Abstract
The introduction of the seven-valent pneumococcal conjugate vaccine (PCV7) in Portugal led to extensive serotype replacement among carriers of pneumococci, with a marked decrease of PCV7 types. Although antimicrobial resistance was traditionally associated with PCV7 types, no significant changes in the rates of nonsusceptibility to penicillin, resistance to macrolides, or multidrug resistance were observed. This study aimed to investigate the mechanisms leading to maintenance of antimicrobial resistance, despite marked serotype replacement. We compared, through molecular typing, 252 antibiotic-resistant pneumococci recovered from young carriers in 2006 and 2007 (era of high PCV7 uptake) with collections of isolates from 2002 and 2003 (n=374; low-PCV7-uptake era) and 1996 to 2001 (n=805; pre-PCV7 era). We observed that the group of clones that has accounted for antimicrobial resistance since 1996 is essentially the same as the one identified in the PCV7 era. The relative proportions of such clones have, however, evolved substantially overtime. Notably, widespread use of PCV7 led to an expansion of two Pneumococcal Molecular Epidemiology Network (PMEN) clones expressing non-PCV7 capsular variants of the original strains: Sweden15AST63 (serotypes 15A and 19A) and Denmark14ST230 (serotypes 19A and 24F). These variants were already in circulation in the pre-PCV7 era, although they have now become increasingly abundant. Emergence of novel clones and de novo acquisition of resistance contributed little to the observed scenario. No evidence of capsular switch events occurring after PCV7 introduction was found. In the era of PCVs, antimicrobial resistance remains a problem among the carried pneumococci. Continuous surveillance is warranted to evaluate serotype and clonal shifts leading to maintenance of antimicrobial resistance.
INTRODUCTION
Streptococcus pneumoniae is a Gram-positive bacterium that frequently colonizes asymptomatically the nasopharynx of young children. However, it is also an important human pathogen that can cause a wide range of diseases, from otitis media to pneumonia and meningitis. Worldwide, it has been estimated that 14.5 million episodes of serious pneumococcal disease and 826,000 deaths per year occur in children aged less than 5 years (23).
In 2000, a seven-valent pneumococcal conjugate vaccine (PCV7) targeting the seven most common serotypes (4, 6B, 9V, 14, 18C, 19F, and 23F) causing invasive disease among young children in the United States became available. Since then, in countries where the vaccine has been introduced in the universal vaccination plan, a dramatic reduction in the incidence of invasive disease caused by PCV7 serotypes has been observed in all age groups (11, 17, 35). This extended phenomenon beyond the target group, known as herd immunity, has been attributed to decreased transmission of pneumococci from children to other age groups. Indeed, the nasopharynx of children is the main reservoir of pneumococci, and a decrease in carriage of PCV7 serotypes has been observed in vaccinated children (5). In addition to that, a replacement of PCV7 serotypes by non-PCV7 serotypes has been observed in carriage and disease (20, 25, 27, 38).
PCV7 became commercially available in Portugal in June 2001 but has not been introduced in the National Vaccination Program and is not reimbursed by the state. However, available data indicate that vaccination of children through the private sector has been increasing steadily, reaching ca. 70% of the target group by 2007 (this study; Pfizer, Portugal).
We have been studying pneumococcal colonization in Portugal since 1996. In the pre-PCV7 era, we documented that most drug-resistant isolates colonizing young children were associated with PCV7 serotypes (32, 33). Between 2001 and 2003, we conducted a pilot study to evaluate the impact of PCV7 on nasopharyngeal carriage (12). More recently, we described that since PCV7 became available in Portugal, extensive serotype replacement has occurred among vaccinated and unvaccinated children. In addition, the proportion of colonizing isolates resistant to at least one antibiotic did not change (30).
In this study, we aimed to identify the mechanisms responsible for maintenance of antimicrobial resistance levels, despite extensive serotype replacement. We characterized a collection of drug-resistant pneumococcal (DRPn) colonizing isolates obtained from young children in 2006 and 2007 (high-PCV7-uptake era) and compared it to two collections obtained years before: one before the introduction of PCV7 in Portugal (1996 to 2001, pre-PCV7 era) and the other recovered in the 2 years after PCV7 introduction (2002 and 2003, low-PCV7-uptake era).
MATERIALS AND METHODS
Study design and study collection.
Nasopharyngeal samples were obtained from children aged up to 6 years old attending day care centers in Oeiras and/or Lisbon, two contiguous urban areas of Portugal. All samples were collected during the winter months of January to March. Day care centers were selected to include children with different social backgrounds. In each year, one sample was obtained from each child. Information regarding age, gender, recent antimicrobial consumption, and PCV7 vaccination was obtained. Approval for the study was obtained from the Ministry of Health and the directors of the day care centers. Signed informed consent was obtained from the parents or guardians of participating children. This study design for sampling has been maintained for several years, allowing direct comparisons over time (19, 21, 33).
To evaluate clonal changes among antibiotic-resistant pneumococci colonizing healthy children following introduction of PCV7, three time periods were considered: (i) 1996 to 2001, the pre-PCV7 era; (ii) 2002 to 2003, the low-PCV7-uptake era, when 17.5% of the children enrolled in our studies had received at least one dose of the vaccine; and (iii) 2006 and 2007, the high-PCV7 uptake era, when 67.1% of the children had received at least one PCV7 dose. In the last period, 30.7% had received four doses and 20.0% were appropriately vaccinated for age but had not yet reached the four doses.
For each time period, all DRPn isolates originating from the following collections were selected: (i) 805 DRPn isolates recovered during the pre-PCV7 era (out of 2,152 isolates, recovered from 3,370 children), (ii) 374 DRPn isolates recovered during the low-PCV7-uptake era (out of 1,116 isolates, recovered from 1,600 children), and (iii) 272 DRPn isolates recovered during the high-PCV7-uptake era (out of 720 isolates, recovered from 1,109 children). Antibiotic-resistant isolates obtained between 1996 and 2003 were characterized before (19, 21, 33, 37). Isolates obtained in 2006 were partially described (according to their serotypes) before (29), and isolates obtained in 2007 are first described in this study.
