Abstract
Methicillin-susceptible Staphylococcus aureus (MSSA) isolates lacking mecA yet testing positive on the Xpert MRSA assay were recovered from culture for 7.7% of 248 Xpert-positive nasal samples. These “false-positive” Xpert results may be attributed to staphylococcal cassette chromosome (SCC) elements without the mecA gene. Pulsed-field gel electrophoresis (PFGE) analysis revealed a diverse population of MSSA strains.
TEXT
Molecular methods for the detection of methicillin-resistant Staphylococcus aureus (MRSA) carriage have been introduced as a rapid alternative to culture methods that require 24 to 72 h. The Xpert MRSA assay for the GeneXpert real-time PCR instrument (Cepheid, Sunnyvale, CA) allows direct detection of MRSA from nasal swabs within 1 h. To avoid detection of coagulase-negative staphylococci carrying mecA, the Xpert MRSA targets the staphylococcal cassette chromosome mec (SCCmec)-orfX junction created by incorporation of the genetic element carrying mecA into the S. aureus chromosome (15). The BD GeneOhm MRSA assay (formerly designated the IDI-MRSA assay; BD Diagnostics, Sparks, MD) was released earlier and targets the same SCCmec-orfX junction (11). Because these assays do not specifically target the mecA gene, strains that do not contain a functional mecA gene may be detected (2, 5, 13). There is limited information regarding the prevalence of these empty-cassette variants. The objective of this study was to determine the prevalence and the genetic relatedness of methicillin-susceptible S. aureus (MSSA) isolates detected by the Xpert MRSA assay in a Midwest academic medical center during the first year of use.
From January through December 2009, nasal specimens received in the clinical microbiology laboratory with a request for MRSA PCR were tested on the Xpert MRSA assay if collected using the Copan dual swab device (Copan Diagnostics, Murrieta, CA). Because an empty-cassette variant was detected during validation of the Xpert MRSA assay, the following protocol was implemented in the clinical laboratory to allow monitoring for these false positives. Institutional Review Board approval for publication was obtained when the findings of the internal laboratory investigation of Xpert MRSA assay performance were perceived to be of potential interest to other clinical microbiology laboratories.
(This work was presented in part at the 110th American Society for Microbiology General Meeting, 26 May 2010, San Diego, CA [abstract C-2617].)
Dual swab specimens were collected. One swab was used for the Xpert MRSA nasal assay with testing performed and interpreted according to the manufacturer's instructions. If the result was “MRSA positive,” the second swab was cultured by broth enrichment using BBL Trypticase soy broth with 6.5% NaCl (BD Diagnostics, Sparks, MD). After overnight incubation, the broth was subcultured to 2 plates, one containing BBL CHROMagar MRSA and one containing Trypticase soy agar with 5% sheep blood agar (SBA) (BD Diagnostics, Sparks, MD). The CHROMagar plate was read after 20 to 28 h of incubation. Any mauve colonies were confirmed as MRSA with a Staphaurex Plus latex agglutination test (Remel, Lenexa, KS). If mauve colonies were not observed at 24 (± 4) h, the plates were reincubated an additional 24 (± 4) h. If MRSA was not recovered from CHROMagar, the SBA plate was examined for S. aureus. Any S. aureus recovered from the SBA was tested for oxacillin and cefoxitin susceptibility using a BD Phoenix PMIC 102 panel (BD Diagnostics, Sparks, MD).
If MSSA (susceptible to cefoxitin and oxacillin) was recovered in culture, these colonies were tested directly on the Xpert MRSA assay. Instructions recommended by the manufacturer for testing QC organisms were followed when the MSSA isolates were tested. Colonies were suspended in sterile saline and adjusted to a 0.5 McFarland standard (∼108 CFU/ml). A 1:100 dilution was made by transferring 0.1 ml into 9.9 ml of sterile saline (∼106 CFU/ml). Each MSSA isolate confirmed as positive on the Xpert MRSA assay was further evaluated as described below.
The genetic relatedness of MSSA isolates detected as MRSA by the Xpert assay was determined by pulsed-field gel electrophoresis (PFGE) according to previously published procedures after digestion with SmaI (Sigma-Aldrich, St. Louis, MO) (12). After electrophoresis, the gels were stained with ethidium bromide. The PFGE patterns were analyzed using Bionumerics software (Applied Maths, Kortrijk, Belgium). A dendrogram was constructed using the unweighted-pair group method with arithmetic averages and the DICE coefficient (1.0% optimization and 1.0% position tolerance). Isolates with banding patterns differing by 3 bands or fewer were considered closely related and assigned to the same PFGE type designated by a capital letter. Within a PFGE type containing multiple isolates, those with indistinguishable banding patterns were assigned to the same subtype designated by a number following the letter of the PFGE type (12, 14). The PFGE patterns were compared to the profiles of common USA type MRSA strains kindly provided by Brandi Limbago at the Centers for Disease Control and Prevention (10).
