Abstract
The protein association of estrogen receptor α ERα with DNA-bound SP1 and C/EBPβ is essential for the 17β-estradiol (E2)-induced activation of human prolactin receptor (hPRLR) gene transcription. Protein-protein interaction and complex formation at the hPIII promoter of hPRLR was investigated. The basic region and leucine zipper (bZIP) of C/EBPβ, zinc finger (ZF) motifs of SP1, and the DNA binding domain of ERα were identified as regions responsible for the interactions between transfactors. The E2-induced interaction was confirmed by bioluminescence resonance energy transfer (BRET) assays of live cells. The combination of BRET/bimolecular luminescence complementation assay revealed that ERα exists as a constitutive homodimer, and E2 induced a change(s) in ERα homodimer conformation favorable for its association with C/EBPβ and SP1. Chromatin immunoprecipitation and small interfering RNA knockdown of members of the complex in breast cancer cells demonstrated the endogenous recruitment of components of the complex onto the hPIII promoter of the hPRLR gene. SP1 is the preferred transfactor for the recruitment of ERα to the complex that facilitates the C/EBPβ association. The E2/ERα-induced hPRLR transcription was demonstrated in ERα-negative breast cancer cells. This study indicates that the enhanced complex formation of ERα dimer with SP1 and C/EBPβ by E2 has an essential role in the transcriptional activation of the hPRLR gene.
INTRODUCTION
The prolactin receptor (PRLR) is a member of the lactogen/cytokine receptor family which mediates the diverse cellular actions of prolactin (PRL) in several target tissues. PRL is a major factor in the proliferation and differentiation of breast epithelium and is the primary hormone in the stimulation and maintenance of lactation. It is also a tumor promoter in rodents and has been implicated in the development of breast cancer (5). The human PRLR (hPRLR) has several forms, including a long form and several short forms, which are products of alternative splicing with variable lengths in their cytoplasmic domains containing some unique sequences (8, 9). The short forms S1a and S1b are dominant-negative repressors of the function of homodimers of the PRLR long form that mediates all known stimulatory functions of PRL through the Jak-2/Stat5 signaling pathway. The dominant-negative effect of the short forms results from their heterodimerization with the long form, leading to inactivation due to the lack of required cytoplasmic sequences for STAT signaling activation (23, 29). Intramolecular S-S bonds in the extracellular domain of the PRLR short forms were found to be required for their inhibitory action on PRL-induced PRLR long form-mediated STAT5-dependent action (29). The PRLR forms are expressed in normal and tumoral breast tissue, in most human breast cancer cells, and in several other tissues (8, 17, 19). In humans, hPRLR expression is controlled by a complex regulatory system at the transcriptional level which is governed by multiple promoters. These include the preferentially utilized generic promoter 1/exon 1 (PIII/hEI3), which also is present in rat and mouse, and five human-specific exon 1 promoters (hE1N1 to hE1N5) (11, 12). These promoters were found to be utilized in breast cancer tissue and cell lines, including MCF7 and T47D, and variably in other tissues (12). Among these promoters, the preferentially utilized human promoter III (hPIII), the human counterpart of rodent PIII, was functionally characterized in breast cancer cells. 17β-Estradiol (E2) induced an increase in PRLR mRNA transcripts directed by the preferentially utilized hPIII promoter. Also, in transfection studies E2 activated the hPIII promoter, which lacks an estrogen response element (ERE) (16). This promoter contains functional SP1 and C/EBPβ elements that bind SP1/SP3 and C/EBPβ, respectively (10, 12). The abolition of the E2 effect by the mutation of SP1 and C/EBPβ elements within PIII indicated the cooperation in E2-induced transcription of hPRLR. E2 regulates PRLR expression through a nonclassical ERE-independent mechanism in target cells. The E2/estrogen receptor α (ERα) complex translocates to the nucleus and binds transfactors at the hPIII promoter, leading to the recruitment of coactivators (p300, SRC-1, and pCAF) to the complex with consequent region-specific changes in histone acetylation. These hormone/receptor-induced associations and chromatin changes favored TFIIB and RNA polymerase II recruitment, leading to the activation of hPIII-directed transcription and PRLR gene expression (4).
In this study, we have mapped the physical domains involved in interactions between ERα and transcription factors SP1 and C/EBPβ. The constitutive and E2 dependency of the interactions characterized in vitro also were demonstrated in vivo. Bioluminescence resonance energy transfer (BRET)/complementation assays, chromatin immunoprecipitation (ChIP) analyses, and small interfering RNA (siRNA) interference studies have established the participation of the ERα dimer and E2/ERα complexes in the interaction with transcription factors SP1 and C/EBPβ.
MATERIALS AND METHODS
Bacterial expression system.
Coding sequences of human ERα (amino acids [aa] 180 to 595; accession no. NM_000125), SP1 (amino acids 1 to 778; accession no. NM_138473), and C/EBPβ (amino acids 1 to 345; accession no. NM_005194) were PCR amplified and inserted in frame into BamHI and XhoI sites for ERα and C/EBPβ and into EcoRI and XhoI sites for SP1 of the pET-41b(+) vector (Novagen, Madison, WI). The following primers were used for PCR: ERα forward primer, GAT CGG ATC CGA AGG AGA CTC GCT ACT GTG CAG TG; reverse primer, GAT CCT CGA GGA CCG TGG CAG GGA AAC CCT C; SP1 forward primer, GAT CGA ATT CTA TGG ATG AAA TGA CAG CTG TG; reverse primer, GAT CCT C GA GGA AGC CAT TGC CAC TGA TAT TAA TG; C/EBPβ forward primer, GAT CGG ATC CGA TGC AAC GCC TGG TGG CCT G; reverse primer, GAT CCT CGA GGC AGT GGC CGG AGG AGG. The restriction sequences are underlined.
In vitro transcription and translation.
Full-length human SP1, C/EBPβ, and their deletion mutants were generated by PCR using p3XFLAG-CMV-SP1 (Sigma) and pOTB7-C/EBPβ (Invitrogen) as templates, respectively. The primers used for the various SP1 constructs are the following: mS1 (amino acids 76 to 707; nucleotides 226 to 2121) forward, GCC GCC GAA TTC CCC ACC ATG GGC CCG AGTC AGT CAG G; reverse, GCC GCC TCT AGA GCC TCC CTT CTT ATT CTG GTG; mS2 (amino acids 76 to 677; nucleotides 226 to 2031) forward, same as that used for mS1; reverse, GCC GCC TCT AGA CTT CTC ACC TGT GTG TGT ACG; mS3 (amino acids 76 to 648; nucleotides 227 to 1944) forward, same as that used for mS1; reverse, GCC GCC TCT AGA TGG CCT CTC GCC TGT ATG C; mS4 (amino acids 610 to 778; nucleotides 1828 to 2334) forward, GCC GCC GAA TTC CCC ACC ATG TCG GGG GAT CCT GGC; reverse, GCC GCC TCT AGAGAA GCC ATT GCC ACT G; mS5 (amino acids 1 to 610; nucleotides 1 to 1830) forward, GCC GCC GAA TTC CCC ACC ATG GAT GAA ATG ACA GCT GTG; reverse, GCC GCC TCT AGA CGA GCC CCT TCC TTC ACT G. The primers for the C/EBPβs are the following: wild-type (WT) C/EBPβ (amino acids 1 to 345; nucleotides 1 to 1035) forward, TCA AAG CTT GGC CCA CCA TGC AAC GCC TGG TGG CCT G; reverse, TCA CTC GAG GCA GTG GCC GGA GGA GG; mC1 (amino acids 1 to 301; nucleotides 1 to 903) forward, same as that used for the full-length primer; reverse, TCA CTC GAG GTG CTG CGT CTC CAG GTT GCG; mC2 (amino acids 1 to 189; nucleotides 1 to 567) forward, same as that used for the full-length primer; reverse, TCA CTC GAG CTC CTC CTT CCG CTT GCA GTC; mC3 (amino acids 189 to 345; nucleotides 565 to 903) forward, TCA AAG CTT GGC CCA CCA TGG AGG CCG GGG CGC C; reverse, the same as that used for the wild type. The Kozak and/or restriction sequence is underlined at the 3′ end. All PCR products were restricted and cloned into EcoRI and XbaI sites for SP1 and HindIII and XhoI sites for C/EBPβ, respectively, of pcDNA3.1-V5/HIS (Invitrogen, CA) to generate the in vitro-translated protein containing epitope tag V5 at their C termini. The constructs for the expression of ERα were described previously (4).
BRET and BiLC assays.
