Abstract
We evaluated Bruker Biotyper (version 2.0) matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry (MS) for the identification of 305 clinical isolates of staphylococci, streptococci, and related genera by comparing direct colony testing with preparatory extraction. Isolates were previously identified by use of phenotypic testing and/or 16S rRNA gene sequencing. Manufacturer-specified score cutoffs for genus- and species-level identification were used. After excluding 7 isolates not present in the Biotyper library, the Biotyper correctly identified 284 (95%) and 207 (69%) isolates to the genus and species levels, respectively, using extraction. By using direct colony testing, the Biotyper identified 168 (56%) and 60 (20%) isolates to the genus and species levels, respectively. Overall, more isolates were identified to the genus and species levels with preparatory extraction than with direct colony testing (P < 0.0001). The analysis was repeated after dividing the isolates into two subgroups, staphylococci, streptococci, and enterococci (n = 217) and “related genera” (n = 81). For the former subgroup, the extraction method resulted in the identification of 213 (98%) and 171 (79%) isolates to the genus and species levels, respectively, whereas the direct colony method identified 136 (63%) and 56 (26%) isolates to the genus and species levels, respectively. In contrast, for the subgroup of related genera, the extraction method identified 71 (88%) and 36 (44%) isolates to the genus and species levels, respectively, while the direct colony method identified 32 (40%) and 4 (5%) isolates to the genus and species levels, respectively. For both subgroups, preparatory extraction was superior to direct colony testing for the identification of isolates to the genus and species levels (P < 0.0001). Preparatory extraction is needed for the identification of a substantial proportion of Gram-positive cocci using the Biotyper method according to manufacturer-specified score cutoffs.
INTRODUCTION
Established methods for the identification of cultured bacteria in the clinical microbiology laboratory include Gram staining, rapid biochemical tests, and long biochemical and molecular tests. The last two approaches may require hours to days, yield results that are difficult to interpret, and, occasionally, fail to differentiate among closely related species. A rapid and reliable identification system is therefore needed.
Matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry (MS) has been described to be a rapid, inexpensive, and accurate method for bacterial identification (3, 5, 6, 16, 17). Using MALDI-TOF MS, the protein spectral profile of an isolate is generated and compared to a reference database for identification. A previous study assessing the performance of MALDI-TOF MS for the identification of Gram-negative and -positive bacteria routinely isolated from clinical samples reported the identification of 84% of isolates to the species level (17).
Two methods have been described for the preparation of bacteria for identification by MALDI-TOF MS (9, 12–14, 17, 19). The direct colony method involves the application of bacterial colonies from growth plates directly onto a steel plate as a thin film before testing is performed by MALDI-TOF MS (16). In contrast, the extraction method involves the lysis of bacterial colonies by chemicals or enzymes to release proteins in a lysate or extract. The extract is applied onto the testing plate for analysis by MALDI-TOF MS. Both methods have been used for Gram-positive and Gram-negative bacteria. The physical disruption of peptidoglycan in the Gram-positive cell wall using the direct colony method may not always be sufficient to adequately prepare proteins for detection by MALDI-TOF MS (18). In addition, the application of a colony directly onto the plate rather than the use of a more pure extracted protein preparation may not produce valid scores because metabolites, pigments, and/or agar material on the surface of the cell may interfere with the crystallization process (8). In one study, investigators spotted bacteria directly onto the target plate for analysis by the Bruker Biotyper system (Bruker Daltonics, Billerica, MA) and achieved spectra with mass ranges from 2 to 25 kDa and >50 peaks for 10 different Gram-negative bacteria. Using this same protocol, those authors were unable to consistently produce quality spectra covering this mass range for fresh whole-cell Gram-positive bacteria and had to lyse the cells with lysozyme (18). Cherkaoui et al. reported that the majority of bacteria not identified by MALDI-TOF MS were Gram-positive bacteria when the direct colony method was used (6). The quality of spectra obtained by use of preparatory extraction is generally superior to that obtained by direct colony testing. For the above-mentioned reasons, we hypothesized that extraction would be a necessary preparatory step to obtain high-percentage identification of Gram-positive bacteria using MALDI-TOF MS with the Biotyper system.