Confirmation of MSSA isolates with positive Xpert MRSA results as S. aureus was performed using a multiplex assay detecting a 16S rRNA staphylococcus genus-specific target, the mecA and the nuc genes (19). Single-target PCR was performed to confirm strains as mecA negative using two different primer pairs, mecA1/mecA2 and mecA 147-F/mecA 112-R (5′ATCAGTATTTCACCTTGTCCG-3′) (19, 21). SCCmec typing was performed using an updated multiplex-PCR assay that identifies SCCmec types I to V, subdivides type IV strains (IVa to IVF), and detects the kdp gene and mercury element (17). Comprehensive SCCmec typing (I to VIII) was also conducted by targeting all currently described ccr and mec gene complexes, including ccr1, ccr2, ccr3, ccr4, ccrC, A mec, B mec, C1 mec, and C2 mec, with the specific primers as described previously (7-9, 18, 20). Each MSSA isolate was also tested using the new GeneXpert MRSA/SA nasal assay (provided by Cepheid, Sunnyvale, CA).
Over a 1-year period, 2,127 nasal samples were tested using the Xpert MRSA assay. Two hundred fifty one of the 2,127 samples (11.8%) tested positive by the Xpert MRSA assay. MRSA was recovered in culture from 166 of the 251 Xpert-positive samples (66.1%). MSSA isolates that tested positive by the Xpert assay were isolated from 23 of the 251 Xpert-positive samples. Xpert-positive results from specimens that were culture negative likely contained nonviable DNA, reflecting the higher sensitivity of molecular detection methods. Two MSSA isolates that represented duplicates from the same patient and a third MSSA isolate not available for additional testing were excluded from the subsequent analysis. The mecA gene was detected in 1 of the 20 MSSA isolates from unique patients. This MSSA isolate contained ccr2 and B mec gene complexes, making it SCCmec type IV. The updated SCCmec typing assay determined that this isolate was type IVc. The remaining 19 MSSA isolates (7.7%) were considered empty-cassette variants causing positive Xpert MRSA results. A ccr gene complex was detected in only 1 of the 19 isolates (ccr2); this isolate also contained the kdp gene.
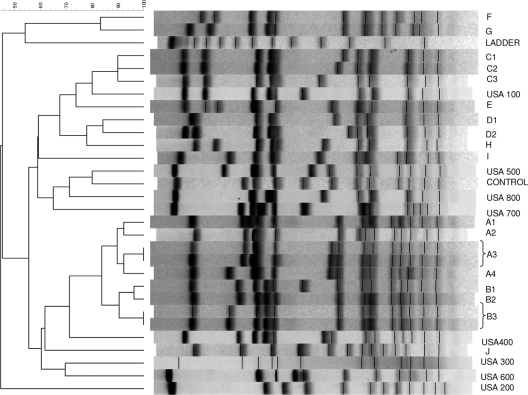
The PFGE analyses of the 20 MSSA isolates (including one with mecA gene) revealed a diverse population (10 PFGE types and 18 subtypes) (Fig. 1). The mecA-containing MSSA isolate was not closely related to the other isolates (PFGE type H). The isolate with the kdp-and-ccr2 gene complex (PFGE C3) was closely related to 2 isolates (PFGE C1 and C2).
Fig. 1.
PFGE profiles of 20 MSSA isolates detected by the Xpert MRSA assay.
Updated and comprehensive SCCmec typing methods for detecting types I to VIII (6) were employed, yet the ccr gene was detected in only 1 of the 19 mecA-negative isolates. The other 18 isolates may contain only the SCCmec-orfX right extremity junction component or likely the novel undescribed SCC elements that cannot be detected by currently available primers. Methods that are beyond the scope of this project, such as sequencing of the entire SCC region or PCR for identifying regions found in the right extremity region of SCCmec (3), would be needed to further understand the reason for reactivity with the Xpert assay.
