Abstract
Using real-time technology, we reliably identified chronic hepatitis C virus (HCV) infection and quantified virus from reflex samples originally submitted for serologic testing. There was no need to process specimens obtained directly for quantitation separately. Whether the initial source is a reflex sample or one obtained directly, a repeat HCV RNA test is needed before starting treatment.
TEXT
Hepatitis C virus (HCV) is a major public health problem in the United States. Acute infection is usually asymptomatic and remains undiagnosed, with 75 to 85% of patients developing chronic infection that decades later may manifest in cirrhosis and hepatocellular carcinoma (3, 14). Diagnosis is dependent on laboratory testing, typically beginning with detection of antibodies to HCV proteins, which can be observed due to current infection with or previous exposure to the virus, as well as to false-positive results. Confirmation of current infection requires detection of HCV RNA in the blood of persons who are anti-HCV positive. According to the most recent National Health and Nutrition Examination Survey published in 2006, there are an estimated 4.1 million anti-HCV-positive persons, of whom 3.2 million are also HCV RNA positive (3).
In most clinical settings, HCV RNA testing is done after a health care provider receives a positive anti-HCV result. A request is then made for measurement on a second sample from the same individual (direct testing). For various reasons, however, this often is not done, and individuals either are not correctly identified as being currently infected or are labeled as infected when they have actually cleared the virus. Reflex testing (HCV RNA testing done automatically on the same, positive anti-HCV sample) can significantly shorten the time to clarifying patient status and prevent diagnostic misclassification based on incomplete information.
The prevalence of chronic HCV infection is higher in certain populations, including those receiving care in the Veterans Health Administration (VHA) (6). The VHA responded to this dilemma in 1998 by implementing CDC guidelines to identify viremic, anti-HCV-positive veterans for appropriate counseling and management (1). To streamline the process, VHA Directive 2009-063 mandated reflex HCV RNA confirmatory testing after a reactive serologic screening. The application and subsequent clinical utility of this directive may be impacted by several laboratory issues. Prior studies analyzing reflex specimens (7, 16, 18) did not evaluate whether there are significant differences in the frequency of HCV RNA detection or in the HCV viral load compared to those in specimens treated more optimally from the time of collection (5, 13). In addition, the viral load is known to fluctuate over time (2, 8, 10, 12, 17).
Through automated, real-time PCR technology, our objective was to assess the reliability of using reflex samples received after serologic testing versus the reliability of using direct samples obtained for HCV quantitation in determining viral status and providing the baseline viral load for treatment at VA Medical Centers in Washington, DC, Baltimore, MD, and Martinsburg, WV. The period for our evaluation was from February 2008 through November 2010.
For reflex samples, peripheral blood was drawn by venipuncture into a serum separator tube and centrifuged within 6 h. The serum was stored at 2 to 8°C for 1 to 5 days before it was tested for the anti-HCV antibody on the Vitros ECiQ immunodiagnostic system (Ortho Clinical Diagnostics, Raritan, NJ) (7, 16, 18). Reactive sera, defined by a signal/cutoff ratio of >9.5 or, if the signal/cutoff ratio is <9.5, an indeterminate or positive recombinant immunoblot assay (RIBA) result, were frozen at −20°C for 1 to 3 days and then at −80°C until quantitative analysis. For direct samples, peripheral blood was drawn by venipuncture into EDTA or a serum separator tube and centrifuged within 6 h. The plasma/serum was frozen at −20°C for 0 to 3 days and then at −80°C until quantitative analysis.
Testing was performed by using the Abbott RealTime HCV assay (an analyte-specific reagent) with the m2000sp platform for sample preparation and the m2000rt for amplification and detection (Abbott Molecular, Inc., Des Plaines, IL). The quantitative range was 20 to 20,000,000 international units per milliliter (IU/ml), or 1.301 to 7.301 log10 IU/ml. For quality control, five RNA levels from pooled patient sera/plasma were assayed on each run. From November 2009 to November 2010, the mean log10 IU/ml values (percent coefficient of variation [CV]) were 1.509 (11.4%), 2.445 (3.0%), 3.772 (2.5%), 5.249 (1.3%), and 6.723 (1.5%) on 38 assays. The slope, intercept, and r2 were determined for statistical analysis by a least-squares linear regression analysis of the log10 IU/ml, and the P value was determined by a two-tailed, paired sample t test.