Enhanced yellow fluorescent protein (EYFP) and renilla luciferase (Rluc) genes were PCR amplified using pEYFP-N1 (BD Biosciences) and pRL (Promega) as templates, respectively, and cloned into either XhoI and AgeI or KpnI and BamHI sites of pcDNA3.1-V5/HIS (Invitrogen) to generate pcDNA–YFP and pcDNA-Rluc. Bioluminescence resonance energy transfer (BRET) constructs of ERα-YFP, ERα-Rluc, C/EBPβ-YFP, C/EBPβ-Rluc, and Rluc-SP1 were generated by ligating the coding sequences of ERα and C/EBPβ into KpnI and BamHI sites of both pcDNA-YFP and pcDNA-Rluc and SP1 into XhoI and AgeI sites of pcDNA-Rluc. For the YFP-SP1 construct, the coding region of SP1 was cloned into EcoRI and KpnI sites of pEYFP-C1 (BD Biosciences). For the bimolecular luminescence complementation (BiLC) assays, the renilla luciferase gene was split into two parts, the N terminus (amino acids 1 to 229) and C terminus (amino acids 174 to 310), where 55 amino acids overlapped. The N terminus (1 to 229 aa) was cloned into XhoI and AgeI and the C terminus (aa 174 to 310) into KpnI and BamHI sites of pcDNA-ERα to generate constructs of ERα-Rluc(1-229 aa) and Rluc(174-310 aa)-ERα, respectively. For flexibility between two fusion proteins, a complementarily synthesized linker sequence of 10 amino acids (GGGGSGGGGS) was annealed, restricted, and inserted between BamHI and XhoI sites of all BRET and complementation constructs except for YFP-SP1, for which the internal sequence was used as the linker. The primers used for the BRET constructs are the following: YFP forward, GTC ACT CGA GAT GGT GAG CAA GGG CGA GGA GCT G; reverse, GTC AAC CGG TTT ACT TGT ACA GCT CGT CCA TGC CGA G; Rluc forward, GTC AGG TAC CCC ACC ATG GCT TCC AAG GTG TAC GAC CCC GAG C or GTC ACT CGA GAT GGC TTC CAA GGT GTA CGA CCC CGA GC; reverse, GTC AGG ATC CCC TGC TCG TTC TTC AGC ACG CGC TCC ACG or GTC AAC CGG TTT ACT GCT CGT TCT TCA GCA CGC GCT CCA CG; ERα forward, GTC AGG TAC CCC ACC ATG ACC ATG ACC CTC CAC ACC AAA GCA TCC; reverse, GTC AGG ATC CCG ACC GTG GCA GGG AAA CCC TCT G; C/EBPβ forward, GTC AGG TAC CCC ACC ATG CAA CGC CTG GTG GCC TGG GAC; reverse, GTC AGG ATC CCG CAG TGG CCG GAG GAG GCG AG; C/EBPβ fused to YFP (deleted of basic region and leucine zipper, aa 1 to 189) forward, GTC AGG TAC CCC ACC ATG CAA CGC CTG GTG GCC TGG GAC; reverse, GTC AGG ATC C CCT CCT TCC GCT TGC AGT CCG CGG GCT C; SP1 fused to Rluc forward, GTC ACT CGA GAT GGA TGA AAT GAC AGC TGT GGT GAA AAT TG; reverse, GTC AAC CGG TTC AGA AGC CAT TGC CAC TGA TAT TAA TGG; SP1 fused to YFP forward, GAT CGA ATT CTA TGG ATG AAA TGA CAG CTG TG; reverse, GAT CGG TAC CTC AGA AGC CAT TGC CAC TG; SP1 fused to YFP (deleted of zinc finger [ZF] domains, aa 1 to 609) forward, GAT CGA ATT CTA TGG ATG AAA TGA CAG CTG TG; reverse, GAT CGG TAC C GCC CCT TCC TTC ACT GTC TTT ACA GTA GGG-3; Rluc(1-229 aa) forward, GTC ACT CGA GAT GGC TTC CAA GGT GTA CGA CCC CGA G; reverse, GTC AAC CGG TTT AGC CTC CCT TAA CGA GAG GGA TCT C; Rluc(174-310 aa) forward, GTC AGG TAC CCC ACC ATG GTG CTT GAG AAT AAC TTC TTC GTC; reverse, GTC AGG ATC CCC TGC TCG TTC TTC AGC ACG CGC TCC ACG; linker, CAG TGG ATC CTC GGT GGA GGC GGT TCA GGC GGA GGT GGC TCT CTC GAG TGA C. The Kozak and/or restriction sequence of each primer is underlined.
In vitro translation.
The in vitro expression of ERα, SP1, and C/EBPβ and their deletion mutants was performed by using in vitro transcription and translation-coupled rabbit reticulocyte extracts in the presence of methionine according to the manufacturer's instructions (TnT coupled reticulocyte lysate system; Promega, Madison, WI). Ten to 20 μl of 50-μl TnT mixtures was used in the study.
GST pulldown assay.
The overnight cultures of Escherichia coli Rosetta (DE3) transformed with pET-41b(+)-ERα, pET-41b(+)-SP1, and pET-41b(+)-C/EBPβ were diluted 1:50 in fresh LB medium and grown at 30°C to an optical density at 600 nm of 0.5. Fusion proteins then were induced with 0.5 mM isopropyl-1-thio-β-d-galactopyranoside (IPTG) at 30°C for 2 h. The IPTG-induced cells were pelleted at 5,000 × g for 10 min at 4°C, suspended in ice-cold phosphate-buffered saline (PBS) containing 1 mM MgCl2, 1 mM EDTA, and 1× protease inhibitor cocktail (Sigma), and lysed by probe sonication. The sonicated sample was clarified at 12,000 × g for 15 min at 4°C, and the supernatant was incubated with glutathione S-transferase (GST) binding resin (Novagen) at 4°C overnight. The resins were washed four times with ice-cold PBS containing 0.1% NP-40. The expression of fusion proteins was analyzed with Coomassie brilliant blue staining and Western blotting. For pulldown assays, resins loaded with the fusion proteins (1 μg) were mixed with 10 or 20 μl of in vitro-expressed protein in a final volume of 1 ml PBS containing 0.1% NP-40. After overnight incubation, resins were collected by centrifugation and washed three times with PBS containing 0.1% NP-40. The resins were boiled in 2× SDS sample buffer, and protein interaction was analyzed by Western blotting.
DNA affinity precipitation assay (DAPA).
Wild and mutant oligonucleotides corresponding to the sequence spanning from bp −392 to −351 of the human prolactin receptor PIII promoter relative to the start site of translation (+1) were synthesized with biotinylation at the 5′ end and annealed complementarily. Ten μl of each in vitro-translated protein (ERα-V5, SP1-V5, and C/EBPβ-V5) in 1 ml of binding buffer (20 mM HEPES, pH 8.0, 50 mM NaCl, 1 mM dithiothreitol [DTT], 0.01% NP-40, 5% glycerol) was precleared using 100 μl of streptavidin-coupled beads (50% slurry) for 1 h at 4°C. The precleared samples were resuspended with 1 ml of binding buffer and incubated with 1 μg of the biotinylated oligonucleotides for 18 h at 4°C. Streptavidin beads then were added and incubated for an additional 2 h at 4°C. Following incubation, the beads were collected and washed three times with binding buffer. The proteins from the biotinylated oligonucleotide/streptavidin bead complexes were analyzed by Western blotting. The nucleotide sequences used as the probes are the following: wild type, GAA CAT AAA TGT TGC AAC ACT GAC TCC TCC TCT CAT GAA GAA; mutant SP1 binding site, GAA CAT AAA TGT TGC AAC ACT GAa TaC AAC TCT CAT GAA GAA; mutant C/EBPβ binding site, GAA CAT AAA Tac cat Agt ACT GAC TCC TCC TCT CAT GAA GAA; double mutant SP1 and C/EBPβ binding sites, GAA CAT AAA Tac cat Agt ACT GAa TaC aaC TCT CAT GAA GAA. Wild-type or mutant C/EBPβ and SP1 binding motifs are underlined, and mutated nucleotides are in lowercase.
Western blot analysis.
Protein samples were resolved on 8 to 16% Tris-glycine gradient SDS gels and transferred to nitrocellulose membranes (Invitrogen). These were blocked in 5% nonfat milk, 135 mM NaCl, 10 mM Tris-HCl (pH 7.2), and 0.05% Tween 20. Immunoblots were carried out with monoclonal anti-ERα (sc-554; 1:1,000; Santa Cruz Biotechnology), monoclonal anti-SP1 (1:5,000; sc-17824; Santa Cruz), polyclonal anti-rabbit C/EBPβ (1:1,000; sc-7962; Santa Cruz), or monoclonal anti-V5 antisera (1:1,000; Invitrogen). The appropriate second antibody was employed (goat anti-rabbit [sc-2054] or anti-mouse [sc-2055] IgG-horseradish peroxidase at a dilution of 1:5,000) and developed by using extended chemiluminescence (Pierce, IL).