We showed that Gram-negative bacteria could be identified by the Biotyper, with only 11% of bacteria requiring preparatory extraction (i.e., 89% were identified by use of direct colony testing) (16). However, our group also demonstrated that, compared with direct colony testing, extraction yielded higher scores for the identification of yeast using the Biotyper (N. Dhiman, personal communication) (7). Direct colony testing is logistically simpler and faster than preparatory extraction, rendering it the preferred approach for clinical laboratories. In this paper, we compared the abilities of the Biotyper to identify Gram-positive cocci, including staphylococci, streptococci, and related genera (identified in our laboratory by conventional methods), using direct colony testing and preparatory extraction. To the best of our knowledge, this is the first study to compare the abilities of the Biotyper to identify Gram-positive cocci using direct colony testing and preparatory extraction. We included both common and uncommon species of staphylococci, streptococci, and related genera, in contrast to previous studies that studied common bacteria or focused on a single bacterium (e.g., Staphylococcus aureus and Streptococcus agalactiae) (8, 14), single groups of bacteria (e.g., viridans group streptococci, Staphylococcus species, coagulase-negative staphylococci, and Corynebacterium species [9, 10, 12, 13]), or Staphylococcus and Streptococcus species tested together with Gram-negative bacteria (1, 2, 6, 11, 15, 17, 19). The referenced studies used Bruker Daltonics systems, except for one that used an Axima MALDI-TOF MS instrument and Shimadzu software (Shimadzu-Biotech Corp., Kyoto, Japan) and the SARAMIS database (AnagnosTec GmbH, Potsdam, Germany) (1), another that used a Micromass MALDI-TOF MS system (Micromass UK Ltd., Manchester, United Kingdom) (8), and a final one that compared the two systems (6). The isolates in this study were obtained from clinical samples. Finally, we limited the number of the common isolates to evaluate infrequent and common isolates, and we did not add new entries to the manufacturer's protein spectrum database. Therefore, the results of this study should reflect the performance of the instrument if implemented directly in the clinical laboratory.
(This work was presented in part at the 111th General Meeting of the American Society for Microbiology, New Orleans, LA, 21 to 24 May 2011.)
MATERIALS AND METHODS
Bacterial isolates.
Three hundred five aerobic Gram-positive cocci collected from multiple clinical sources, including tissue, blood, stool, urine, wound, cerebrospinal fluid, respiratory tract, and other sources, were studied. These isolates represent the species of staphylococci, streptococci, and related genera listed in Koneman's Color Atlas and Textbook of Diagnostic Microbiology, 6th ed. (20). Isolates were cultured overnight or until visible growth was observed on 5% sheep blood agar in 5% CO2 at 35°C, with the exception of Granulicatella and Abiotrophia spp., which were cultured on chocolate agar. Identification was performed by using conventional biochemical analysis and/or partial 16S rRNA gene sequencing (Microseq ID, version 2.0, AB_bacterial500LIB_2.1; Applied Biosystems, Carlsbad, CA). Seven isolates (Globicatella sp., Weissella confusa/Weissella cibaria [two isolates], Micrococcus lylae, Micrococcus terreus, and Arthrobacter cumminsii/Arthrobacter albus [two isolates]) were excluded from further analysis because they were not present in the Biotyper database.
MALDI-TOF mass spectrometry.
For the direct colony method, bacteria were applied as a thin film onto a 24-spot steel plate (Bruker Daltonics) and allowed to dry at room temperature. Subsequently, 2 μl of MALDI matrix (a saturated solution of α-cyano-4-hydroxycinnamic acid [HCCA; Bruker Daltonics] in 50% acetonitrile and 2.5% trifluoroacetic acid) was applied onto the colony and allowed to dry before testing. For the extraction method, 1 to 2 colonies (or a few colonies in the case of a small colony size) were suspended in 300 μl of molecular-grade water (Sigma-Aldrich, St. Louis, MO) and vortexed. Next, 900 μl of 100% ethanol (Sigma-Aldrich) was added, vortexed, and centrifuged (20,800 × g) for 3 min. The supernatant was decanted, and the pellet was dried at room temperature. Fifty microliters of 70% formic acid (Fluka [Sigma-Aldrich], St. Louis, MO) and 50 μl of acetonitrile (Fluka) were added and thoroughly mixed by pipetting, followed by centrifugation (20,800 × g) for 2 min. Two microliters of supernatant was spotted onto the 24-spot plate and allowed to dry at room temperature before the addition of 2 μl of matrix. For each plate, a bacterial test standard (Bruker Daltonics) was included to calibrate the instrument and validate the run.