When tested using the new GeneXpert MRSA/SA nasal assay (Xpert SA nasal complete), 19 of the 20 MSSA isolates were found to be negative for MRSA and positive for S. aureus. The MSSA isolate harboring the mecA gene tested positive for MRSA and S. aureus. According to the package insert, primers for the MRSA/SA nasal assay target the spa gene, the mecA gene, and the SCCmec-attB junction. The increased specificity of the MRSA/SA nasal assay in regard to empty-cassette variants is reassuring; however, there is the potential for false-positive “MRSA” results due to the presence of MSSA and mecA-positive coagulase-negative staphylococci in the same specimen.
The few studies attempting to determine the prevalence of MSSA strains containing SCC elements that can cause positive MRSA PCR results have produced variable results. A French study testing multidrug-resistant MSSA isolates (n = 247, resistant to at least 2 non-β-lactam antibiotic classes) from diverse genetic backgrounds found that 68% were detected by the IDI-MRSA assay due to the presence of SCCmec elements lacking mecA (3). This contrasted sharply with the 4.6% prevalence of MSSA empty-cassette variants reported by a 2004 study that did not select for multidrug resistance among the 569 MSSA isolates tested from throughout the world (5).
A limited number of studies evaluating the performance of PCR assays for the detection of MRSA colonization have included techniques for detecting positive results due to MSSA empty-cassette variants. In Kentucky, only 2 of 64 positive BD GeneOhm MRSA assay results (3.1%) from nasal specimens showed evidence of being positive due to MSSA empty-cassette variants (13). A Canadian study reported the recovery of 38 MSSA isolates that tested positive with the IDI-MRSA assay (repeat testing on pure culture) for 25% of 150 patients with initially positive PCR results from nasal/rectal pooled specimens after overnight incubation in a selective broth (1). The 38 MSSA isolates represented 17 PFGE genotypes, consistent with the diversity found in the current study. Nearly one-third of the 38 MSSA isolates in the Canadian study (1) were variants of common MRSA clones (USA500 [11 isolates] and USA100 [2 isolates]), while our study found only 3 MSSA isolates (16%) that were possibly related to a USA clone (USA100). The dendrogram shows similarity to USA100 of PFGE C1 to C3 just at the 80% cutoff, but there was a 5-band difference in pattern. The isolate appearing most similar to the banding pattern of USA100 (Fig. 1, PFGE C3) contained the ccr2 gene complex that is found in the majority of USA100 strains (SCCmec type II). A recent study (16) reported 24 MSSA isolates obtained from geographically diverse areas (6 different states and 4 Canadian hospitals) with positive BD GeneOhm assay results that were closely related to USA100 (n = 7) or USA400 (n = 17), markedly different from the genetic diversity found in our single-center study.
An evaluation of the BD GeneOhm MRSA assay performed on Baltimore jail inmates reported 12.1% of 123 positive PCR results that grew only MSSA isolates that tested positive when run directly on the assay (4). A study in the Chicago area reported that 7.4% of cultures for 215 BD GeneOhm MRSA PCR positive nasal specimens yielded only MSSA isolates with probable remnants of SCCmec (11). Although the Xpert MRSA assay is described as a second-generation test, our study found nearly the same rate of false-positive results (7.7%) caused by mecA dropout strains as that reported for the BD GeneOhm assay study conducted in Chicago (11). The only U.S. multicenter evaluation of the Xpert MRSA assay that we are aware of did not screen for empty-cassette variants (15).
It is reassuring that the prevalence of MSSA isolates with remnants of the SCCmec cassette detected by the Xpert MRSA assay in the current study was below 10%. Because the prevalence and genetic diversity of MSSA causing false-positive results appears highly variable, institutions using an MRSA PCR assay without additional primers specific for mecA should consider screening for these empty-cassette strains. Ongoing studies comparing culture to molecular test results are needed to detect changes in the MSSA and MRSA population that may affect the accuracy of molecular assays.
Acknowledgments
Twenty-five GeneXpert MRSA/SA nasal assay cartridges were provided by Cepheid.
D.J.D. has received research funding from Merck, Pfizer, Schering-Plough, Astellas, Forest Laboratories, and bioMérieux. S.S.R. has received research funding from Abbott Laboratories, BD Diagnostics, Forest Laboratories, and Schering-Plough. K.Z. has received financial support from Alberta Health Services and the Alberta Heritage Foundation for Medical Research. All other authors have no conflicts of interest to disclose.
Footnotes
Published ahead of print on 15 June 2011.