Nasopharyngeal swabs and isolation of S. pneumoniae.
Nasopharyngeal sampling, transport, pneumococcal isolation, identification, and preparation of frozen stocks were done as previously described (29, 32).
Antimicrobial susceptibility testing.
MICs to penicillin were determined with the Etest (AB Biodisk, Solna, Sweden) according to the manufacturer's instructions. The interpretation criteria used were ≤0.06 μg/ml, 0.1 to 1 μg/ml, and ≥2 μg/ml for classification of susceptibility, intermediate resistance, and resistance to penicillin, respectively. Testing of susceptibility to erythromycin, clindamycin, tetracycline, chloramphenicol, and sulfamethoxazole-trimethoprim (SXT) was performed by disk diffusion, according to the recommendations and definitions of the Clinical and Laboratory Standards Institute (6). Multidrug resistance was defined as resistance to three or more classes of antimicrobial agents.
Capsular typing.
Capsular assignment was performed by a combination of multiplex PCRs targeting serotypes 1, 3, 6A, 6B, 6C, 7F, 9N/L, 9V, 10A, 11A, 14, 15A, 15B/C, 16F, 17F, 18A/B/C/F, 19A, 19F, 22F, 23A, 23F, 31, 33F/33A/37, 34, 35F, and 38 using primers previously described (2, 24; www.cdc.gov). Strains that could not be typed by this method were serotyped by the Quellung reaction using specific antisera (Statens Serum Institute, Copenhagen, Denmark) (36). Nontypeable (NT) strains were defined by the absence of the cpsA gene (screened by PCR using primers previously described [2]) and a negative reaction with omniserum (Statens Serum Institute, Copenhagen, Denmark).
DNA fingerprinting by PFGE.
Preparation of total DNA, digestion with SmaI endonuclease, and separation of DNA fragments by pulsed-field gel electrophoresis (PFGE) were carried out as previously described (33). PFGE patterns were analyzed with Bionumerics software (version 5; Applied Maths, Ghent, Belgium), and a dendrogram was generated using the Dice similarity coefficient with an optimization of 1.0% and a tolerance of 1.5%. PFGE clusters were defined by a similarity of 80% or higher on the dendrogram (4, 37). Representatives of Pneumococcal Molecular Epidemiology Network (PMEN) clones were also used for comparison of molecular types (www.sph.emory.edu/PMEN).
MLST.
Multilocus sequence typing (MLST) was undertaken essentially as described previously by amplifying internal fragments of seven housekeeping genes: aroE, gdh, gki, recP, spi, xpt, and ddl (9). Sequencing reactions were conducted at Macrogen, Inc. (Seoul, South Korea). Sequencing analysis was done with DNAStar software (Lasergene). Allele number and sequence type (ST) assignments were done at the MLST database for S. pneumoniae (www.mlst.net).
Selection of isolates for MLST was based on analysis of the dendrogram generated by clustering of PFGE patterns: for each PFGE cluster with five or fewer isolates, at least one isolate was selected for MLST; for larger clusters, at least one-fifth of the strains were selected for MLST. Within a cluster, strains were selected to cover the diversity of PFGE patterns obtained.
Comparison between typing methods.
The Simpson index of diversity (SID) was used to measure the diversity of the populations. Sets of partitions were compared by using the adjusted Rand and Wallace values as described previously using the online tool for quantitative assessment of classification agreement available at www.comparingpartitions.info (4).
RESULTS
Resistance patterns, serotypes, and clonal types among antibiotic-resistant pneumococci recovered in 2006 and 2007, high-PCV7-uptake era.
Of the 272 antibiotic-resistant pneumococcal isolates available for characterization, close to one-third were associated with PCV7 serotypes 14 (11.8%), 23F (8.1%), 19F (7.7%), and 6B (1.1%). The non-PCV7 serotypes associated with antibiotic resistance were, in decreasing order of prevalence, 19A (23.2%), 15A (12.1%), 6C (9.6%), 6A (7.4%), 11A (4.4%), 24F (2.6%), 33F (2.6%), 23B (1.1%), 3 (0.3%), 22F (0.3%), and 34 (0.3%); 7.4% of the isolates were NT.
The antibiotic resistance patterns and associated serotypes are summarized in Table 1. Multidrug resistance was detected in vaccine types 6B, 19F, and 23F and nonvaccine types 6A, 6C, 15A, 19A, 24F, and 33F and NT isolates. In particular, nonsusceptibility to penicillin associated with resistance to erythromycin, clindamycin, and tetracycline was the most common antibiotype, being detected in one-third of the drug-resistant isolates. High-level resistance to penicillin (MIC ≥ 2 μg/ml) was identified in isolates of serotypes 14 (n=20), 15A (n=1), and 19A (n=3) and an NT isolate (n=1).
Table 1.