MALDI-TOF MS was performed with the MicroFlex LT mass spectrometer (Bruker Daltonics) according to the manufacturer's suggested recommendations. Spectra were analyzed by using MALDI Biotyper automation control and the Bruker Biotyper 2.0 software and library (version 2.0, 3,740 entries; Bruker Daltonics). Identification score criteria used were those recommended by the manufacturer: a score of ≥2.000 indicated species-level identification, a score of 1.700 to 1.999 indicated identification to the genus level, and a score of <1.700 was interpreted as no identification. Isolates that failed to produce a score of ≥1.700 with direct colony or extraction methods were retested.
Discrepant results.
Discrepant results generated between conventional methods and the Biotyper were resolved by additional biochemical analysis and/or partial 16S rRNA gene sequencing (Microseq ID, version 2.0, AB_bacterial500LIB_2.1), as needed. Further analysis indicated that seven isolates had been misidentified by conventional methods but were correctly identified by the Biotyper, as determined by additional genotypic testing; these isolates were classified as having been correctly identified by the Biotyper.
Data analysis.
Because biochemical or phenotypic analysis fails to differentiate some subsets of closely related species, 62 isolates were reported as groups, including the Streptococcus mitis group, Streptococcus bovis group, Streptococcus anginosus group, and Streptococcus salivarius group. Identification was considered correct if the Biotyper identified a species in the corresponding group. In addition, 20 isolates were considered to represent one of two possible species because we did not definitely differentiate between the two, including two Staphylococcus intermedius/S. pseudointermedius, one Staphylococcus pasteuri/S. warneri, six Rothia dentocariosa/R. aeria, six Micrococcus luteus/M. yunnanensis, one Enterococcus avium/E. raffinosus, and four Enterococcus casseliflavus/E. gallinarum isolates. The identification of these isolates by the Biotyper was considered correct if it identified one of the two species. Nine bacteria reported as being group C, G, or F streptococci were considered correctly identified by the Biotyper if the identified bacteria were known to carry the respective antigens.
Statistical analysis.
Comparisons of Biotyper genus- or species-level identification using preparatory extraction versus direct colony methods overall and stratified by two groups of isolates (staphylococci, streptococci, and enterococci versus “related genera”) were made by using McNemar's test of paired proportions. Comparisons of genus- and species-level identification percentages between the two groups of isolates were made by using chi-square tests. P values of less than 0.05 were considered statistically significant. All analyses were performed by using SAS, version 9.1 (SAS Institute, Cary, NC).
RESULTS
Identification using direct colony and extraction methods.
Using the extraction method, the Biotyper correctly identified 284 (95%) isolates to the genus level and 207 (69%) isolates to the species level (Table 1). In comparison, the Biotyper correctly identified 168 (56%) isolates to the genus level and 60 (20%) isolates to the species level by use of the direct colony method (Table 1). The difference between the two methods was statistically significant at the genus and species levels (P < 0.0001). Testing of 25 isolates was repeated by the extraction method, resulting in the identification of 9 isolates to the genus level and 6 isolates to the species level. Using the direct colony method, 81 isolates were repeat tested, resulting in the identification of 15 isolates to the genus level and 2 isolates to the species level.
Table 1.