BRET and BiLC assays.
COS1 cells were seeded at a density of 200,000 cells on 12-well tissue culture plates. After 16 h in culture, the cells were transfected alone or with various combinations of BRET constructs (ERα-YFP, ERα-Rluc, Rluc-SP1, YFP-SP1, C/EBPβ-YFP, and C/EBPβ-Rluc) and/or BiLC constructs [ERα-Rluc(1-229 aa) and Rluc(174-310 aa)-ERα] using Lipofectamine 2000. After 24 h of transfection, the cells were detached, suspended in phenol-free Dulbecco's modified essential medium (DMEM) containing 10% charcoal-treated serum (CTS), distributed on black transparent 96-well plates at a density of 50,000 cells, and incubated for an additional 24 h. The cells then were incubated with 1% CTS-DMEM with or without 100 nM 17β-estradiol. After 2 h of treatment, cells were washed twice with PBS and incubated with a final concentration of 5 μM coelenterazine in PBS, and readings were taken immediately for the light emitted between 400 and 600 nm using a Mithras LB940 (Berthold Technologies). The BRET ratio was calculated using the following formula: (emission at 535 nm) − (emission at 485 nm) × Cf/(emission at 485 nm), where Cf corresponds to (emission at 535 nm)/(emission at 485 nm) for the receptor-Rluc construct expressed alone. The total fluorescence and luminescence were used as relative measures of the expression level of the acceptor (YFP) and donor (Rluc) proteins, respectively. After the cells were distributed in 96-well microplates, the total fluorescence of the cells was measured using an excitation filter of 485 nm and an emission filter of 530 nm. The same cells were kept in the dark for 18 min with coelenterazine H incubation for the determination of Rluc expression, as assessed by luminescence. Saturation curves were plotted with GraphPad Prism4 software using a nonlinear repression curve assuming one-site binding. The total luminescence activity also was determined for BiLC assay.
hPIII promoter activity analysis.
The activity of the generic PIII promoter (bp −480/−112) of the human prolactin receptor gene, which contains putative C/EBPβ and SP1 binding sites, was examined using a luciferase reporter system with human breast cancer cell lines MCF-7A2 (ER positive) (gift from E. Berleth, C. Roswell Park Cancer Institute, New York, NY) (28) and MDA-MB-231 (ER negative) (ATCC). MCF-7A2 was maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) and antibiotics under 95% air and 5% CO2. MDA-MB-231 was maintained in Leibovitz's L-15 medium containing 10% FBS and antibiotics under 100% air without CO2. Cells were cultured in medium containing 5% CTS for 2 to 3 days and 1% CTS for 2 to 3 days to eliminate endogenous estrogen (see Fig. 8 to 10). The length of CTS culture conditions varied slightly depending on the experimental design. Cells were transiently transfected with the pGL2 reporter gene plasmid with the insertion of hPIII promoter/exon 1 (bp −480/−112) (pGL2-PIII; 1 μg), the wild-type or mutated SP1 or C/EBPβ site alone or in combination (7), and the β-galactosidase gene (0.1 μg; for normalization) using Lipofectamine 2000. After 16 h, cells were treated with 17β-estradiol (100 nM) for 24 h and harvested for the determination of luciferase activity (Promega). For estrogen receptor-negative MDA-MB-231 cells, the plasmid (0.5 μg) expressing ERα (hERpcDNA 3.1) (4) also was included in the study.
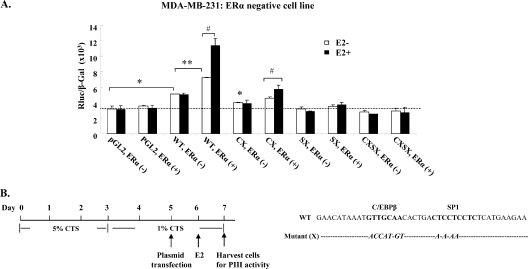
Fig. 8.
Effect of estrogen/ERα on hPRLR hPIII promoter activity in ERα-negative (MDA-MB-231) breast cancer cells. (A) hPIII promoter activity was analyzed in ERα-negative breast cancer cells transiently expressing human ERα (ERα+) in the presence or absence of E2 (100 nM; 16 h). ERα−, cells not expressing ERα. WT, wild-type pGL2 with hPIII promoter sequence (−480/−112) containing SP1 (S) and C/EBPβ (C) cis elements. X, mutated element. (B) Diagram of experimental procedure and sequences of mutated SP1 (SX) or C/EBPβ (CX). Data were presented as relative luciferase activities (Rluc) normalized to the activities of cotransfected β-galactosidase (β-gal). Results are the means ± standard errors from three individual experiments in triplicate. *, **, and #, P < 0.05.
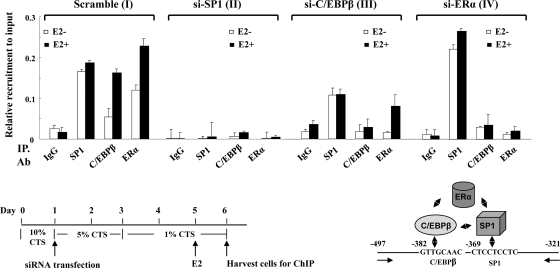
Fig. 10.
Recruitment of endogenous SP1-C/EBPβ-ERα onto hPIII promoter by ChIP analysis. The upper graphs show CHIP performed with chromatin from MCF-7A2 cells transiently transfected with the coding region of siSP1 (II), siC/EBPβ (III), or siERα (IV) (30 nM) followed by treatment with or without E2 (100 nM). Scramble siRNA (I) was used as the negative control. The promoter region of hPIII containing SP1-C/EBPβ (−497/−321) was analyzed by PCR following immunoprecipitation (IP) with the antibody indicated. IgG was used as a negative control. The lower panels show the experimental procedure and a diagram of protein complex interaction onto the hPIII promoter.
Real-time PCR quantitation of hPRLR mRNA.
MCF-7 cells 48 h after transfection with scramble siRNA (control) or with validated siRNA that had been designed to knock down the endogenous expression of SP1 or C/EBPβ (singly or combined) and also of ERα (as described below) were treated with 17β-estradiol (100 nM) for 24 h. Total RNA was isolated using an RNeasy (Qiagen) isolation kit. One μg of total RNA was used for reverse transcription (RT) reactions with the Superscript II first-strand RT synthesis kit (Invitrogen). One-step real-time quantitative RT-PCR was carried out essentially as previously described (4) using SYBR green quantitative PCR master mix and an ABI 7500 sequence detection system (Applied Biosynthesis, Foster City, CA). The primers utilized for PCR to detect hPRLR mRNA and to detect hPRLR mRNA transcribed from promoter hPIII were a specific forward primer from noncoding exon 1 (−135/−114) and a reverse primer from sequences located in common coding exon hE3 (+49/+71) (4). Results from two individual experiments of each sample were assayed in triplicate and normalized to the level of β-actin.
RNA interference.
siRNA against SP1, C/EBPβ, ERα, or scramble RNA (30 nM) (Ambion or Qiagen) were transiently transfected into MCF-7A2 cells using siPORT plus transfection reagent (Ambion) by following the company's protocol. After 16 h, the cells were incubated with 5% CTS (2 days) followed by 1% CTS (2 days) for hPRLR mRNA levels (Fig. 1D), promoter activity (see Fig. 9), and ChIP analyses (see Fig. 10). Cells were transiently transfected in 1% CTS with the designated expressing plasmid (0.5 μg) (see Fig. 9 and 10), pGL2-PIII (1 μg), and the β-galactosidase gene (0.1 μg, for normalization) using Lipofectamine followed by treatment with E2 or vehicle alone (0.01% alcohol; control) for hPIII promoter activity measurements. The siRNA for the coding region of SP1 (GCAACAUGGGAAUUAUGAATT), C/EBPβ (GGCCCU GAGUAAUCGCUUTT), and ERα (CAGGCACAUGAGUAACAAATT) were purchased from Ambion. siRNA derived from the noncoding region of SP1 (TTGGGTAAGTGTGTTGTTTAA), C/EBPβ (CCCGCCCGTGGTGTTATTTAA), and control scramble siRNA (AATTCTCCGAACGTGTCACGT) were purchased from Qiagen.