Antimicrobial-resistant pneumococci isolated in 2006 and 2007, a high-PCV7-uptake eraa
| Serotype | No. of isolates resistant to: |
Most common antibiotype | |||
|---|---|---|---|---|---|
| At least one ATB tested | Pen | Ery | Multiple drugs | ||
| 3 | 1 | 0 | 1 | 0 | Ery, Cli |
| 6A | 20 | 1 | 6 | 4 | Ery, Cli, SXT |
| 6B | 3 | 2 | 3 | 2 | — |
| 6C | 26 | 20 | 23 | 19 | Pen (I), Ery, Cli, Tet |
| 14 | 32 | 29 | 3 | 0 | Pen, SXT |
| 11A | 12 | 0 | 0 | 0 | Tet |
| 15A | 33 | 33 | 33 | 33 | Pen (I), Ery, Cli, Tet |
| 19A | 63 | 49 | 54 | 54 | Pen (I), Ery, Cli, Tet |
| 19F | 21 | 7 | 21 | 20 | Ery, Cli, Tet |
| 22F | 1 | 1 | 0 | 0 | Pen (I) |
| 23B | 3 | 0 | 0 | 0 | SXT |
| 23F | 22 | 22 | 2 | 3 | Pen (I) |
| 24F | 7 | 7 | 7 | 7 | Pen (I), Ery, Cli, Tet |
| 33F | 7 | 0 | 7 | 2 | Ery, Cli |
| 34 | 1 | 0 | 0 | 0 | SXT |
| NT | 20 | 20 | 20 | 20 | Pen (I), Ery, Cli, Tet |
ATB, antibiotic; Pen, penicillin; Ery, erythromycin; Cli, clindamycin; Tet, tetracycline; SXT, sulfamethoxazole-trimethoprim; I, intermediate resistance to penicillin (MIC ≥ 0.1 μg/ml and ≤1 μg/ml); —, the three 6B isolates had three distinct antibiotypes.
For molecular analysis, 20 isolates exhibiting resistance to SXT only (of serotypes 6A [n=14], 23B [n=3], 19A [n=2], and 34 [n=1]) were excluded. Isolates with resistance to SXT only from previous study periods were not systematically characterized, and, thus, their inclusion would hinder comparisons between the three time periods. All remaining isolates (n=252) were characterized by PFGE, and close to one-third (n=80) were also characterized by MLST. The PFGE profiles of PMEN representative strains (clones 1 to 26) were determined and used in the clustering analysis.
Thirty-six PFGE clusters and 37 STs were identified (Table 2). An excellent correlation between PFGE and related STs (defined as MLST profiles that were identical or single-locus variants [SLVs] of each other) was obtained, as indicated by a Wallace coefficient of 1.000. In other words, for any two strains classified within the same PFGE cluster, the STs obtained were either identical or SLVs. These observations supported the suitability of our results based on PFGE/MLST analysis for comparison with MLST data from other studies. Of note, MLST could not discriminate between two PFGE clusters of serotype 6C (both with ST3396), two PFGE clusters of serotype 23F (ST338), and six PFGE clusters of NT isolates (ST344).
Table 2.
Clonality of antimicrobial-resistant pneumococci isolated in 2006 and 2007, a high-PCV7-uptake eraa
| Serotype-PFGE cluster | No. (%) of isolates | MLST | PMEN clone | No. of drug-resistant carriers who received PCV7b |
P value | |
|---|---|---|---|---|---|---|
| 0 doses (n=86) | 1-4 doses (n=152) | |||||
| PCV7 serotypes | ||||||
| 6B-1 | 1 (0.4) | 5217 | 1 | 0 | 0.183 | |
| 6B-2 | 1 (0.4) | 315 | Poland6BST315 | 0 | 1 | 0.451 |
| 6B-3 | 1 (0.4) | 469 | 1 | 0 | 0.183 | |
| 14-4 | 28 (11.1) | 557 (SLV), 5219 (DLV), 4585 (SLV) | Spain9VST156 | 8 | 16 | 0.763 |
| 14-5 | 3 (1.2) | 9 | England14ST9 | 3 | 0 | 0.020 |
| 14-ND | 1 (0.4) | 4584 | 0 | 1 | 0.451 | |
| 19F-6 | 17 (6.7) | 179 (SLV) | Portugal19FST177 | 8 | 9 | 0.330 |
| 19F-7 | 2 (0.8) | 88 | 1 | 0 | 0.183 | |
| 19F-8 | 2 (0.8) | 5218 | 1 | 1 | 0.682 | |
| 23F-9 | 15 (6.0) | 338 | Colombia23FST338 | 13 | 2 | <0.001 |
| 23F-10 | 6 (2.4) | 338 | Colombia23FST338 | 3 | 1 | 0.102 |
| 23F-11 | 1 (0.4) | 81 | Spain23FST81 | 1 | 0 | 0.183 |
| Non-PCV7 serotypes | ||||||
| 3-12 | 1 (0.4) | 180 | Netherlands3ST180 | 1 | 0 | 0.183 |
| 6A-13 | 5 (2.0) | 490 | 2 | 3 | 0.856 | |
| 6A-14 | 1 (0.4) | 395 | 0 | 1 | 0.451 | |
| 6C-15 | 3 (1.2) | 2689 | 1 | 1 | 0.682 | |
| 6C-16 | 2 (0.8) | 2185 | 1 | 1 | 0.682 | |
| 6C-17 | 11 (4.4) | 3396 | 6 | 5 | 0.193 | |
| 6C-18 | 6 (2.4) | 3396 | 1 | 5 | 0.315 | |
| 6C-19 | 4 (1.6) | 3673 | 2 | 2 | 0.560 | |
| 11A-20 | 12 (4.8) | 4582 | 5 | 6 | 0.510 | |
| 15A-21 | 33 (13.1) | 2105 (SLV), 63 | Sweden15AST63 | 7 | 24 | 0.092 |
| 19A-22 | 24 (9.5) | 276 (SLV), 4267 (SLV) | Denmark14ST230 | 5 | 19 | 0.099 |
| 19A-23 | 11 (4.4) | 416 (DLV) | Netherlands15BST199 | 3 | 8 | 0.531 |
| 19A-24 | 7 (2.8) | 4302, 5221 | 2 | 5 | 0.672 | |
| 19A-21 | 19 (7.5) | 63 | Sweden15AST63 | 2 | 15 | 0.029 |
| 22F-25 | 1 (0.4) | 433 | 1 | 0 | 0.183 | |
| 24F-26 | 7 (2.8) | 230, 1708 (SLV) | Denmark14ST230 | 0 | 7 | 0.043 |
| 33F-27 | 7 (2.8) | 717 | 0 | 7 | 0.043 | |
| NT-28 | 9 (3.6) | 344, 4586 (SLV) | NorwayNTST344 | 3 | 6 | 0.858 |
| NT-29 | 2 (0.8) | 344 | NorwayNTST344 | 1 | 1 | 0.682 |
| NT-30 | 1 (0.4) | 344 | NorwayNTST344 | 0 | 1 | 0.451 |
| NT-31 | 1 (0.4) | 344 | NorwayNTST344 | 0 | 1 | 0.451 |
| NT-32 | 1 (0.4) | 4583 | 1 | 0 | 0.183 | |
| NT-33 | 2 (0.8) | 1156 | 2 | 0 | 0.059 | |
| NT-34 | 1 (0.4) | 5220 (SLV) | NorwayNTST344 | 0 | 0 | |
| NT-35 | 1 (0.4) | 344 | NorwayNTST344 | 0 | 1 | 0.451 |
| NT-36 | 2 (0.8) | 344 | NorwayNTST344 | 0 | 2 | 0.285 |
ND, not determined; DLV, double-locus variant of prototype strain of PMEN clone. Boldface data indicate statistically significant differences.