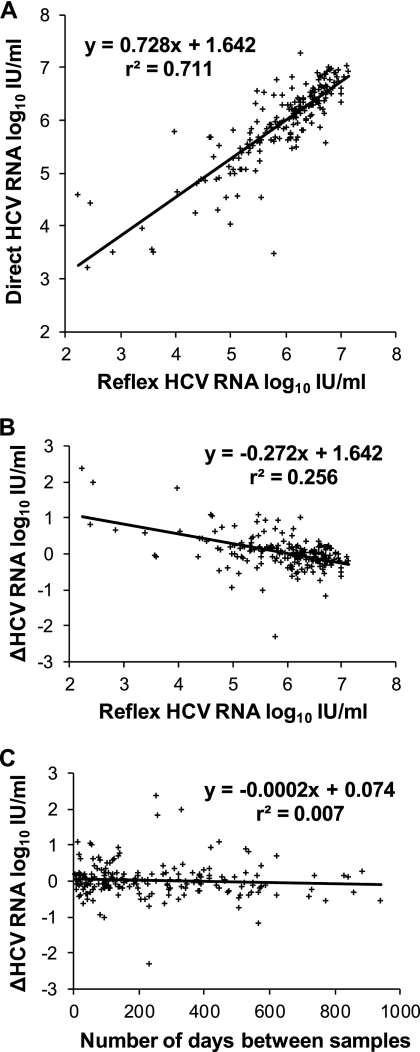
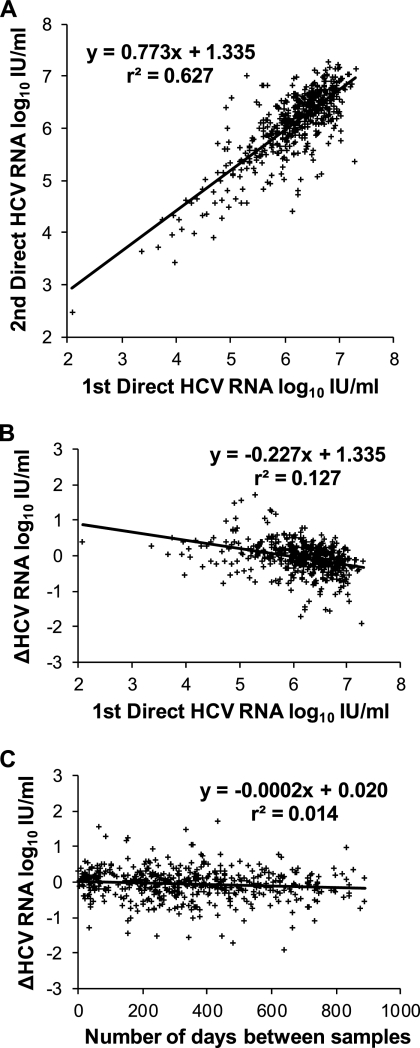
Quantitative HCV RNA testing of anti-HCV-positive patients is important for two main reasons. The absence of the detectable virus identifies individuals who do not require treatment. Based on 1,435 reflex samples, our frequency of detection of 80.5% was within the expected range (3, 14). Of the remaining 280 veterans, 67 had subsequent direct tests and 64 (96%) were again not viremic. In addition, the rate of HCV RNA decrease is a predictor of the sustained viral response to antiviral treatment (4, 9). For that reason, we wanted to explain the larger differences, of >0.5 log10 IU/ml, between the reflex sample results and the direct sample results for 19% of the chronically infected population (Table 1; Fig. 1A). Based on four small experiments, preanalytical factors did not account for this variation (Table 2). The most likely cause was biological fluctuation, matching the pattern observed in a parallel analysis of serial direct specimens (Table 1; Fig. 2A).
Table 1.
Variability of HCV RNA viral loads in serial clinical samplesa
| Serial specimen types used (n) | No. of days between sample collection | HCV RNA range (log10 IU/ml) | ΔHCV RNA (log10 IU/ml) |
P value | |ΔHCV RNA| |
|||
|---|---|---|---|---|---|---|---|---|
| Mean | Range | Mean log10 IU/ml | % (no.) of samples with result of >0.5 | % (no.) of samples with result of >1.0 | ||||
| Reflex, direct (198) | 6-941 | 2.228-7.127 | 0.030 | −2.310-2.365 | 0.382 | 0.306 | 19 (37) | 5 (10) |
| Direct, direct (493) | 6-887 | 2.079-7.301 | −0.061 | −1.914-1.727 | 0.002 | 0.319 | 19 (95) | 4 (21) |
Patients were treatment naïve with viral loads within the quantitative range. If there were more than two results for one individual, only the first and last were compared.