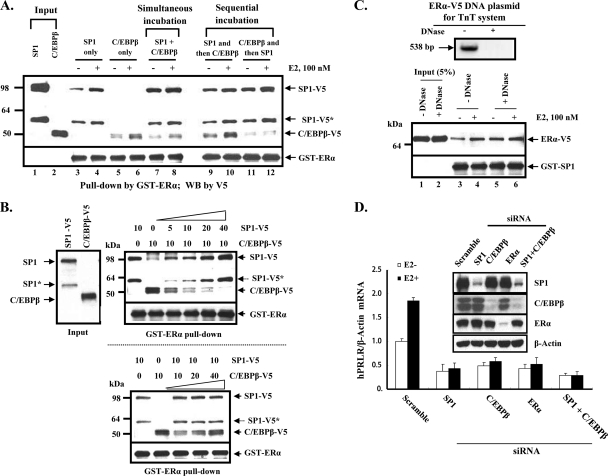
Fig. 1.
(A to C) In vitro protein complex formation of ERα with SP1 and C/EBPβ. (D) Inhibition of E2-induced PRLR mRNA by knockdown of SP1, C/EBPβ, and ERα. (A) Western blot of individual, simultaneous, or sequential incubation of in vitro-translated V5-tagged human SP1 and/or human C/EBPβ with GST-ERα (human; aa 185 to 595 with the deletion of the AF1 domain) in the absence or presence of E2 (100 nM). Results are representative of three independent experiments. (B) Dose-dependent effect of SP1 on C/EBPβ or of C/EBPβ on SP1 association with GST-ERα in the simultaneous incubation study. Increasing doses of TnT reticulocyte extracts (top, 5 to 40 μl for SP1-V5 with constant 10 μl C/EBPβ; bottom, 10 to 40 μl of C/EBPβ with constant 10 μl SP1) were incubated with GST-ERα. Following pulldown assays (see Materials and Methods) for the detection of SP1-V5 or C/EBPβ-V5 association with ERα, V5 antibody was used in the Western blot analysis. *, SP1 protein variant translated from an alternative ATG codon. (C) Protein-protein interaction was independent of the DNA template used for in vitro transcription/translation. The upper gel shows human ERα-V5 plasmid (1 μg), used for the TnT coupled reticulocyte lysate system, treated with DNase (100 U, 1 h, 25°C) and resolved in a 1% agarose gel. Five hundred thirty-eight bp of ERα-V5 was amplified by PCR with a primer (+1/537 bp; NM_000125). The lower gel shows 10 μl of TnT reticulocyte extract used in the study for the Western blotting of ERα-V5 pulldown by GST-SP1. (D) Effect of E2 on the expression of hPRLR mRNA in MCF7 cells with the knockdown of transcription factors (individually and combined). Cells were transfected with SP1 or C/EBPβ siRNA singly or combined or with ERα siRNA or scramble siRNA (control). After 48 h of transfection, cells were left untreated or were treated with 17β-estradiol (100 nM) for 24 h. hPRLR mRNA expression was determined by real-time PCR analyses of hPRLR transcripts from total RNA. The inset shows a Western blot of endogenous SP1, C/EBPβ, or ERα in cell lysates after siRNA treatment. Results are normalized to results for β-actin and are means ± standard errors of duplicate experiments in triplicate.
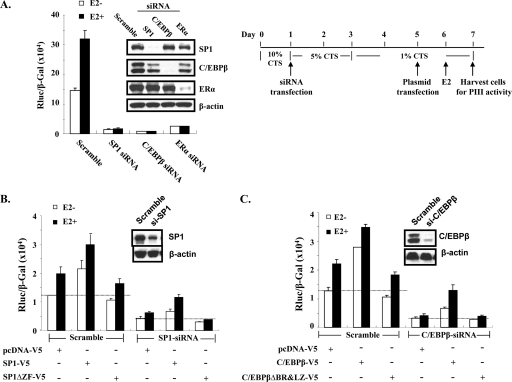
Fig. 9.
In vivo evidence of zinc fingers of SP1 and the basic leucine zipper of C/EBPβ required for protein complex formation to activate hPRLR hPIII promoter activity in ERα-positive (MCF-7A2) cells. (A) On the left, hPIII promoter activity was determined in MCF-7A2 cells transiently transfected with siRNA (30 nM) from coding region of SP1, C/EBPβ, or ERα. On the right is a diagram of the experimental procedure. (B) hPIII promoter activity was determined in MCF-7A cells transiently transfected with siRNA from the noncoding region of SP1 followed by vector alone (pcDNA-V5), wild-type SP1-V5, or the deletion of three zinc finger motifs (Sp1ΔZF-V5). (C) hPIII promoter activity was determined in MCF-7A cells transiently transfected with siRNA from the noncoding region of C/EBPβ followed by vector alone (pcDNA-V5), wild-type C/EBPβ-V5, or the deletion of both the basic region (BR) and the leucine zipper (LZ) (C/EBPβΔBR&LZ-V5). Scramble siRNA was used as a negative control. The inset shows a Western blot of endogenous SP1, C/EBPβ, or ERα in cell lysates after 4 days of transfection. β-Actin was used as the loading control.
ChIP.
ChIP assays were performed using the MAGnify kit (Invitrogen) with a minor modification of the company's protocol. Briefly, MCF7-A2 cells transfected with scramble siRNA (control) or other small interfering RNAs were washed, collected, and counted. A total of 2 × 106 cells were suspended in 1× PBS and cross-linked by 1% formaldehyde for 10 min at room temperature, followed by quenching with glycine. Cells were washed twice with 1× PBS and resuspended in 100 μl of lysis buffer and sonicated 12 times using a Misonix sonicator 4000 (Misonix) with a cup horn (75% power, 20-s pulse on and 60 s off) at 4°C. The shearing of chromatin was checked by visualizing small amounts of the sonicated sample on 1% agarose gels, and the samples of chromatin-sheared fragments between 200 and 1,000 bp were used for the subsequent immunoprecipitation step. Antibodies or IgG (2 to 4 μg) were incubated with the sonicated samples overnight at 4°C. After the reversal of cross-linked immunoprecipitated chromatin by heating the samples at 55°C for 15 min, DNA was purified with 50 μl of elution buffer. Chromatin purified from an equivalent number of cells not subjected to the immunoprecipitation step was used as the input control. ChIP samples were analyzed by SYBR green real-time quantitative PCR with standard conditions (20-μl reaction volume and 50 cycles) (ABI 7500). The sequences of the primers to amplify the hPIII promoter used in this study were the following: forward (−497), CTT CGC AGG ATT CCA GCT CCC CCA AC; reverse, (−321) GAA GCT CAA CTC GGT GCA CTT GTT C. Results were expressed as a relative value of recruitment to the input.
Statistical analysis.
The significance of the differences in the study between groups in the presence and absence of E2 was determined by Tukey's multiple-comparison test (one-way analysis of variance).
RESULTS
ERα forms a complex with SP1 and C/EBPβ in vitro: requirement of SP1 and C/EBPβ for E2-induced PRLR mRNA gene expression.
To examine the association of ERα with SP1 and C/EBPβ in vitro, we performed GST pulldown assays using purified GST-ERα expressed in bacterial cells. When GST-ERα was incubated with in vitro-translated V5-tagged SP1 or C/EBPβ, it was able to interact with either protein, and E2 increased the signal of this interaction (Fig. 1A, lane 3 versus 4 and lane 5 versus 6). However, when both SP1 and C/EBPβ were incubated simultaneously with GST-ERα, although ERα interacted with both proteins its association with C/EBPβ was significantly reduced in the absence or presence of E2 treatment (Fig. 1A, lane 5 versus 7 and lane 6 versus 8). The reduced association of C/EBPβ in the simultaneous reaction is indicative of a preferred interaction of SP1 with ERα. To further address this aspect, we utilized a sequential incubation approach (Fig. 1A, lanes 9 to 12). Preincubation with SP1 recruited more C/EBPβ to GST-ERα/SP1 complex with increased interaction by E2 compared to that observed with C/EBPβ in the initial incubation (Fig. 1A, lanes 9 and 10 versus lanes 11 and 12). A dose-related inhibitory effect of SP1 on C/EBPβ binding to the ERα in the simultaneous incubation study confirmed SP1 as the preferred interacting partner (Fig. 1B, top). In contrast, increasing concentrations of C/EBPβ showed no effect on the recruitment of SP1 to ERα (Fig. 1B, lower). Protein-protein interactions elucidated in this (Fig. 1A) and subsequent studies (Fig. 2 to 4) were specific and independent of protein-DNA interaction (the template used for in vitro translation), in contrast to a previous report showing interference by DNA association (15). The addition of DNase to the incubation of purified GST-SP1 with in vitro-translated ERα-V5 showed unchanged protein-protein interactions in either the absence or presence of E2 (Fig. 1C, lanes 3 and 4 versus lanes 5 and 6). These results suggest that SP1 is the preferred transfactor for recruiting ERα, and that this association facilitates the recruitment of C/EBPβ to the complex. Further studies demonstrated that endogenous PRLR mRNA induced by estradiol exposure (100 nM; 1-fold concentration) is significantly inhibited/abolished by the knockdown of SP1 or C/EBPβ or the combination of both (Fig. 1D). Comparable inhibition was observed in cells with the knockdown of ERα. Thus, the estradiol effect was abolished in cells exposed to all siRNA treatments. siRNAs were found to be highly effective in reducing/abolishing the expression of transcription factors SP1, C/EBPβ, and ERα (Fig. 1D, inset). These results demonstrate the endogenous relevance of SP1 and C/EBPβ in E2-induced PRLR gene expression in breast cancer cells and also indicate the cooperation of these transfactors in E2-induced hPRLR gene expression.