For 14 carriers, information on PCV7 was not available.
In 2006 and 2007, the four most frequent drug-resistant clones accounted for over half (52.4%) of the antibiotic-resistant isolates, and all were PMEN clones: Sweden15AST63 (expressing serotypes 15A [13.1%] and 19A [7.5%]), Denmark14ST230 (expressing serotypes 19A [9.5%] and 24F [2.8%]), Spain9VST156 (serotype 14 variant [11.1%]), and Colombia23FST338 (8.4%).
Overall, 93.7% of the antibiotic-resistant isolates either belonged to PMEN clones or had identical or closely related STs associated with international isolates previously described in the MLST database (Table 2).
Association of drug-resistant clones with use of PCV7 among carriers.
As detailed above, in 2006 and 2007, over two-thirds of the participants had been vaccinated with PCV7. We investigated whether there was an association between carriage of specific drug-resistant clones and vaccination status. We observed that clone Colombia23FST338 was significantly more prevalent among nonvaccinated children, while clones Sweden15AST63 (serotype 19A variant), Denmark14ST230 (serotype 24F variant), and 33F-ST717 were significantly more prevalent among vaccinated children (Table 2).
Evolution of characteristics of antimicrobial-resistant pneumococci.
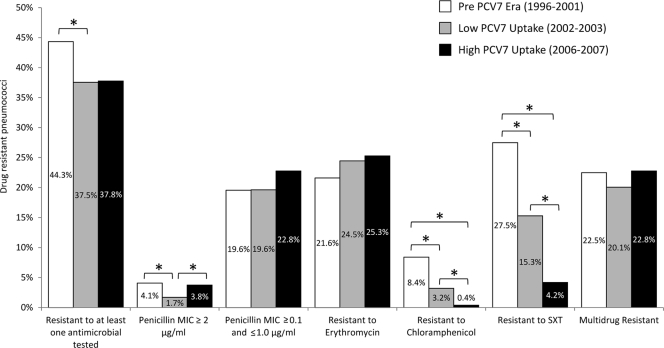
Figure 1 compares antimicrobial resistance rates over the three time periods. The proportion of antimicrobial-resistant pneumococci (defined as resistance to at least one antimicrobial tested) decreased significantly from the pre-PCV7 era to the low-PCV7-uptake period (44.3% and 37.5%, respectively; P < 0.001). This could be attributed to a significant decrease in the rates of pneumococcal resistance to SXT and chloramphenicol. Of note, the levels of high resistance rates to penicillin (MIC ≥ 2 μg/ml) were significantly lower in the low-PCV7-uptake era (1.7%) than in the preceding and following periods (4.1% and 3.8%, respectively). Rates of resistance to macrolides and low-level resistance to penicillin, as well as multidrug resistance, did not suffer significant changes overtime.
Fig. 1.
Antimicrobial resistance rates over time. *, significant changes (P < 0.05).
Clonal diversity, measured by the SID, was comparable in all three periods (Table 3). However, changes occurred in the population, as the clonal diversity among drug-resistant pneumococci of PCV7 serotypes was lower in the period of high-PCV7 uptake, while it remained stable among drug-resistant pneumococci of non-PCV7 serotypes. Regarding the major serotypes recovered in 2006 and 2007 (i.e., types 6A, 6C, 14, 15A, 19A, 19F, 23F, and NT), following introduction of PCV7, a significant decrease in clonal diversity was observed for serotypes 14 and 19F. This effect was not observed for serotype 23F, since the clonal diversity decreased in the low-PCV7-uptake period but increased again in the high-PCV7-uptake period. For non-PCV7 serotypes, the clonal diversity of serotype 6C and nontypeable strains increased. For serotypes 6A and 19A, no significant changes in clonal diversity were observed during the three periods. Still, in the case of serotype 6A, the reduced number of isolates obtained in the latter period may have hindered the observation of a reduction in diversity due to the large confidence interval obtained (Table 3).
Table 3.
