).
The above findings have relevant implications for the therapeutic management of active CMV infection in Allo-SCT recipients. Threshold values of CMV plasma DNAemia for initiation of preemptive antiviral therapy in the Allo-SCT setting vary between centers, but they are usually set at around 500 to 1,000 copies/ml (23). In this context, our data underscore the fact that the choice of the automated extraction method and the QRT-PCR assay for the surveillance of active CMV infection may critically determine the decision to initiate or defer preemptive antiviral therapy and thus may ultimately influence the patient's clinical outcome. Thus, the CMV plasma DNA cutoff level triggering the implementation of antiviral therapy must be set taking into consideration the intrinsic performance of both the nucleic acid extraction method and the QRT-PCR assay employed. The recent advent of the 1st World Health Organization International Standard for CMV for Nucleic Acid Amplification (NAT)-Based Assays (8) should allow researchers to work out the equivalencies between CMV DNA loads measured by different QRT-PCR methods coupled with distinct nucleic acid extraction methods, ultimately permitting the establishment of clinically safe CMV DNA thresholds triggering therapeutic intervention.
In summary, our data indicated that there are substantial differences in nucleic acid extraction efficiency between automated systems and in CMV DNA loads measured using different commercially available QRT-PCR assays, which may impact on therapeutic decisions and should be taken into consideration for the interpretation of data from clinical studies. In addition, our results support the feasibility of comparing published clinical and virological data from patients who were monitored using the Abbott CMV PCR kit and the Roche LightCycler CMV Quant kit or their equivalent commercial products.
ACKNOWLEDGMENTS
We thank Julia Garcia, Mónica Reig, and Matilde Pastor for their technical assistance.
Footnotes
Published ahead of print on 22 June 2011.
REFERENCES
- 1. Abbate I., et al. 2008. Evaluation of an automated extraction system in combination with Affigene® CMV Trender for CMV DNA quantitative determination: comparison with nested PCR and pp65 antigen test. J. Virol. Methods 151: 61–65 [DOI] [PubMed] [Google Scholar]
- 2. Allice T., et al. 2008. Evaluation of a novel real-time PCR system for cytomegalovirus DNA quantitation on whole blood and correlation with pp65-antigen test in guiding pre-emptive antiviral treatment. J. Virol. Methods 148: 9–16 [DOI] [PubMed] [Google Scholar]
- 3. Bland J. M., Altman D. G. 1986. Statistical method for assessing agreement between two methods of clinical measurement. Lancet i: 307–310 [PubMed] [Google Scholar]
- 4. Boom R., et al. 2002. Human cytomegalovirus DNA in plasma and serum specimens of renal transplant recipients is highly fragmented. J. Clin. Microbiol. 40: 4105–4113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Caliendo A. M., et al. 2007. Evaluation of real-time PCR laboratory-developed test using analyte-specific reagents for cytomegalovirus quantification. J. Clin. Microbiol. 45: 1723–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Caliendo A. M., et al. 2001. Comparison of quantitative and qualitative PCR assays for cytomegalovirus DNA in plasma. J. Clin. Microbiol. 39: 1334–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Espy M. J., et al. 2006. Real-time PCR in clinical microbiology: applications for routine laboratory testing. Clin. Microbiol. Rev. 19: 165–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fryer J. F., Heath A. B., Anderson R., Minor P. D., the collaborative study group 2010. Collaborative study to evaluate the proposed 1st WHO International Standard for human cytomegalovirus (HCMV) for nucleic acid amplification (NAT)-based assays. WHO ECBS report no. WHO/BS/10.2138. WHO, Geneva, Switzerland: http://whaqlibdoc.who.int/hq/2010/WHO_BS_10.2138_eng.pdf [Google Scholar]
- 9. Gimeno C., et al. 2008. Quantification of DNA in plasma by an automated real-time PCR assay (cytomegalovirus PCR kit) for surveillance of active cytomegalovirus infection and guidance of preemptive therapy for allogeneic hematopoietic stem cell transplant recipients. J. Clin. Microbiol. 46: 3311–3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gouarin S., et al. 2007. Multicentric evaluation of a new commercial cytomegalovirus real-time PCR quantification assay. J. Virol. Methods 146: 147–154 [DOI] [PubMed] [Google Scholar]