Fig. 2.
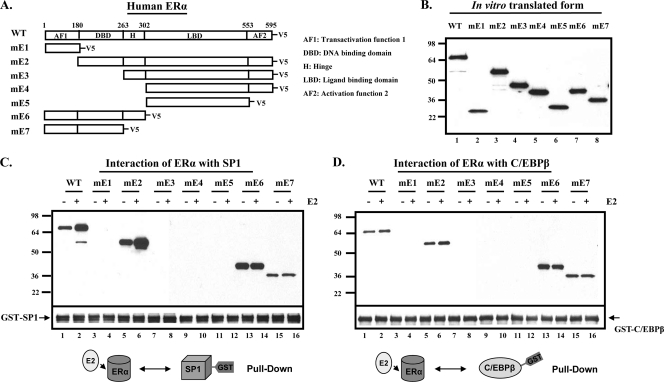
Identification of domain of ERα responsible for the interaction with SP1 or C/EBPβ. (A) A schematic presentation of the V5 epitope-tagged wild-type (WT) and deletion forms (mE1 to mE7) of human ERα. (B) In vitro-translated proteins from various forms of ERα were used in a pulldown assay. GST-SP1 (C) or GST-C/EBPβ (D) purified by GST-affinity resin was incubated with the in vitro-translated wild-type or deleted form of ERα-V5 in the presence or absence of 17β-estradiol (E2; 100 nM). The inputs and interacting forms of ERα-V5 were detected by Western blot analysis using V5 antibody. Results are representative of three independent experiments.
Fig. 4.
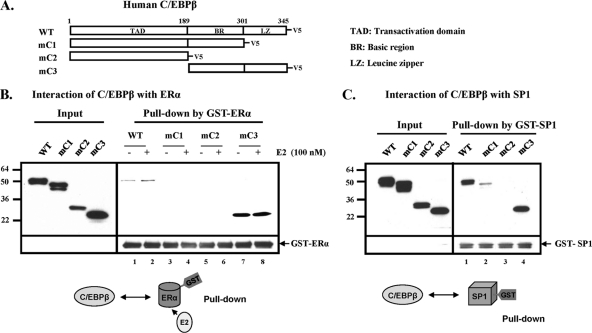
Identification of domains of C/EBPβ responsible for interaction with ERα or SP1. (A) A schematic presentation of V5 epitope-tagged wild-type (WT) or deletion forms (mC1 to mC3) of human C/EBPβ. GST-ERα (B) or GST-SP1 (C) was incubated with each form of in vitro-translated C/EBPβ-V5 in the presence or absence of 17β-estradiol (E2; 100 nM). The inputs and interacting forms of C/EBPβ-V5 were detected by Western blot analysis using V5 antibody. Results are representative of three independent experiments.
ERα domain responsible for the interaction with SP1 or C/EBPβ.
To identify the specific domain(s) of ERα required for the interaction with SP1 or C/EBPβ, in vitro-translated deletion constructs of V5-tagged ERα (Fig. 2B) were incubated with GST-SP1 or GST-C/EBPβ in the absence or the presence of E2 and subjected to pulldown assays. Consistently with results shown in Fig. 1, the direct constitutive interaction of WT ERα with each transcription factor was observed. Its association with SP1 was markedly enhanced by E2 (Fig. 2C, lane 1 versus 2), and this was less marked for C/EBPβ (Fig. 2D, lane 1 versus 2). The AF1 domain of ERα (mE1) showed no association with either SP1 or C/EBPβ (Fig. 2C and D, lanes 3 and 4). Furthermore, the deletion of the N terminus containing the AF1 domain (mE2) from the ERα did not affect its association with either transcription factor, and the interaction, as was the case for the wild type, was enhanced by E2 (Fig. 2C and D, lanes 5 versus 6). ERα constructs lacking the DBD but containing H, LBD, and AF2 (mE3); LBD and AF2 (mE4); or LBD alone (mE5) showed no interaction with SP1 or C/EBPβ (Fig. 2C and D, lanes 7 to 12). Strong interaction was present with shorter constructs containing DBD, AF1, and H but lacking the LBD and AF2 regions (mE6) (Fig. 2C and D, lanes 13 and 14). It is important to note that while the interaction was still present, marked reduction was observed in the absence of the H region (mE7) (Fig. 2C and D, lanes 15 and 16). The effect of E2 was evident with wild-type or mE2 mutant containing LBD, while mE6 and mE7, both lacking the LBD but containing the DBD, did not show an E2-dependent increase. These results clearly demonstrated that the DBD domain of ERα is required for the interaction with either SP1 or C/EBPβ. The Hinge region of the receptor, which is immediately adjacent to the C terminus of the DBD, seems to indirectly contribute to the ERα interaction with SP1 and C/EBPβ.
SP1 region of importance for interaction with ERα or C/EBPβ.
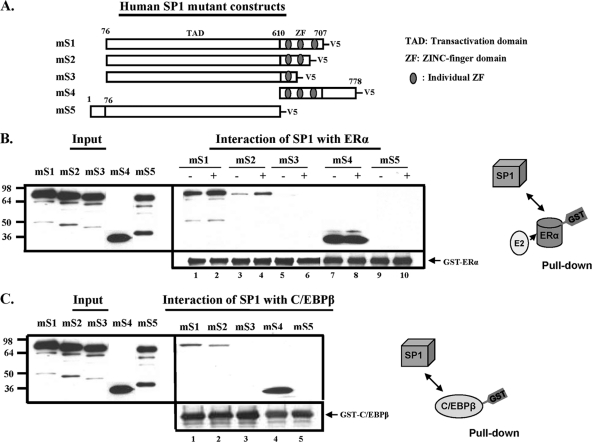
In subsequent studies, we identified the SP1 domain(s) involved in its interaction with ERα or C/EBPβ utilizing pulldown assays of in vitro-translated V5-tagged SP1 deletion constructs incubated with GST-ERα or GST-C/EBPβ. The SP1 construct (mS1; aa 76 to 707) with the three Zn finger (ZF) regions (aa 610 to 707) that lack the N-terminal (aa 1 to 76) and C-terminal (aa 708 to 778) domains displayed marked interaction with ERα (Fig. 3B, lane 1) that was enhanced with E2 (lane 2). The deletion that excluded the third ZF (mS2) and immediate adjacent sequences showed significant reduced interaction with ERα that was enhanced by E2 (Fig. 3B, lanes 3 and 4). The additional ablation of the second ZF (mS3) with immediately adjacent sequences completely abolished the SP1 interaction with ERα (Fig. 3B, lanes 5 and 6). Furthermore, the ZF region with the C-terminal sequence (mS4) showed marked interaction with ERα (Fig. 3B, lanes 7 and 8). On the other hand, the deletion of the ZF region (aa 610 to 707) along with the C terminus of SP1 completely abolished its interaction with ERα (mS5) (Fig. 3B, lanes 9 and 10). Similarly, the deletion of the ZF3 region of SP1 (mS2) decreased its interaction with C/EBPβ compared to that with mS1 (containing all ZFs) (Fig. 3C, lane 2 versus 1), and no detectable signal was observed with mS5 containing only the first ZF (mS5), while a strong interaction was observed with the short SP1 sequence containing the three zinc fingers and the C-terminal domain (mS4). These studies revealed that the zinc fingers of SP1 are important for the interaction with ERα or C/EBPβ. Although the third ZF contributes to the interaction, the second ZF appears to be an essential contributor.
Fig. 3.
Identification of domain of SP1 responsible for interaction with ERα or C/EBPβ. (A) A schematic presentation of V5 epitope-tagged deletion forms (mS1 to mS5) of human SP1. GST-ERα (B) or GST-C/EBPβ (C), purified by GST affinity resin, was incubated with various in vitro-translated deletion forms of SP1 in the presence or absence of 17β-estradiol (E2; 100 nM). The inputs and interacting forms of SP1-V5 were detected by Western blot analysis using V5 antibody. Results are representative of three independent experiments.