Bruker Biotyper identification of staphylococci, streptococci, and related genera to the genus and species levels by use of direct colony testing and preparatory extraction
| Organism | No. of isolates | No. of isolates (% correct) identified by: |
No. of spectra in database | |||
|---|---|---|---|---|---|---|
| Extraction |
Direct colony |
|||||
| Genus level | Species level | Genus level | Species level | |||
| Staphylococci, streptococci, and enterococci | ||||||
| Staphylococcus aureus | 20 | 20 | 19 | 18 | 8 | 12 |
| Staphylococcus lugdunensis | 8 | 8 | 8 | 7 | 1 | 5 |
| Staphylococcus epidermidis | 14 | 14 | 11 | 11 | 1 | 9 |
| Staphylococcus saprophyticus | 6 | 3 | 0 | 0 | 0 | 8 |
| Staphylococcus capitis | 6 | 6 | 6 | 4 | 2 | 6 |
| Staphylococcus haemolyticus | 5 | 5 | 5 | 2 | 0 | 8 |
| Staphylococcus hominis | 5 | 5 | 4 | 4 | 2 | 6 |
| Staphylococcus pettenkoferi | 5 | 5 | 0 | 1 | 0 | 4 |
| Staphylococcus schleiferi | 4 | 4 | 4 | 0 | 0 | 6 |
| Staphylococcus intermedius/S. pseudointermedius | 2 | 2 | 1 | 0 | 0 | 3 |
| Staphylococcus simulans | 2 | 2 | 0 | 0 | 0 | 5 |
| Staphylococcus pasteuri/S. warneri | 1 | 1 | 1 | 1 | 0 | 2 |
| Streptococcus pyogenes | 10 | 10 | 10 | 4 | 1 | 8 |
| Streptococcus agalactiae | 7 | 7 | 7 | 1 | 1 | 10 |
| Group C beta-hemolytic streptococci | 6 | 6 | 6 | 1 | 1 | NA |
| Group F beta-hemolytic streptococci | 1 | 1 | 1 | 1 | 0 | NA |
| Group G beta-hemolytic streptococci | 2 | 2 | 1 | 0 | 0 | NA |
| Streptococcus pneumoniae | 10 | 10 | 10 | 6 | 3 | 9 |
| Streptococcus mitis group | 10 | 10 | 3 | 10 | 2 | 10 |
| Streptococcus mutans group | 10 | 10 | 8 | 9 | 6 | 10 |
| Streptococcus salivarius group | 11 | 11 | 7 | 6 | 0 | 6 |
| Streptococcus anginosus group | 11 | 10 | 5 | 9 | 4 | 5 |
| Streptococcus bovis group | 13 | 13 | 10 | 9 | 2 | 11 |
| Streptococcus dysgalactiae | 1 | 1 | 1 | 0 | 0 | 3 |
| Streptococcus equi subsp. zooepidemicus | 1 | 1 | 1 | 1 | 0 | 5 |
| Streptococcus halichoeri | 1 | 1 | 1 | 0 | 0 | 1 |
| Enterococcus faecium | 15 | 15 | 15 | 14 | 12 | 9 |
| Enterococcus faecalis | 15 | 15 | 15 | 11 | 7 | 8 |
| Enterococcus avium | 6 | 6 | 6 | 1 | 0 | 2 |
| Enterococcus raffinosus | 2 | 2 | 0 | 0 | 0 | 1 |
| Enterococcus avium/E. raffinosus | 1 | 1 | 1 | 0 | 0 | 3 |
| Enterococcus gallinarum | 1 | 1 | 1 | 1 | 1 | 3 |
| Enterococcus casseliflavus/E. gallinarum | 4 | 4 | 2 | 4 | 2 | 8 |
| Enterococcus gilvus | 1 | 1 | 1 | 0 | 0 | 1 |
| Total | 217 | 213 (98) | 171 (79) | 136 (63) | 56 (26) | |
| Related genera | ||||||
| Rothia mucilaginosa | 11 | 11 | 1 | 1 | 0 | 3 |
| Rothia dentocariosa/R. aeria | 6 | 5 | 4 | 2 | 0 | 5 |
| Micrococcus luteus/M. yunnanensis | 6 | 6 | 5 | 4 | 0 | 2 |
| Micrococcus luteus | 3 | 2 | 1 | 1 | 1 | 2 |
| Granulicatella adiacens | 7 | 5 | 0 | 0 | 0 | 1 |
| Granulicatella elegans | 1 | 0 | 0 | 0 | 0 | 1 |
| Abiotrophia defectiva | 6 | 6 | 6 | 5 | 0 | 2 |
| Aerococcus sanguinicola | 12 | 11 | 4 | 4 | 0 | 2 |
| Aerococcus urinae | 7 | 7 | 5 | 3 | 0 | 3 |
| Aerococcus viridans | 1 | 1 | 0 | 0 | 0 | 5 |
| Gemella morbillorum | 2 | 0 | 0 | 0 | 0 | 1 |
| Gemella sp. | 2 | 2 | 0 | 2 | 0 | 2 |
| Kocuria kristinae | 3 | 3 | 1 | 0 | 0 | 3 |
| Kocuria rosea | 1 | 1 | 1 | 1 | 0 | 5 |
| Kocuria palustris | 1 | 1 | 0 | 0 | 0 | 1 |
| Helcococcus kunzii | 2 | 2 | 2 | 2 | 0 | 2 |
| Lactococcus lactis | 4 | 4 | 4 | 4 | 2 | 5 |
| Leuconostoc citreum | 1 | 1 | 1 | 1 | 1 | 2 |
| Pediococcus sp. | 1 | 1 | 1 | 1 | 0 | 2 |
| Facklamia languida | 1 | 1 | 0 | 1 | 0 | 1 |
| Facklamia ignava | 1 | 0 | 0 | 0 | 0 | 1 |
| Arthrobacter cumminsii | 1 | 1 | 0 | 0 | 0 | 1 |
| Macrococcus caseolyticus | 1 | 0 | 0 | 0 | 0 | 1 |
| Total | 81 | 71 (88) | 36 (44) | 32 (40) | 4 (5) | |
Identification of staphylococci, streptococci, and enterococci compared to identification of related genera.