Clonal diversity of antimicrobial-resistant pneumococci
| Serotype | Pre-PCV7 era (1996 to 2001, n=805) |
Low-PCV7 uptake (2002 and 2003, n=374) |
High-PCV7 uptake (2006 and 2007, n=252) |
|||
|---|---|---|---|---|---|---|
| No. of clones defined by PFGE | SID (95% CIb) | No. of clones defined by PFGE | SID (95% CI) | No. of clones defined by PFGE | SID (95% CI) | |
| Alla | 74 | 0.924 (0.917-0.931) | 48 | 0.936 (0.925-0.947) | 36 | 0.919 (0.910-0.936) |
| PCV7 | 49 | 0.894 (0.884-0.903) | 23 | 0.882 (0.860-0.904) | 11c | 0.782 (0.727-0.837) |
| Non-PCV7 | 34 | 0.916 (0.896-0.937) | 29 | 0.919 (0.898-0.940) | 25 | 0.873 (0.838-0.907) |
| 19A | 9 | 0.644 (0.493-0.794) | 6 | 0.641 (0.528-0.754) | 4 | 0.714 (0.663-0.765) |
| 15A | 2 | 0.173 (−0.027-0.374) | 2 | 0.500 (0.067-0.933) | 1 | 0.00 (0.00-0.00) |
| 6C | 2 | 0.200 (0.104-0.504) | 1 | 0.00 (0.00-0.00) | 5 | 0.754 (0.649-0.859) |
| 6A | 7 | 0.818 (0.727-0.909) | 5 | 0.857 (0.704-1.010) | 2 | 0.330 (−0.072-0.739) |
| 14 | 6 | 0.538 (0.495-0.581) | 7 | 0.616 (0.510-0.720) | 2 | 0.181 (0.094-0.352) |
| 19F | 15 | 0.743 (0.664-0.822) | 7 | 0.711 (0.634-0.789) | 3 | 0.343 (0.980-0.588) |
| 23F | 8 | 0.501 (0.440-0.562) | 3 | 0.093 (−0.006-0.193) | 3 | 0.480 (0.301-0.660) |
| Nontypeable | 3 | 0.351 (0.185-0.517) | 5 | 0.699 (0.583-0.814) | 9 | 0.795 (0.628-0.961) |
In the pre-PVC7 era there were nine PFGE clones that contained both PCV7 serotypes and non-PCV7 serotypes; in the low-PVC7-uptake era, there were four PFGE clones that contained both PCV7 serotypes and non-PCV7 serotypes.
95% CI, 95% confidence interval.
Boldface data indicate statistically significantly differences.
Replacement and evolution of antimicrobial-resistant clones.
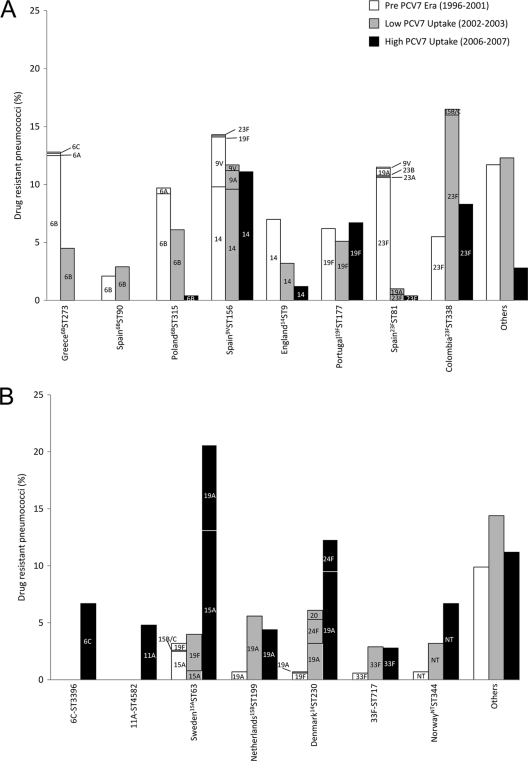
A clear serotype replacement of antimicrobial-resistant clones was observed over time: while in the pre-PCV7 era, 80.0% of the antimicrobial-resistant pneumococci were associated with PCV7 serotypes, in the low-PCV7-uptake period and high-PCV7-uptake period, the corresponding values were 66.9% and 31.0%, respectively (Fig. 2).
Fig. 2.
Prevalence of drug-resistant clones over time. (A) PCV7 serotype clones; (B) non-PCV7 serotype clones. The serotypes expressed by each clone are indicated.
Out of eight major PCV7-type drug-resistant PMEN clones in circulation before PCV7 introduction, five either were not detected in 2006 and 2007 or were present in low proportions (Fig. 2A). These were associated with serotypes 6B (Greece6BST273, Spain6BST90, Poland6BST315), 14 (England14ST9), and 23F (Spain23FST81). Although variants of Greece6BST273 associated with non-PCV7 types 6A and 6C had been detected in the pre-PCV7 era, they were not found in the following periods. Similarly, non-PCV7 type variants of Spain23FST81 detected before PCV7 introduction did not appear to thrive (Fig. 2A). Of particular interest, serotype 19A variants of this clone, previously detected, were not found in the period of high PCV7 uptake.
The three PCV7 clones that remained in circulation in the era of high-PCV7 uptake had different properties: clone Spain9VST156 (expressing serotype 14) has high resistance to penicillin and is typically susceptible to macrolides. Clone Portugal19FST177 is typically multiresistant, displaying resistance to macrolides, lincosamides, streptogramins, and tetracyclines, often accompanied by low-level resistance to penicillin. Clone Colombia23FST338 is typically associated with low-level resistance to penicillin. Apparently, PCV7 introduction did not significantly affect the prevalence of these clones. The reasons for such are not clear but do not appear to be associated with serotype or a specific antibiotype.
Five of the seven most frequent non-PCV7 serotype clones found in 2006 and 2007 were already in circulation in the previous periods (Fig. 2B). All increased in prevalence among drug-resistant isolates following PCV7 introduction. In particular, three clones, Netherlands15BST199 (serotype 19A variant), 33F-ST717, and NorwayNTST344, were always associated with the same serotypes over the three periods (19A, 33F, and NT, respectively). In contrast, for clones Sweden15AST63 and Denmark14ST230, the serotypes most commonly expressed changed over time (Fig. 2B). In particular, serotype 19A accounted for a significant proportion of isolates from both clones in the era of high PCV7 uptake. In addition, two new clones emerged in the high-PCV7-uptake period. Clone 6C-ST3396 was associated with serotype 6C and was multiresistant (resistant to macrolides, lincosamides, streptogramins, and tetracycline). Clone 11A-ST4582 was associated with serotype 11A and was resistant to tetracycline only (Fig. 2B).