Fig. 1.
Comparison of HCV RNA assay results for 198 pairs of serial clinical samples, reflex and direct. Patients were treatment naïve. If there were more than two results for one individual, only the first and last were compared.
Table 2.
Interassay comparisons: effects of preanalytical factors on HCV RNA
| Specimen type (n) comparison | HCV RNA range (log10 IU/ml) | Linearity |
ΔHCV RNA (log10 IU/ml) |
P value | Mean |ΔHCV RNA| (log10 IU/ml) | |||
|---|---|---|---|---|---|---|---|---|
| Slope | Intercept | r2 | Mean | Range | ||||
| Sample 1 vs. sample 2a (29) | 1.477b-7.053 | 1.002 | −0.034 | 0.992 | −0.025 | −0.415b-0.222 | 0.352 | 0.103 |
| Serum 1 vs. serum 2c (10) | 5.013-6.586 | 1.151 | −0.910 | 0.973 | −0.010 | −0.205-0.205 | 0.782 | 0.103 |
| Serum vs. plasmad (10) | 4.783-6.911 | 0.968 | 0.103 | 0.976 | −0.089 | −0.320-0.054 | 0.102 | 0.106 |
| Reflex vs. directe (10) | 5.182-6.848 | 0.991 | 0.137 | 0.875f | 0.080 | −0.202-0.487f | 0.203 | 0.139 |
Following analysis, plasma/serum was refrozen at −80°C, thawed, and analyzed again in January 2008.
An initial viral load of 1.892 was followed by 1.477 log10 IU/ml (78 → 30 IU/ml); the next greatest decrease was −0.307 log10 IU/ml.
Following analysis, serum was refrozen at −80°C, thawed, stored for 5 days at 2 to 8°C, and analyzed again in November 2010.
Serum and EDTA-plasma were drawn at the same time, handled as direct samples (serum sample centrifuged 2 to 4 h before plasma), and analyzed separately from November 2010 to February 2011.
Reflex sera and direct EDTA-plasma samples were drawn at the same time, processed, and analyzed separately in November 2010 and January 2011.
The next greatest increase in ΔHCV RNA log10 IU/ml after 0.487 was 0.183. Excluding this outlier, the r2 was 0.946.
Fig. 2.
Comparison of HCV RNA assay results for 493 pairs of serial clinical samples, both direct. Patients were treatment naïve. If there were more than two results for one individual, only the first and last were compared.
Utilizing a real-time assay with high sensitivity and broad dynamic range, the variability of serial HCV RNA that we observed was consistent with earlier findings (2, 8, 10, 12, 17), with no clinically significant differences between values from an initial reflex and subsequent direct sample and from two direct samples (Table 1; Fig. 1A and 2A). Viral loads remained stable at levels of <0.5 log change for most patients, but fluctuations of >1.0 log did occur. These differences did not correlate with the initial test result (Fig. 1B and 2B) or the number of days between samples (Fig. 1C and 2C). Spontaneous loss of virus from circulation is virtually never seen beyond the first 6 months (19), and the level of viremia is not an indicator of disease activity (17) or progression (11, 15). Based on our findings, whether an initial value is obtained from reflex or direct samples, a repeat HCV RNA test is needed just before starting treatment to reduce the impact of random fluctuation on the evaluation of treatment effectiveness.
We were able to reliably identify chronic hepatitis C virus infection and quantify virus from reflex samples originally submitted to the serology laboratory for anti-HCV antibody testing. Separate handling for quantitation was not necessary. Our results are applicable to the methods used here. They may not pertain to other anti-HCV assays, since the Vitros system uses disposable tips to prevent carryover. We believe that our results do apply to alternative HCV RNA testing methods, because the Abbott RealTime results are similar to those of other studies. The convenience and reliability of reflex testing make it a preferred option for confirming HCV infection and determining viral load.
Acknowledgments
We thank Rebecca Shinol, Karen Rexroth, Maria Earp, and Barbara Harris for their technical support and Judith Myers and Bonnie Charon for their clinical perspectives.
The IRB and R & D Committee at this VA Medical Center reviewed and approved the manuscript. The views expressed are those of the authors and do not reflect the views or policies of the Department of Veterans Affairs. The authors report no conflict of interest.
Footnotes
Published ahead of print on 8 June 2011.