Basic region and leucine zipper of C/EBPβ participate in the interaction with ERα or SP1.
To determine the interacting domain of C/EBPβ with ERα or SP1, in vitro-translated V5-tagged C/EBPβ wild type and deletion mutants were used for GST-ERα or GST-C/EBPβ pulldown assays (Fig. 4). C/EBPβ with the deletion of the basic region and/or leucine zipper (LZ; mC1 and mC2) failed to interact with ERα, whereas C/EBPβ lacking the N-terminal transactivation domain (mC3) displayed strong interaction (Fig. 4B, lanes 7 and 8 versus lanes 3 to 6). This indicated that the leucine zipper of C/EBPβ participated in the interaction with ERα. The mC3 mutant containing only the basic region and leucine zipper did not show the increase in binding activity by E2 that was observed with the WT. Further studies revealed C/EBPβ interaction with SP1 (Fig. 4C). In this case, the deletion of the transactivation domain did not affect C/EBPβ interaction with SP1 (mC3), while the deletion of the LZ region significantly reduced the interaction (mC1), and the deletion of both the basic region and leucine zipper completely eliminated the interaction. Thus, while the LZ region is important for interaction, the basic region appears to be essential for C/EBPβ interaction with SP1.
Transfactor-bound DNA required for recruitment of ERα to hPIII promoter.
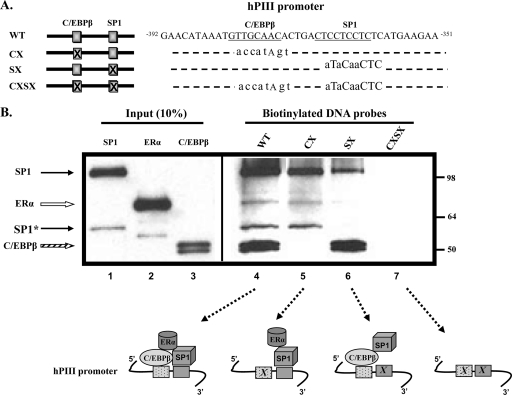
Because ERα is tethered to SP1 and C/EBPβ, which are bound to their cognate sites at the PIII promoter of the hPRLR gene (4, 10, 12), the ERα association with C/EBPβ and SP1 that we observed in GST pulldown assays could differ from that found in the presence of the DNA sequence containing both relevant sites. Thus, we next examined ERα recruitment to C/EBPβ and SP1 (V5 tagged) bound to the biotinylated double-stranded DNA oligonucleotide containing the sequence spanning from −392 to −351 of hPIII of PRLR, which included both C/EBPβ and SP1 binding sites (Fig. 5A). Also, the effect of mutations of each or both sites was analyzed. As shown in Fig. 5B, the wild-type oligonucleotide-bound C/EBPβ and SP1 proteins recruited ERα, while the SP1 mutation (SX) severely impaired/abolished ERα recruitment, as shown by its complete absence from Western blots, while it did not affect C/EBPβ binding (lane 6 versus 5). The minor SP1 band observed in this case likely resulted from SP1 direct association with C/EBPβ bound to its DNA binding site (Fig. 4C). This indicated that SP1 bound to DNA is required for the full recruitment of ERα, and that C/EBPβ bound to DNA did not recruit ERα in the absence of SP1-bound DNA. We also observed that when the C/EBPβ site was mutated (CX), the unbound C/EBPβ did not associate with DNA-bound SP1, as shown by its complete absence from Western blots (Fig. 5B, lane 5). In this case, ERα recruitment by SP1 bound to DNA was somewhat reduced (lane 5). This observation suggested that DNA-bound C/EBPβ stabilized the DNA binding of SP1 associated with ERα. The double mutation of C/EBPβ and SP1 sites (CXSX; negative control) showed no associated proteins. Taken together, the data of DAPA studies (Fig. 5B) and GST pulldown interactions (Fig. 1) suggest that SP1 bound to DNA is required for recruiting ERα to the complex at the PIII promoter, and C/EBPβ contributes to the stabilization of the complex.
Fig. 5.
Association of ERα, C/EBPβ, and SP1 with the E2-responsive sequence of hPIII promoter of human prolactin receptor gene. (A) A schematic presentation of biotinylated DNA probes used in the DAPA experiment. DNA sequences within the hPIII promoter and putative C/EBPβ and SP1 binding motifs are indicated. (B) Wild-type and mutant probes (X; sequences are indicated in lowercase letters) were incubated together with in vitro-translated wild-type human SP1-V5, ERα-V5, and C/EBPβ-V5. Proteins associated with each DNA probe were detected with Western blot analysis using V5 antibody. SP1-V5, solid and dotted arrows; ERα-V5, open arrow; C/EBPβ-V5, striped arrow. *, SP1 protein variant translated from an alternative ATG codon. Results are representative of at least three separate experiments.
Interactions between ERα, C/EBPβ, and SP1 occur in living cells.
Following the determination of the domains responsible for interactions between ERα, C/EBPβ, and SP1 in vitro, we next examined the interactions in vivo by employing the BRET assay for the detection of protein-protein interaction, and the maximal BRET ratio obtained for a given pair (BRETmax) and the relative affinity of the acceptor/donor ratio required to reach half-maximal BRET (BRET50) values were derived from saturation curves (Fig. 6A to F). In this study, we used luciferase (Rluc) as the donor and yellow fluorescent protein (YFP) as the acceptor and prepared expression plasmids for full-length ERα, C/EBPβ, and SP1, which were tagged at their C or N terminus with Rluc or YFP. The Rluc and YFP fusion constructs were tested for their expression, luminescence, or fluorescence activities. Since the orientation of Rluc and YFP on the fusion proteins could affect the efficiency of energy transfer between interacting proteins, all possible pairs of N- and C-terminal fusion proteins were screened, and those showing optimal BRET signals were employed in BRET assays (see Materials and Methods).
Fig. 6.
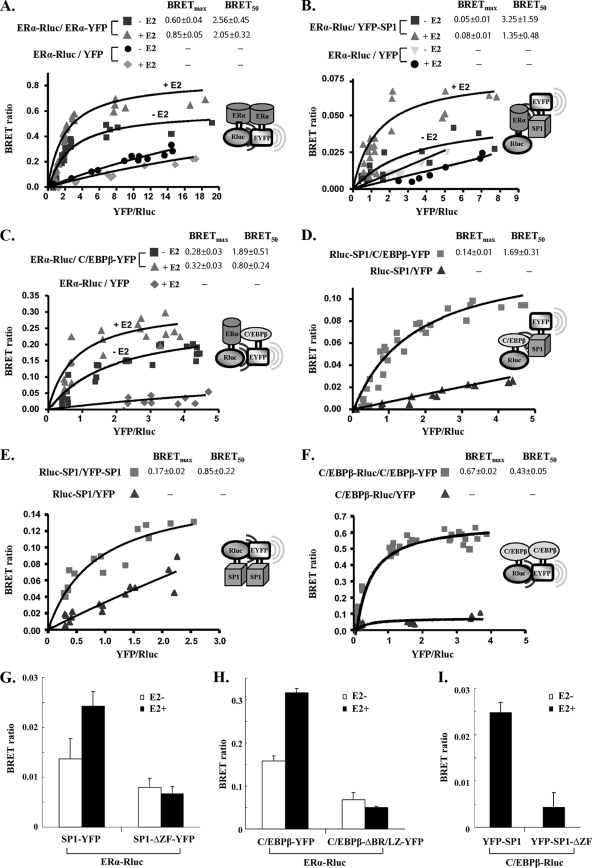
Interactions of ERα-ERα, ERα-C/EBPβ, ERα-SP1, SP1-C/EBPβ, SP1-SP1, and C/EBPβ-C/EBPβ in living mammalian cells. A fixed amount of plasmids for Rluc-tagged donor (0.2 μg of ERα-Rluc, Rluc-SP1, or C/EBPβ-Rluc) was cotransfected into COS1 cells with the following increasing doses of YFP-tagged acceptor plasmid: ERα-YFP of 0.025 to 1 μg with ERα-Rluc (A); YFP-SP1 of 0.1 to 0.8 μg with ERα-Rluc (B); C/EBPβ-YFP of 0.1 to 0.8 μg with ERα-Rluc (C); C/EBPβ-YFP of 0.05 to 1 μg with Rluc-SP1 (D); YFP-SP1 of 0.05 to 2 μg with Rluc-SP1 (E); and C/EBPβ-YFP of 0.05 to 2 μg with C/EBPβ-Rluc (F). In all cases, YFP alone as the acceptor together with the respective Rluc fusion plasmid as the donor were used as the negative control. BRET levels were plotted as a function of the ratio of the expression level of the YFP construct to the Rluc construct (YFP/Rluc). BRETmax, the maximal BRET ratio obtained for a given pair; BRET50, relative affinity of the acceptor/donor ratio required to reach a half-maximal BRET value. (G to I) The BRET ratio was determined when fixed amounts of ERα-Rluc (0.1 μg) (G and H) or C/EBPβ-Rluc (0.1 μg) (I) were cotransfected into COS1 cells with the YFP-tagged wild type (0.2 μg) or corresponding mutant (0.2 μg). SP1-ΔZF-YFP, zinc finger motif-deleted mutant; C/EBPβ-ΔBR/LZ-YFP, basic region and leucine zipper-deleted mutant. Data are expressed as means ± standard deviations from four independent assays.