REFERENCES
- 1. Desjardins M., Guibord C., Lalonde B., Toye B., Ramotar K. 2006. Evaluation of the IDI-MRSA assay for the detection of methicillin-resistant Staphylococcus aureus from nasal and rectal specimens pooled in a selective broth. J. Clin. Microbiol. 44:1219–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Donnio P.-Y., et al. 2005. Partial excision of the chromosomal cassette containing methicillin-resistance determinant results in methicillin-susceptible Staphylococcus aureus. J. Clin. Microbiol. 43:4191–4193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Donnio P.-Y., et al. 2007. Molecular and epidemiological evidence for spread of multiresistant methicillin-susceptible Staphylococcus aureus strains in hospitals. Antimicrob. Agents Chemother. 51:4342–4350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Farley J. E., et al. 2008. Comparison of the BD GeneOhm assay to culture by use of BBL CHROMagar MRSA for detection of MRSA in nasal surveillance cultures form an at-risk community population. J. Clin. Microbiol. 46:743–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Huletsky A., et al. 2004. New real-time PCR assay for rapid detection of methicillin-resistant Staphylococcus aureus directly from specimens containing a mixture of staphylococci. J. Clin. Microbiol. 42:1875–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements (IWG-SCC) 2009. Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob. Agents Chemother. 53:4961–4967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ito T., et al. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1323–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kondo Y., et al. 2007. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob. Agents Chemother. 51:264–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McClure J., Conly J. M., Elsayed S., Zhang K. 2010. Multiplex PCR assay to facilitate identification of the recently described staphylococcal cassette chromosome mec type VIII. Mol. Cell. Probes 24:229–232 [DOI] [PubMed] [Google Scholar]
- 10. McDougal L. K., et al. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 41:5113–5120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Paule S. M., et al. 2007. Performance of the BD GeneOhm methicillin-resistant Staphylococcus aureus test before and during high-volume clinical use. J. Clin. Microbiol. 45:2993–2998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pfaller M. A., Caliendo A. M., Versalovic J. 2010. Chromosomal restriction fragment analysis by pulsed-field gel electrophoresis: application to molecular epidemiology, p. 12.4.5.1–124.5.7. In Garcia L. S. (ed.), Clinical microbiology procedures handbook, 3rd ed. ASM Press, Washington, DC [Google Scholar]
- 13. Snyder J. W., Munier G. K., Johnson C. L. 2010. Comparison of the BD GeneOhm methicillin-resistant Staphylococcus aureus (MRSA) PCR assay to culture by use of BBL CHROMagar MRSA for detection of MRSA in nasal surveillance cultures from intensive care unit patients. J. Clin. Microbiol. 48:1305–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tenover F. C., et al. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wolk D. M., et al. 2009. Multicenter evaluation of the Cepheid Xpert methicillin-resistant Staphylococcus aureus (MRSA) test as a rapid screening method for detection of MRSA in nares. J. Clin. Microbiol. 47:758–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wong H., Louie L., Lo R. Y. C., Simor A. E. 2010. Characterization of Staphylococcus aureus with a partial or complete absence of staphylococcal cassette chromosome elements. J. Clin. Microbiol. 48:3525–3531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang K., McClure J., Elsayed S., Conly J. 2010. Enhanced multiplex PCR assay for typing of staphylococcal cassette chromosome mec (SCCmec) types I to V in methicillin-resistant Staphylococcus aureus (MRSA), abstr. C-2611, p. 186 Abstr. 110th Gen. Meet. Am. Soc. Microbiol. American Society for Microbiology, San Diego, CA. [Google Scholar]
- 18. Zhang K., McClure J., Elsayed S., Louie T., Conly J. M. 2005. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 43:5026–5033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang K., McClure J., Elsayed S., Louie T., Conly J. M. 2008. Novel PCR assay for simultaneous identification of community-associated methicillin-resistant Staphylococcus aureus strains USA300 and USA400 and detection of mecA and Panton-Valentine leukocidin genes, with discrimination of Staphylococcus aureus from coagulase-negative staphylococci. J. Clin. Microbiol. 46:1118–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang K., McClure J., Elsayed S., Louie T., Conly J. M. 2009. Novel staphylococcal cassette chromosome mec type, tentatively designated type VIII, harboring class A mec and type 4 ccr gene complexes in a Canadian epidemic strain of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 53:531–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang K., et al. 2004. New quadriplex PCR assay for detection of methicillin and mupirocin resistance and simultaneous discrimination of Staphylococcus aureus from coagulase-negative staphylococci. J. Clin. Microbiol. 42:4947–4955 [DOI] [PMC free article] [PubMed] [Google Scholar]