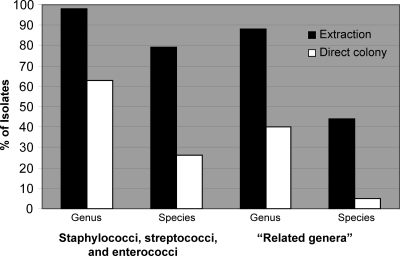
To examine the abilities of the direct colony and extraction methods to identify staphylococci, streptococci, and enterococci versus “related genera,” the isolates were divided into Staphylococcus, Streptococcus, and Enterococcus species (217 isolates) and related genera (81 isolates). The extraction method identified 213 (98%) isolates of the staphylococci, streptococci, and enterococci to the genus level and 171 (79%) isolates to the species level, compared to the identification of 136 (63%) and 56 (26%) isolates to the genus and species levels, respectively, by the direct colony method (Fig. 1). For the isolates of related genera, the extraction method identified 71 (88%) to the genus level and 36 (44%) to the species level, compared with 32 (40%) and 4 (5%) isolates identified to the genus and species levels, respectively, by the direct colony method (Fig. 1). The extraction method identified significantly more staphylococcal, streptococcal, and enterococcal isolates than isolates of related genera to the genus level (213 [98%] versus 71 [88%] isolates, respectively; P < 0.0001) and the species level (171 [79%] versus 36 [44%] isolates, respectively; P < 0.0001). The direct colony method correctly identified staphylococci, streptococci, and enterococci better than isolates of related genera to both the genus (136 [63%] versus 32 [40%] isolates; P < 0.0003) and species (56 [26%] versus 4 [5%] isolates; P < 0.0001) levels.
Fig. 1.
Percent identification of staphylococci, streptococci, enterococci, and “related genera” to the genus and species levels by the Bruker Biotyper system.
Erroneous identifications.
Four of the ten S. mitis group isolates (40%) were misidentified as Streptococcus pneumoniae by the extraction method. Optochin susceptibility and bile solubility confirmed that the isolates belonged to the S. mitis group. All 10 S. pneumoniae isolates were correctly identified. Two Enterococcus casseliflavus/E. gallinarum isolates were misidentified as Enterococcus phoeniculicola by the extraction method (and one was misidentified by the direct colony method); the second best identification for these isolates was E. casseliflavus. 16S rRNA gene sequencing confirmed that these isolates were E. casseliflavus.
Erroneous phenotypic identification.
Phenotypic identification was erroneous for seven isolates, including four isolates of Staphylococcus epidermidis that were identified as Staphylococcus capitis by the Biotyper, two isolates of Enterococcus raffinosus that were identified as Enterococcus gilvus and Enterococcus avium by the Biotyper, and one E. casseliflavus isolate that was identified as Enterococcus gallinarum by the Biotyper. Further testing of these isolates by 16S rRNA gene sequencing confirmed that the Biotyper identification was correct.
MALDI-TOF MS identification in relationship to database redundancy.
We evaluated the relationship between the number of individual-species spectra in the Bruker database and the likelihood of species-level identification using preparatory extraction. When the database contained ≥9 spectra of a given species, 75 to 100% of the isolates of that species were identified to the species level (Table 1). When the library contained six to eight spectra of a given species, 64 to 100% of isolates of that species (except for Staphylococcus saprophyticus) were identified to the species level. When the database contained ≤5 spectra of a given species, 0 to 100% of the isolates of that species were identified to the species level.