REFERENCES
- 1. Alter M. J., Kuhnert W. L., Finelli L. 2003. Guidelines for laboratory testing and result reporting of antibody to hepatitis C virus. MMWR Recommend. Rep. 52(RR03):1–16 [PubMed] [Google Scholar]
- 2. Arase Y., et al. 2000. Fluctuation patterns of HCV-RNA serum level in patients with chronic hepatitis C. J. Gastroenterol. 35:221–225 [DOI] [PubMed] [Google Scholar]
- 3. Armstrong G., et al. 2006. The prevalence of hepatitis C virus infection in the United States, 1999 through 2002. Ann. Intern. Med. 144:705–714 [DOI] [PubMed] [Google Scholar]
- 4. Davis G., et al. 2003. Early virologic response to treatment with peginterferon alfa-2b plus ribavirin in patients with chronic hepatitis C. Hepatology 38:645–652 [DOI] [PubMed] [Google Scholar]
- 5. de Moreau de Gerbehaye A., Bodéus M., Robert A., Horsmans Y., Goubau P. 2002. Stable hepatitis C virus RNA detection by RT-PCR during four days storage. BMC Infect. Dis. 2:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dominitz J., et al. 2005. Elevated prevalence of hepatitis C infection in users of United States veterans medical centers. Hepatology 41:88–96 [DOI] [PubMed] [Google Scholar]
- 7. Dufour D., Talastas M., Fernandez M., Harris B. 2003. Chemiluminescence assay improves specificity of hepatitis C antibody detection. Clin. Chem. 49:940–944 [DOI] [PubMed] [Google Scholar]
- 8. Fanning L., et al. 2000. Natural fluctuations of hepatitis C viral load in a homogeneous patient population: a prospective study. Hepatology 31:225–229 [DOI] [PubMed] [Google Scholar]
- 9. Fried M. W., et al. 2002. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347:975–982 [DOI] [PubMed] [Google Scholar]
- 10. Halfon P., et al. 1998. Assessment of spontaneous fluctuations of viral load in untreated patients with chronic hepatitis C by two standardized quantitation methods: branched DNA and Amplicor Monitor. J. Clin. Microbiol. 36:2073–2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hu S., Kyulo N., Xia V., Hillebrand D., Hu K. 2009. Factors associated with hepatic fibrosis in patients with chronic hepatitis C: a retrospective study of a large cohort of U.S. patients. J. Clin. Gastroenterol. 43:758–764 [DOI] [PubMed] [Google Scholar]
- 12. Kuramoto I., Moriya T., Schoening V., Holland P. 2002. Fluctuation of serum HCV-RNA levels in untreated blood donors with chronic hepatitis C virus infection. J. Viral Hepat. 9:36–42 [DOI] [PubMed] [Google Scholar]
- 13. Leckie G., et al. 2004. Performance attributes of the LCx HCV RNA quantitative assay. J. Virol. Methods 115:207–215 [DOI] [PubMed] [Google Scholar]
- 14. Lim J. 2001. Natural history of hepatitis C infection: a concise review. Yale J. Biol. Med. 74:229–237 [PMC free article] [PubMed] [Google Scholar]
- 15. McCormick S., Goodman Z., Maydonovitch C., Sjogren M. 1996. Evaluation of liver histology, ALT elevation, and HCV RNA titer in patients with chronic hepatitis C. Am. J. Gastroenterol. 91:1516–1522 [PubMed] [Google Scholar]
- 16. Oethinger M., Mayo D., Falcone J., Barua P., Griffith B. 2005. Efficiency of the Ortho VITROS assay for detection of hepatitis C virus-specific antibodies increased by elimination of supplemental testing of samples with very low sample-to-cutoff ratios. J. Clin. Microbiol. 43:2477–2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pontisso P., et al. 1999. Hepatitis C virus RNA profiles in chronically infected individuals: do they relate to disease activity? Hepatology 29:585–589 [DOI] [PubMed] [Google Scholar]
- 18. Seo Y. S., et al. 2009. Significance of anti-HCV signal-to-cutoff ratio in predicting Hepatitis C viremia. Korean. J. Intern. Med. 24:302–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yokosuka O., et al. 1999. Spontaneous negativation of serum hepatitis C virus RNA is a rare event in type C chronic liver diseases: analysis of HCV RNA in 320 patients who were followed for more than 3 years. J. Hepatol. 31:394–399 [DOI] [PubMed] [Google Scholar]