- 11. Gracia-Ahufinger I., et al. 2010. Differences in cytomegalovirus plasma viral loads measured in allogeneic hematopoietic stem cell transplant recipients using two commercial real-time PCR assays. J. Clin. Virol. 48: 142–146 [DOI] [PubMed] [Google Scholar]
- 12. Hanson K. E., et al. 2007. Comparison of the Digene Hybrid capture system cytomegalovirus (CMV) DNA (version 2.0), Roche CMV UL54 analyte-specific reagent, and Qiagen RealArt CMV LightCycler PCR reagent test using AcroMetrix OptiQuant CMV DNA quantification panels and specimens from allogeneic-stem-cell transplant recipients. J. Clin. Microbiol. 45: 1972–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kerschner H., et al. 2011. Clinical evaluation of a fully automated CMV PCR assay. J. Clin. Virol. 50: 281–286 [DOI] [PubMed] [Google Scholar]
- 14. Mengelle C., Mansuy J. M., Da Silva I., Davrinche C., Izopet J. 2011. Comparison of 2 highly automated nucleic acid extraction systems for quantitation of human cytomegalovirus in whole blood. Diagn. Microbiol. Infect. Dis. 69: 161–166 [DOI] [PubMed] [Google Scholar]
- 15. Michelin B. D., et al. 2008. Detection of cytomegalovirus (CMV) DNA in EDTA whole-blood samples: evaluation of the quantitative artus CMV LightCycler PCR kit in conjunction with automated sample preparation. J. Clin. Microbiol. 46: 1241–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Miller S., Seet H., Khan Y., Wright C., Nadarajah R. 2010. Comparison of QIAGEN automated nucleic acid extraction methods for CMV quantitative PCR testing. Am. J. Clin. Pathol. 133: 558–563 [DOI] [PubMed] [Google Scholar]
- 17. Pang X. L., Chui L., Fenton J., LeBlanc B., Preiksaitis J. K. 2003. Comparison of LightCycler-based PCR, COBAS Amplicor CMV monitor, and pp65 antigenemia assays for quantitative measurement of cytomegalovirus viral load in peripheral blood specimens from patients after solid organ transplantation. J. Clin. Microbiol. 41: 3167–3174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pang X. L., et al. 2009. Interlaboratory comparison of cytomegalovirus viral load assays. Am. J. Transpl. 9: 258–268 [DOI] [PubMed] [Google Scholar]
- 19. Piiparinen H., Höckerstedt K., Grönhagen-Riska C., Lautenschlager I. 2004. Comparison of two quantitative CMV PCR tests, Cobas Amplicor CMV Monitor and TaqMan assay, and pp65-antigenemia assay in the determination of viral loads from peripheral blood of organ transplant patients. J. Clin. Virol. 30: 258–266 [DOI] [PubMed] [Google Scholar]
- 20. Pillet S., Bourlet T., Pozzetto B. 2009. Comparative evaluation of a commercially available automated system for extraction of viral DNA from whole blood: application to monitoring of Epstein-Barr virus and cytomegalovirus load. J. Clin. Microbiol. 47: 3753–3755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pumannova M., Roubalova K., Vitek A., Sajdova J. 2006. Comparison of quantitative competitive PCR-enzyme-linked immunosorbent assay with LightCycler-based PCR for measuring cytomegalovirus DNA in patients after hematopoietic stem cell transplantation. Diagn. Microbiol. Infect. Dis. 54: 115–120 [DOI] [PubMed] [Google Scholar]
- 22. Razonable R. R., et al. 2001. Comparative quantitation of cytomegalovirus (CMV) DNA in solid organ transplant recipients with CMV infection by using two high-throughput automated systems. J. Clin. Microbiol. 39: 4472–4476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Solano C., Navarro D. 2010. Clinical virology of cytomegalovirus infection following hematopoietic transplantation. Future Virol. 5: 111–124 [Google Scholar]
- 24. Vincent E., et al. 2009. Detection of cytomegalovirus in whole blood using three different real-time PCR chemistries. J. Mol. Diagn. 11: 54–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wirgart B. Z., Andersson P., Grillner L. 2005. Evaluation of the ReSSQ assay in relation to the COBAS AMPLICOR CMV MONITOR test and an in-house nested PCR method for detection of cytomegalovirus DNA. J. Clin. Microbiol. 43: 4057–4063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yerly S., et al. 2007. Cytomegalovirus quantification in plasma by an automated real-time PCR assay. J. Clin. Virol. 38: 298–303 [DOI] [PubMed] [Google Scholar]