To further evaluate the effect of the number of spectra on identification, we added four new Rothia mucilaginosa spectra (derived from the extracted organism) to the library that had three spectra of this species, and we tested five different isolates before and after the addition of the spectra. Three of the five isolates had scores of ≥1.7 and ≤2.0 and two had scores of ≤1.7 before the addition of the spectra. After the addition of the spectra, all five isolates had scores of ≥2.1.
MALDI-TOF MS identification scores.
We analyzed percent identifications using cutoffs different from those recommended by the manufacturer for genus-level (i.e., scores of ≥1.6, ≥1.5, and ≥1.4) and species-level (i.e., scores of ≥1.9, ≥1.8, and ≥1.7) identifications using the direct colony and extraction methods (Table 2). Decreasing the genus-level identification score for the extraction method from ≥1.7 to ≥1.6, ≥1.5, and ≥1.4 increased genus-level identification from 284 (95%) isolates to 291 (98%), 293 (98%), and 295 (99%) isolates, respectively (Table 2). For the direct colony method, the identification increased from 168 (56%) isolates to 191 (64%), 214 (72%), and 226 (76%) isolates, respectively. Decreasing the species-level identification score for the extraction method from ≥2.0 to ≥1.9, ≥1.8, and ≥1.7 increased species-level identification from 207 (69%) isolates to 224 (75%), 248 (83%), and 276 (93%) isolates, respectively (Table 2). For the direct colony method, the identification increased from 60 (20%) isolates to 95 (32%), 129 (43%), and 161 (54%) isolates, respectively. Using the extraction method, no further misidentifications resulted from changing the genus or species score cutoffs. However, using the direct colony method, one S. haemolyticus isolate was misidentified as an Escherichia species isolate at scores of 1.4 and 1.5, and one Enterococcus raffinosus isolate was misidentified as a Sporosarcina species isolate at a score of 1.4.
Table 2.
Numbers and percentages of isolates identified with lowered scores assigned for genus and species identifications using extraction and direct colony methods
| Score cutoff | No. (%) of isolates identified |
|
|---|---|---|
| Extraction | Direct colony | |
| Genus | ||
| ≥1.4 | 295 (99) | 226 (76) |
| ≥1.5 | 293 (98) | 214 (72) |
| ≥1.6 | 291 (98) | 191 (64) |
| ≥1.7 | 284 (95) | 168 (56) |
| Species | ||
| ≥1.7 | 276 (93) | 161 (54) |
| ≥1.8 | 248 (83) | 129 (43) |
| ≥1.9 | 224 (75) | 95 (32) |
| ≥2.0 | 207 (69) | 60 (20) |
DISCUSSION
The extraction method has been used previously for the identification of Gram-positive bacteria by the Biotyper system either as the sole method of identification (9, 13, 14) or when the direct colony method fails to achieve a reliable identification (2, 19). One study indicated that extraction increased the level of identification for 37% of 19 isolates of nonfermenting Gram-negative bacteria tested but was not generally needed for Gram-positive bacteria (19). Another study showed that 75% of Gram-positive and -negative bacteria were identified by use of the direct colony method and that an extraction step was needed for the identification of the remaining 25% of isolates (2). In that study, the percent species identification by the direct colony method was low for common Gram-positive bacteria, including S. aureus (79%), Enterococcus faecalis (82%), S. epidermidis and S. agalactiae (58%), S. pneumoniae (73%), E. gallinarum (71%), and Streptococcus anginosus and Staphylococcus lugdunensis (33%), and was 0% for Streptococcus constellatus, E. casseliflavus, and S. capitis.
Our analysis shows that extraction resulted in more high-level identifications than did direct colony testing. The extraction method identified 95% of the isolates to the genus level and 69% of isolates to the species level, whereas the direct colony method identified 56% of isolates to the genus level and 20% of isolates to the species level, using manufacturer-suggested cutoffs. Compared to direct colony testing, extraction requires additional processing time, reagents, and equipment.