Capsular switch.
Capsular variants of major DRPn clones have been observed in all three study periods (Fig. 2). Although there have been changes in the dominant serotype expressed by a clone, we found no evidence for the emergence of novel capsular variants following introduction of PCV7. Although not shown in Fig. 2B, serotype 19A variants of Sweden15AST63 were detected in 2001 to 2003 in another study conducted in Portugal (37).
DISCUSSION
In this study we described the variations observed in the population structure of drug-resistant colonizing pneumococci following widespread use of PCV7 in Portugal. The study was triggered by the observation that antimicrobial resistance rates remained essentially unchanged in the years following an increased use of PCV7 in Portugal, despite the dramatic decrease in the proportion of carriers of pneumococci expressing PCV7 types (30). This was, somehow, unexpected, as in the prevaccine era most antimicrobial resistance was associated with PCV7 serotypes. Furthermore, this observation contrasted with data from other countries, where introduction of PCV7 has been associated with a reduction not only in vaccine serotypes but also in antibiotic resistance prevalence (7, 34). Still, such declines have mostly been observed in countries that documented introduction of PCV7 with a concomitant reduction of antibiotic consumption. On the other hand, maintenance of antibiotic resistance levels among isolates carried in the PCV7 era has also been noted in studies from the United States (16, 26).
The mechanisms leading to the maintenance of rates of resistance to antimicrobials despite serotype replacement could be expansion of existing clones, capsular switch, introduction of new clones, and de novo acquisition of resistance. To determine which mechanisms were in place and the relative extent of each, molecular typing of pre- and post-PCV7 drug-resistant isolates was required.
We observed that expansion of drug-resistant clones such as Sweden15AST63 and Denmark14ST230 (mainly expressing non-PCV7 serotypes 15A, 19A, and 24F) was an important cause for the maintenance of antimicrobial resistance levels. These clones were already in circulation in Portugal in the years preceding introduction of PCV7 and in 2006 and 2007 accounted for 32.9% of all drug-resistant pneumococci. Both clones have been found elsewhere in recent years: in 2007, clone Sweden15AST63 (serotype 15A and 19A variants) was the fourth major clone found among young carriers from Massachusetts (15) and was the major clone (serotype 15A variant) recovered from disease in the United States during 2007 among penicillin-nonsusceptible isolates of non-PCV7 serotypes other than 19A and 6A (13). Clone Denmark14ST230 (expressing serotype 19A) has been detected in several countries, such as France, Spain, and Israel (7, 8, 34). In Portugal, clone Denmark14ST230 is expanding not only in the rate of colonization but also as a major cause of pneumococcal infections (1), although expansion of clones Netherlands15BST199 (serotype 19A variant), NorwayNTST344, and 33F-ST717 was also observed to a lesser extent.
Even though capsular variants of PMEN drug-resistant clones became increasingly abundant in the PCV7 era, we found no evidence that PCV7 could have triggered or enhanced capsular switch events. Indeed, all capsular variants detected in 2006 and 2007 had been detected in studies preceding widespread use of PCV7. Of note, 19A variants currently thriving have genetic backgrounds originally associated not only with PCV7 types (as in the case of Denmark14ST230) but also with non-PCV7 types (as in the case of Netherlands15BST199 and Sweden15AST63). Our interpretations regarding the time of capsular switch events were possible only due to the large collection of drug-resistant isolates from the pre-PCV7 era (1996 to 2001) that was available for comparison. On the other hand, one could argue that lack of detection of truly novel capsular switch events in the PCV7 era might be due to limitations in the number of samples analyzed in that period. Still, if that would have been the case, those events should not have achieved a large magnitude, as they remained undetected in two consecutive years of study, despite the large serotype and clonal diversity identified. Overall, our findings contrast with those of other studies, which suggested that serotype escape variants might have emerged in the United States following introduction of PCV7 (3, 15).
Introduction of new clones appeared to contribute to a small fraction (11.6%) of the drug-resistant population. In particular, a multidrug-resistant clone of serotype 6C (ST3396) was first detected in 2006 and 2007. A previous study on the epidemiology of serotype 6C colonizing isolates in Portugal (22) did not identify this clone or related susceptible counterparts in earlier collections, suggesting a novel introduction in the population. For the novel detection of a clone of serotype 11A (ST4582), resistant to tetracycline only, whether its presence results from a novel introduction in the population or de novo acquisition of resistance is not clear, as the epidemiology of serotype 11A isolates in our population has not been systematically studied. In any case, de novo acquisition of resistance appears to have contributed very little to the maintenance of resistance levels in the era of PCV7 use.
Of note, close to one-third (31.0%) of the drug-resistant isolates were still of PCV7 serotypes, in contrast to 1.5% among drug-susceptible isolates from the same time period (30; data not shown). This discrepancy is suggestive of a selective pressure that makes some drug-resistant PCV7 clonal types more refractory to extinction than their susceptible counterparts. Even if we have no definitive evidence for the nature of such pressure, which most likely is multifactorial, antibiotic use may play an important role. Although antimicrobial use has been declining in the populations that we have been monitoring, it is still high: in 2006 and 2007, 18.2% of the participants had received antibiotics in the month preceding the sampling and 14.9% had received three or more courses of antibiotics in the previous 6 months (30; this study). Other studies have shown that outpatient antimicrobial consumption in Portugal has been declining since 2002, even though it is still high (22.61/1,000 inhabitants/day in 2008) (http://app.esac.ua.ac.be/public), and a positive correlation between antimicrobial consumption and resistance has been described by several authors (10, 14, 28). However, this does not explain why some PCV7-type clones thrived while others of the same serotype did not (for instance, Colombia23FST338 versus Spain23FST81). We were also unable to identify a common resistant phenotype among those persistent clones that could explain their maintenance. Clearly, other factors beyond the capsule and the resistance determinants which may be concealed in the wider genome background of such clones may play a role. Such factors may be related to increased fitness and transmission and enhanced capacity to evade the host immune system or escape vaccine pressure (31).