We initially examined the interaction of the ERα-YFP/ERα-Rluc pair transfected into mammalian cells (HEK293 or COS1) in the absence or presence of E2. The saturation curves showed a significant BRETmax ratio in the absence of E2 that was enhanced by the addition of the hormone (Fig. 6A). No significant changes in BRET50 were observed. The saturation curve indicated that ERα was able to dimerize independently of E2, and an increase in ERα homodimerization was induced by E2 in vivo. Similar BRET ratios were observed for the interaction of ERα-Rluc-YFP-SP1 (Fig. 6B) and ERα-Rluc-C/EBPβ-YFP (Fig. 6C) in the absence of E2, and a significant increased in BRETmax was induced by E2. No changes in BRET50 were observed. Lastly, we assessed the C/EBPβ-YFP and Rluc-SP1 interactions. As shown in Fig. 6D, the BRETmax reached a BRET ratio of 0.10 at saturation. These observations have demonstrated in vivo interactions between ERα, C/EBPβ, and SP1 and between C/EBPβ and SP1. The findings have further proven and strengthened the initial findings derived from the in vitro interactions between the proteins. SP1, like ERα and C/EBPβ, forms a constitutive homocomplex in vivo (Fig. 6E). As expected, C/EBPβ shows strong activity of homo-complexing (homodimerization) in vivo (Fig. 6F).
These binary interactions were further confirmed using mutant C/EBPβ and SP1 BRET constructs in which the participating interacting regions identified in the GST pull-down study were deleted (Fig. 2 to 4). BRET ratios of ERα-Rluc with mutant Sp1-ΔZF-YFP (Fig. 6G) or C/EBPβ-ΔBR/LZ-YFP (Fig. 6H) were significantly reduced compared to those in the absence of E2 treatment and compared to those from the wild type (SP1-YFP or C/EBPβ-YFP). E2 treatment did not cause further increases in the BRET signal of mutant constructs or the wild type (Fig. 6G and H). A significant decrease in BRET signal also was observed for cells transfected with C/EBPβ-Rluc and the zinc finger-deleted SP1 mutant (YFP-SP1-ΔZF) compared to the level for wild-type YFP-SP1 (Fig. 6I). These in vivo studies validate interacting domains (basic region and leucine zipper of C/EBPβ and zinc finger domains of SP1) between three transfactors mapped in the in vitro GST pulldown study (Fig. 2 to 4).
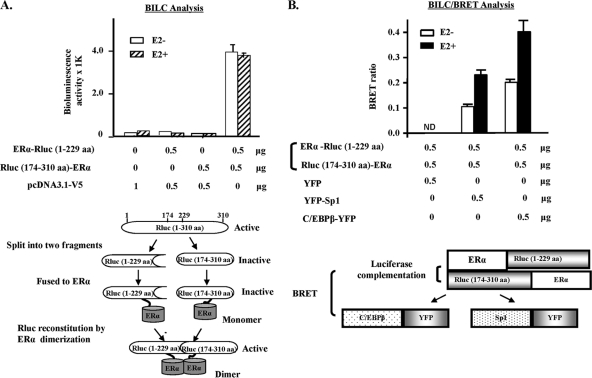
ERα exists as a dimer and E2 enhances ERα interaction with C/EBPβ or SP1 through stabilizing ERα homodimer.
BRET studies indicated the presence of ERα homodimerization in the absence of E2 (Fig. 6A). To further establish the presence of ERα constitutive dimers and the effect of E2, we utilized a protein complementation assay. This assay is based on the detection of protein complex formation in which the reporter protein (Rluc in this study) was split into two fragments and fused to each of two interacting proteins. The interaction of two proteins brings the split fragments into close proximity to allow the reconstitution of its enzymatic activity. A pair of two reporter fragments containing an overlapping region was prepared, because these could facilitate the reconstitution of two fragments (6). The fragments of aa 174 to 310 and aa 1 to 229, which share 55 overlapping amino acids (aa 174 to 229), were used for the study. As was the case for untransfected control cells, no luminescent activity was observed when cells were transfected with individual complementation constructs (Fig. 7A). The significant induction of luminescent activity was observed with the pair of Rluc(1-229 aa)-ERα/ERα-Rluc(174-310 aa). E2 did not exhibit an effect in any of the pairs tested for luminescent activity. The complementation construct [Rluc(1-229aa)-ERα/ERα-Rluc(174-310 aa)] was used further for BRET analyses to assess if Rluc reconstituted due to ER dimerization would interact with C/EBPβ-YFP or YFP-SP1 (Fig. 6B and C). A significant BRET ratio of this reconstituted Rluc resulting from the ERα dimer interacting with C/EBPβ-YFP or YFP-SP1 was observed, and these interactions were significantly increased by E2 (Fig. 7B). These findings have led us propose that E2 stabilizes preformed ERα dimer and ultimately favors its interaction with SP1 and/or C/EBPβ.
Fig. 7.
ERα exists as a dimer form, and E2 stimulates its interaction with SP1 or C/EBPβ. (A) Bioluminescent activities of Rluc complementation constructs in frame with human ERα when transfected into COS1 cells. (B) BRET assays for interaction of luciferase-complemented ER constructs [ERα-Rluc(1-229 aa) + Rluc(174-310 aa)-ERα] with C/EBPβ-YFP or YFP-SP1 when cotransfected into COS1 cells in the presence or absence of 100 nM E2. ND, nondetectable. Data are expressed as means ± standard deviations from three independent assays.
Mutual interactions between the three transfactors on the PIII promoter: prerequisite to E2 inducibility.
To further elucidate the physiological relevance of findings derived from in vitro GST pulldown and in vivo BRET analysis of COS1 cells, the E2/ERα activation of the human PRLR gene via SP1-C/EBPβ by binding to the respective cis element (SP1 and C/EBPβ) in the hPIII promoter was examined in PRLR target breast cancer ERα-negative cells (MDA-MB-231) transiently expressing ERα or empty control vector. Significant hPIII promoter activity was observed in the MDA-MB-231 cells transfected with the hPRLR PIII promoter in the absence of ERα (Fig. 8A, WT, ERα− versus pGL2, ERα−) with no further stimulation by E2, suggesting the presence of basal E2/ERα-independent PIII promoter activity. Upon the cotransfection of an ERα-expressing plasmid in these ER-negative cells alone, hPIII promoter activity was significantly increased compared to that of cells lacking ER (Fig. 8A, WT, ERα+ versus WT, ERα−). PIII promoter activity was further enhanced by E2 treatment, which is consistent with the earlier study of the estrogen-induced transcriptional activation of the hPRLR gene found in ER-positive MCF7 cells (4). The mutation of the SP1 (SX) binding element alone or in combination with C/EBPβ (CXSX) showed no promoter activity in cells (of a pGL2 vector-negative control group) regardless of the presence or absence of ERα expression (Fig. 8A, SX, ERα+ and SX, ERα− or SXCX, ERα+ and SXCX, ERα−). In contrast, minimal PIII promoter activity was noted in the cells transfected with the hPIII-luciferase construct with intact SP1 but mutated in C/EBPβ in the absence of ERα (Fig. 8A, CX, ERα−), and E2 caused no further activation of hPIII promoter activity. In cells expressing ERα (Fig. 8A, CX, ERα+), E2-induced PIII promoter activity was significantly enhanced compared to that of cells in the absence of E2 treatment. These results support the relative greater importance of SP1 compared to that of C/EBPβ to tether ERα to the complex at the promoter required for E2-stimulated transcriptional activity (Fig. 1 and 8 to 10).