The inability to identify all isolates to the species level even with preparatory extraction may be explained by multiple reasons. First, the manufacturer's spectral database contains a variable number of entries per species, usually being higher for common isolates. A higher number of entries for the same species will likely better reflect diversity within the species due to variations in protein expression between strains (14) and potentially under different conditions. In our study, using the extraction method, increased identification to the species level appeared to be associated with the number of protein spectra in the database. This observation is in agreement with a previous report that accurate MALDI-TOF MS identifications correlated with ≥10 reference spectra for a given species in the database (17). These reference spectra should not be established from related isolates but should represent the diversity of isolates of the same species (14). Second, bacteria that produce tiny or mucoid colonies may yield a small amount of protein insufficient for ideal spectra (4). Tiny colonies were noted especially with some enterococci, viridans group streptococci, and Granulicatella species. Mucoid colonies were noted with Rothia species, S. anginosus, and Streptococcus salivarius. It is possible that the creation of a spectral library based on spectra generated using the direct colony method would improve identification using direct colony testing.
All S. pneumoniae isolates were correctly identified. Previously reported studies that tested S. pneumoniae by MALDI-TOF MS have revealed conflicting results. Two studies reported the correct identification of all tested pneumococci at the genus or species level (39 isolates) (11, 19), while one study reported the misidentification S. pneumoniae isolates as Streptococcus parasanguinis for 46% of 24 tested isolates (17). This was explained by inadequate database entries for both microorganisms, and the addition of S. pneumoniae spectra resolved this discrepancy (17). In our study, 4 of 10 Streptococcus mitis group isolates were misidentified as S. pneumoniae isolates (using the extraction method). This finding has been reported by multiple studies for various percentages of isolates tested (6, 11, 15, 19). This is likely related to the similar compositions of ribosomal proteins, the main target for MALDI-TOF MS. Even 16S rRNA gene sequencing has limitations in differentiating these species, as the 16S rRNA gene sequences of these species (e.g., S. mitis, Streptococcus oralis, Streptococcus pseudopneumoniae, and S. pneumoniae) share more than 99% identity (11, 12). Since the Biotyper fails to differentiate between these species, additional testing (e.g., bile solubility, optochin susceptibility, pneumococcal antigen detection) is necessary to exclude S. pneumoniae.
Three E. casseliflavus/E. gallinarum isolates were misidentified as E. phoeniculicola isolates (two by the extraction method and a third by the direct colony method). E. phoeniculicola has been isolated from birds, and it is not known to cause human disease (13). This misidentification was previously reported by two studies (4, 19). In one of those reports, the correct identification of E. casseliflavus was achieved when the Biotyper library, version 2.0, was updated to a new version (4). We tested one of our isolates by the extraction method after implementing an update to our library (version 3.0, 3,995 entries) and also confirmed the E. casseliflavus identification. In the updated library, five E. casseliflavus isolates and one E. phoeniculicola isolate were deleted, and another six E. casseliflavus isolates and one E. phoeniculicola isolate were added.
Two studies used a score of > or ≥1.9 instead of the manufacturer's recommended score of ≥2.0 for species-level identification (15, 17). In our study, decreasing the score for the extraction method from ≥2.0 to ≥1.9, ≥1.8, or ≥1.7 increased species identification by an additional 6, 14, or 24%, respectively. The increases were 12, 23, and 34%, respectively, for the direct colony method. Similarly, decreasing the genus score from ≥1.7 to ≥1.6, ≥1.5, or ≥1.4 resulted in increased identification by an additional 3, 3, and 4%, respectively, using the extraction method, and 8, 16, and 20%, respectively, using the direct colony method. Misidentifications occurred only at a score of ≤1.5. Overall, the application of a genus-level score of 1.6 and a species-level score of 1.7 may be more appropriate than the manufacturer-recommended cutoffs. Further studies are necessary to support this approach.
In conclusion, preparatory extraction is superior to direct colony testing for the identification of Gram-positive cocci by MALDI-TOF MS using the Bruker system. The addition of protein spectra to the database to reflect the diversity of closely related species and a decrease of the identification scores for species- and genus-level identifications may further improve identification.
ACKNOWLEDGMENTS
We thank the outstanding clinical microbiology laboratory technologists at Mayo Clinic, Rochester, MN, for their help in collecting and analyzing isolates for this study. We thank Ryan T. Saffert for his thoughtful review of the manuscript.
Footnotes
Published ahead of print on 25 May 2011.