Regarding the diversity of clones in circulation, we observed that it decreased among drug-resistant pneumococci of PCV7 serotypes in the period of high-PCV7 uptake and remained stable among drug-resistant pneumococci of non-PCV7 serotypes. Our observations are in general agreement with those described by Lipsitch et al (18) that suggested that PCV7 acts as a serotype filter, in the sense that the introduction of PCV7 did not change the diversity of the non-PCV7 serotype population.
In conclusion, with this study we were able to describe the mechanisms leading to maintenance of antimicrobial resistance among pneumococcal colonizing isolates in the PCV7 era. We observed that the group of clones that have accounted for antimicrobial resistance since 1996 is essentially the same as the one identified in the PCV7 era. The relative proportions of such clones have, however, evolved substantially over time. Widespread use of PCV7 led to an expansion of PMEN clones which express non-PCV7 capsular variants of the original strains. These variants were already in circulation in the pre-PCV7 era, although they have now become increasingly abundant. Emergence of novel clones and de novo acquisition of resistance seems to contribute little to the observed scenario. In the years to come, following the introduction of novel conjugate vaccines with expanded coverage, continuous surveillance of the pneumococcal population is essential to evaluate serotype and clonal shifts that can shed light on the vaccine's effect, guide future vaccine development, and increase our understanding of the mechanisms of evolution of the pneumococcal population.
ACKNOWLEDGMENTS
We are grateful to the directors and staff of the day care centers and the parents and children who collaborated in the study. We thank I. Crisóstomo, N. Frazão, and A. Tavares for participating in studies that led to the isolation of strains in 2006 and 2007 and acknowledge the excellent skills of the pediatric nurses, A. Gonçalves and D. Trindade, who collected the nasopharyngeal samples.
This work was supported by projects PDTC/SAU-ESA/65048/2006 and PDTC/BIA-MIC/64010/2006 from the Fundação para a Ciência e Tecnologia (FCT), Portugal, and projects PREVIS (LSHM-CT-2003-503413) and PNEUMOPATH (HEALTH-F3-2009-222983) from the European Commission. A.S.S. and S.N. were supported by grants from FCT (SFRH/BD/27325/2006 and SFRH/BD/40706/2007, respectively).
Footnotes
Published ahead of print on 1 June 2011.
REFERENCES
- 1. Aguiar S. I., et al. 2010. Denmark14-230 clone as an increasing cause of pneumococcal infection in Portugal within a background of diverse serotype 19A lineages. J. Clin. Microbiol. 48: 101–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brito D. A., Ramirez M., de Lencastre H. 2003. Serotyping Streptococcus pneumoniae by multiplex PCR. J. Clin. Microbiol. 41: 2378–2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brueggemann A. B., Pai R., Crook D. W., Beall B. 2007. Vaccine escape recombinants emerge after pneumococcal vaccination in the United States. PLoS Pathog. 3: e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Carriço J. A., et al. 2006. Illustration of a common framework for relating multiple typing methods by application to macrolide-resistant Streptococcus pyogenes. J. Clin. Microbiol. 44: 2524–2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. CDC. 2005. Direct and indirect effects of routine vaccination of children with 7-valent pneumococcal conjugate vaccine on incidence of invasive pneumococcal disease—United States, 1998-2003. MMWR Morb. Mortal. Wkly. Rep. 54: 893–897 [PubMed] [Google Scholar]
- 6. CLSI. 2009. Performance standards for antimicrobial susceptibility testing; 19th informational supplement M100-S19. CLSI, Wayne, PA [Google Scholar]
- 7. Cohen R., et al. 2010. Dynamic of pneumococcal nasopharyngeal carriage in children with acute otitis media following PCV7 introduction in France. Vaccine 28: 6114–6121 [DOI] [PubMed] [Google Scholar]
- 8. Dagan R., Givon-Lavi N., Leibovitz E., Greenberg D., Porat N. 2009. Introduction and proliferation of multidrug-resistant Streptococcus pneumoniae serotype 19A clones that cause acute otitis media in an unvaccinated population. J. Infect. Dis. 199: 776–785 [DOI] [PubMed] [Google Scholar]
- 9. Enright M. C., Spratt B. G. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144: 3049–3060 [DOI] [PubMed] [Google Scholar]
- 10. Ferech M., et al. 2006. European Surveillance of Antimicrobial Consumption (ESAC): outpatient antibiotic use in Europe. J. Antimicrob. Chemother. 58: 401–407 [DOI] [PubMed] [Google Scholar]
- 11. Foster D., et al. 2011. Reduction in invasive pneumococcal disease following implementation of the conjugate vaccine in the Oxfordshire region, England. J. Med. Microbiol. 60: 91–97 [DOI] [PubMed] [Google Scholar]
- 12. Frazão N., et al. 2005. Effect of the seven-valent conjugate pneumococcal vaccine on carriage and drug resistance of Streptococcus pneumoniae in healthy children attending day-care centers in Lisbon. Pediatr. Infect. Dis. J. 24: 243–252 [DOI] [PubMed] [Google Scholar]
- 13. Gertz R. E., Jr, et al. 2010. Increased penicillin nonsusceptibility of nonvaccine-serotype invasive pneumococci other than serotypes 19A and 6A in post-7-valent conjugate vaccine era. J. Infect. Dis. 201: 770–775 [DOI] [PubMed] [Google Scholar]
- 14. Goossens H., Ferech M., Vander Stichele R., Elseviers M. 2005. Outpatient antibiotic use in Europe and association with resistance: a cross-national database study. Lancet 365: 579–587 [DOI] [PubMed] [Google Scholar]
- 15. Hanage W. P., et al. 2011. Carried pneumococci in Massachusetts children: the contribution of clonal expansion and serotype switching. Pediatr. Infect. Dis. J. 30: 302–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huang S. S., et al. 2005. Post-PCV7 changes in colonizing pneumococcal serotypes in 16 Massachusetts communities, 2001 and 2004. Pediatrics 116: e408–e413 [DOI] [PubMed] [Google Scholar]
- 17. Lexau C. A., et al. 2005. Changing epidemiology of invasive pneumococcal disease among older adults in the era of pediatric pneumococcal conjugate vaccine. JAMA 294: 2043–2051 [DOI] [PubMed] [Google Scholar]
- 18. Lipsitch M., et al. 2007. Strain characteristics of Streptococcus pneumoniae carriage and invasive disease isolates during a cluster-randomized clinical trial of the 7-valent pneumococcal conjugate vaccine. J. Infect. Dis. 196: 1221–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mato R., et al. 2005. Natural history of drug-resistant clones of Streptococcus pneumoniae colonizing healthy children in Portugal. Microb. Drug Resist. 11: 309–322 [DOI] [PubMed] [Google Scholar]
- 20. Munoz-Almagro C., et al. 2009. Emergence of invasive pneumococcal disease caused by multidrug-resistant serotype 19A among children in Barcelona. J. Infect. 59: 75–82 [DOI] [PubMed] [Google Scholar]
- 21. Nunes S., et al. 2005. Trends in drug resistance, serotypes, and molecular types of Streptococcus pneumoniae colonizing preschool-age children attending day care centers in Lisbon, Portugal: a summary of 4 years of annual surveillance. J. Clin. Microbiol. 43: 1285–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nunes S., Valente C., Sá-Leão R., de Lencastre H. 2009. Temporal trends and molecular epidemiology of recently described serotype 6C of Streptococcus pneumoniae. J. Clin. Microbiol. 47: 472–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. O'Brien K. L., et al. 2009. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet 374: 893–902 [DOI] [PubMed] [Google Scholar]
- 24. Pai R., Gertz R. E., Beall B. 2006. Sequential multiplex PCR approach for determining capsular serotypes of Streptococcus pneumoniae isolates. J. Clin. Microbiol. 44: 124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pai R., et al. 2005. Postvaccine genetic structure of Streptococcus pneumoniae serotype 19A from children in the United States. J. Infect. Dis. 192: 1988–1995 [DOI] [PubMed] [Google Scholar]
- 26. Park S. Y., et al. 2008. Impact of conjugate vaccine on transmission of antimicrobial-resistant Streptococcus pneumoniae among Alaskan children. Pediatr. Infect. Dis. J. 27: 335–340 [DOI] [PubMed] [Google Scholar]
- 27. Richter S. S., et al. 2009. Changing epidemiology of antimicrobial-resistant Streptococcus pneumoniae in the United States, 2004-2005. Clin. Infect. Dis. 48: e23–e33 [DOI] [PubMed] [Google Scholar]
- 28. Riedel S., et al. 2007. Antimicrobial use in Europe and antimicrobial resistance in Streptococcus pneumoniae. Eur. J. Clin. Microbiol. Infect. Dis. 26: 485–490 [DOI] [PubMed] [Google Scholar]
- 29. Sá-Leão R., et al. 2008. High rates of transmission of and colonization by Streptococcus pneumoniae and Haemophilus influenzae within a day care center revealed in a longitudinal study. J. Clin. Microbiol. 46: 225–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sá-Leão R., et al. 2009. Changes in pneumococcal serotypes and antibiotypes carried by vaccinated and unvaccinated day-care centre attendees in Portugal, a country with widespread use of the seven-valent pneumococcal conjugate vaccine. Clin. Microbiol. Infect. 15: 1002–1007 [DOI] [PubMed] [Google Scholar]
- 31. Sá-Leão R., et al. 2011. Analysis of invasiveness of pneumococcal serotypes and clones circulating in Portugal before widespread use of conjugate vaccines reveals heterogeneous behavior of clones expressing the same serotype. J. Clin. Microbiol. 49: 1369–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sá-Leão R., et al. 2000. Carriage of internationally spread clones of Streptococcus pneumoniae with unusual drug resistance patterns in children attending day care centers in Lisbon, Portugal. J. Infect. Dis. 182: 1153–1160 [DOI] [PubMed] [Google Scholar]
- 33. Sá-Leão R., et al. 2000. Genetic diversity and clonal patterns among antibiotic-susceptible and -resistant Streptococcus pneumoniae colonizing children: day care centers as autonomous epidemiological units. J. Clin. Microbiol. 38: 4137–4144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sanchez-Tatay D., et al. 2008. Antibiotic susceptibility and molecular epidemiology of nasopharyngeal pneumococci from Spanish children. Clin. Microbiol. Infect. 14: 797–801 [DOI] [PubMed] [Google Scholar]
- 35. Simonsen L., et al. 2011. Impact of pneumococcal conjugate vaccination of infants on pneumonia and influenza hospitalization and mortality in all age groups in the United States. mBio 2: e00309–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sorensen U. B. 1993. Typing of pneumococci by using 12 pooled antisera. J. Clin. Microbiol. 31: 2097–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sousa N. G., et al. 2005. Properties of novel international drug-resistant pneumococcal clones identified in day-care centers of Lisbon, Portugal. J. Clin. Microbiol. 43: 4696–4703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vestrheim D. F., Hoiby E. A., Aaberge I. S., Caugant D. A. 2010. Impact of a pneumococcal conjugate vaccination program on carriage among children in Norway. Clin. Vaccine Immunol. 17: 325–334 [DOI] [PMC free article] [PubMed] [Google Scholar]