REFERENCES
- 1. Benagli C., Rossi V., Dolina M., Tonolla M., Petrini O. 2011. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for the identification of clinically relevant bacteria. PLoS One 6:e16424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bizzini A., Durussel C., Bille J., Greub G., Prod'hom G. 2010. Performance of matrix-assisted laser desorption ionization–time of flight mass spectrometry for identification of bacterial strains routinely isolated in a clinical microbiology laboratory. J. Clin. Microbiol. 48:1549–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bizzini A., Greub G. 2010. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry, a revolution in clinical microbial identification. Clin. Microbiol. Infect. 16:1614–1619 [DOI] [PubMed] [Google Scholar]
- 4. Bizzini A., et al. 2011. Matrix-assisted laser desorption ionization–time of flight mass spectrometry as an alternative to 16S rRNA gene sequencing for identification of difficult-to-identify bacterial strains. J. Clin. Microbiol. 49:693–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carbonnelle E., et al. 2011. MALDI-TOF mass spectrometry tools for bacterial identification in clinical microbiology laboratory. Clin. Biochem. 44:104–109 [DOI] [PubMed] [Google Scholar]
- 6. Cherkaoui A., et al. 2010. Comparison of two matrix-assisted laser desorption ionization–time of flight mass spectrometry methods with conventional phenotypic identification for routine identification of bacteria to the species level. J. Clin. Microbiol. 48:1169–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dhiman N., Hall L., Wohlfiel S., Buckwalter S., Wengenack N. 2011. Performance and cost analysis of matrix-assisted laser desorption ionization–time of flight mass spectrometry for routine identification of yeast. J. Clin. Microbiol. 49:1614–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Du Z., Yang R., Guo Z., Song Y., Wang J. 2002. Identification of Staphylococcus aureus and determination of its methicillin resistance by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal. Chem. 74:5487–5491 [DOI] [PubMed] [Google Scholar]
- 9. Dubois D., et al. 2010. Identification of a variety of Staphylococcus species by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 48:941–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dupont C., et al. 2010. Identification of clinical coagulase-negative staphylococci, isolated in microbiology laboratories, by matrix-assisted laser desorption/ionization–time of flight mass spectrometry and two automated systems. Clin. Microbiol. Infect. 16:998–1004 [DOI] [PubMed] [Google Scholar]
- 11. Eigner U., et al. 2009. Performance of a matrix-assisted laser desorption ionization–time-of-flight mass spectrometry system for the identification of bacterial isolates in the clinical routine laboratory. Clin. Lab. 55:289–296 [PubMed] [Google Scholar]
- 12. Friedrichs C., Rodloff A. C., Chhatwal G. S., Schellenberger W., Eschrich K. 2007. Rapid identification of viridans streptococci by mass spectrometric discrimination. J. Clin. Microbiol. 45:2392–2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Konrad R., et al. 2010. Matrix-assisted laser desorption/ionisation time-of-flight (MALDI-TOF) mass spectrometry as a tool for rapid diagnosis of potentially toxigenic Corynebacterium species in the laboratory management of diphtheria-associated bacteria. Euro Surveill. 15(43):pii=19699 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19699 [DOI] [PubMed] [Google Scholar]
- 14. Lartigue M. F., et al. 2009. Identification of Streptococcus agalactiae isolates from various phylogenetic lineages by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 47:2284–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Risch M., et al. 2010. Comparison of MALDI TOF with conventional identification of clinically relevant bacteria. Swiss Med. Wkly. 140:w13095. [DOI] [PubMed] [Google Scholar]
- 16. Saffert R. T., et al. 2011. Comparison of Bruker Biotyper MALDI-TOF mass spectrometer to BD Phoenix automated microbiology system for identification of Gram-negative bacilli. J. Clin. Microbiol. 49:887–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Seng P., et al. 2009. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Infect. Dis. 49:543–551 [DOI] [PubMed] [Google Scholar]
- 18. Smole S. C., King L. A., Leopold P. E., Arbeit R. D. 2002. Sample preparation of Gram-positive bacteria for identification by matrix assisted laser desorption/ionization time-of-flight. J. Microbiol. Methods 48:107–115 [DOI] [PubMed] [Google Scholar]
- 19. van Veen S. Q., Claas E. C., Kuijper E. J. 2010. High-throughput identification of bacteria and yeast by matrix-assisted laser desorption ionization–time of flight mass spectrometry in conventional medical microbiology laboratories. J. Clin. Microbiol. 48:900–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Winn J., et al. 2006. Koneman's color atlas and textbook of diagnostic microbiology, 6th ed. Lippincott Williams & Wilkins, Baltimore, MD [Google Scholar]