To further explore the physiological significance of the ERα-SP1-C/EBPβ complex requirement for E2-induced PIII promoter activity, we investigated endogenous complex formation and its effect on transcription in ERα-positive human breast cancer MCF-7 cells using an RNA interference approach (Fig. 9). Basal and E2-activated PIII promoter activity (scramble group) was almost abolished in ER-positive MCF-7 cells when endogenous SP1 or C/EBPβ protein was depleted by specific siRNA knockdown (Fig. 9A, scramble versus SP1 siRNA or C/EBPβ siRNA). Residual hPIII promoter activity remained in cells of knocked down ERα only (ERα siRNA), which was consistent with the earlier finding of minimal basal promoter activity in ER-negative cells (Fig. 8A, WT, ERα). The interaction domain of SP1 or C/EBPβ demonstrated in BRET analysis (Fig. 6G to I) also was assessed through hPIII promoter activity in MCF7 cells overexpressing the wild type or mutant (Fig. 9B and C). In the scramble control group, basal and E2-activated hPIII promoter activities (pcDNA-V5) were significantly increased in cells overexpressing wild-type SP1 (SP1-V5) (Fig. 9B) or C/EBPβ (C/EBP-V5) (Fig. 9C). hPIII promoter activity remained at the control endogenous level (pcDNA-V5) for cells expressing deleted zinc fingers of SP1 (SP1ΔZF-V5) or the basic region and leucine zipper of C/EBPβ (C/EBPΔBR/LZ-V5).
Similar results were found in a recovery study when cells were first knocked down of endogenous SP1 (SP1-siRNA) or C/EBPβ (C/EBP-siRNA) (Fig. 9B and C) using a noncoding region of the respective siRNA without interfering with the expression of the wild type or a mutant plasmid transfected on day 5 (Fig. 9A, right). Significant basal and E2-stimulated hPIII promoter activity was observed in wild-type SP1 or C/EBPβ-expressing cells, whereas no activity was observed in the mutant constructs (Fig. 9A and B). Both the wild type and mutant were expressed at the predicted protein size (Fig. 3 and 4).
The combination of ChIP and RNA interference approaches was further used to determine if endogenous ERα-SP1-C/EBPβ complex could form onto the PIII promoter to activate hPRLR gene transcription by E2. In the control cells with scramble RNA treatment, the recruitment of C/EBPβ or ERα but not SP1 was inducible by E2 (Fig. 10, I). When SP1 was depleted by si-SP1 RNA in MCF-7 cells (Fig. 10, II), neither C/EBPβ nor ERα was recruited to the endogenous PIII promoter. Cells with the depletion of C/EBPβ (si-C/EBPβ) showed the recruitment of SP1 and ERα (Fig. 10, III). Only SP1 was recruited to the PIII promoter when ERα was knocked down by si-ERα (Fig. 10, IV). E2 does not display any effect on SP1 recruitment in either case (Fig. 10, III and IV). These studies indicate a major role of SP1 in the formation of ERα-SP1-C/EBPβ and the consequent transcriptional activity of the hPRLR gene in breast cancer cells.
DISCUSSION
These studies have demonstrated direct interactions and complex formation among the transcription factors ERα, SP1, and C/EBPβ, which were previously found to participate in the ERE-independent mechanism (4) by which estrogen regulates hPRLR gene transcription through its generic promoter III. The DNA binding domain (DBD) of ERα, basic region and leucine zipper (bZIP) of C/EBPβ, and zinc finger (ZF) motifs of SP1 were mapped as the physical interacting domains. BRET saturation analysis demonstrated E2-stimulated protein-protein interaction in a live cell system. Bimolecular luminescence complementation (BiLC) assay further revealed the existence of ERα as a dimer in the absence of its ligand. The complex formation of ERα dimer with SP1 or C/EBPβ was highly enhanced by the addition of E2. Together, PIII promoter activity, assessed in breast cancer cells by the combination of the knockdown of individual and combined members of the activating complex and ChIP analysis (Fig. 8 to 10), and the endogenous expression of PRLR have provided definitive physiological evidence for the requirement of transfactors C/EBPβ and SP1 in the E2 activation of hPRLR transcription through the PIII promoter.
ERα-estrogen activation via association with transcription factors (13, 21, 26) bound to DNA sites is of functional importance for the regulation of transcription of certain target genes in the absence of an ERE. In the case of PRLR, we reported earlier on the participation of SP1 and C/EBPβ bound to its cognate site at the PRLR promoter in the transcriptional E2/ERα activation of the gene. E2/ERα association with SP1 and C/EBPβ bound to their cognate elements with the recruitment of the coactivators TFIIB and polymerase II is required for the E2-activated transcription of hPRLR through the PIII promoter (4). In the present studies, GST-ERα pulldown analyses of either the simultaneous or sequential addition of SP1 and/or C/EBPβ (Fig. 1) have provided initial evidence on the assembly of these transfactors. Compared to SP1, C/EBPβ was a weak interacting partner with ERα within the complex, as shown in the simultaneous incubation (Fig. 1A, lane 7). The efficiency of the recruitment of C/EBPβ was highly increased when ERα/SP1 complexes were preformed in sequential studies (Fig. 1, lane 9). However, in experiments with simultaneous addition where both SP1 and C/EBPβ were equally available to be pulled down by ERα, minor C/EBPβ association was observed (Fig. 1, 1ane 7). The relevance of DNA-bound SP1 as the preferred interacting partner of ERα (Fig. 5, 8 to 10) was further supported by its dose-dependent inhibitory effect on C/EBPβ association with ERα (Fig. 1C, top). This led us to propose that the association of ERα and SP1 enhances the recruitment of C/EBPβ into the complex. DAPA experiments further indicated the requirement of SP1-bound DNA for ERα recruitment (Fig. 5, lane 6 versus lanes 5 and 4), since the SP1 direct association with C/EBPβ, as represented by the weak band shown in Western blots using the promoter sequence with a mutated site (SX), was unable to recruit ERα. Because both transcription factor' cis elements are required for basal and E2-stimulated PRLR promoter activity (4, 10, 12), the binding of C/EBPβ to its cognate element could be strengthened after its recruitment to the preformed ERα/SP1 complex. In addition to the C/EBPβ association to the complex through ERα, our studies have indicated a strong direct interaction between C/EBPβ and SP1, which may serve to stabilize the ERα-SP1-C/EBPβ complex.
BRET saturation profiles provided further evidence for the complex formation of ERα/SP1, ER-C/EBPβ, and SP1-C/EBPβ in living cells (Fig. 6). Saturation analyses with a hyperbolic curve in each given pair indicated the specificity of constitutive interaction. However, the maximal BRET level presented in this study cannot be used as a quantitative measure of dimers to extrapolate findings from the in vitro pulldown analyses between ERα/SP1 and ERα-C/EBPβ, since the BRET value relates to the dimer numbers and the distance between donor and acceptor of the expressing fusion protein (20). The distance might vary from 10 to 100 Å, and therefore the efficiency of energy transfer would be different for each pair. This variable cannot be determined by current BRET technology. The BRET value also could be influenced by the relative position of YFP and Rluc in the individual fusion protein and also by its expression level in the cells (22). The ERα construct with a C-terminal fusion with Rluc or YFP (ERα-Rluc and ERα-YFP) delivered a robust BRET signal paired with the acceptor or donor of interest (SP1 and C/EBPβ) compared to the signal of the N-terminal design. In the case of C/EBPβ and SP1, C-terminal YFP fused to C/EBPβ (C/EBPβ-YFP) and N-terminal YFP or Rluc fused to SP1 (YFP-SP1 and Rluc-SP1) were proven as the most efficient resonance energy transfer partners (not shown). A significant E2 effect on the complex formation of SP1 with ERα or C/EBPβ with ERα was observed. The effect of E2 in the ERα-SP1 interaction was similar to that reported for fluorescence energy transfer (FRET) (13). These studies support the concept of ERα's functional participation in the transcriptional activation of the PRLR gene.
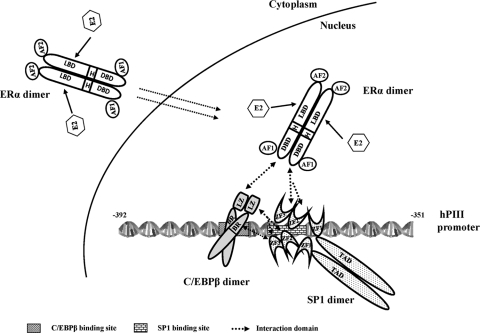
Our BRET studies also demonstrated that ERα dimerization is independent of ligand binding, with findings that were consistent with those of a study using FRET analysis (27). However, it is of interest that E2 caused a significant increase in BRET signal without changing the affinity for forming dimers. This is in contrast to the proposal of the strength of ERα dimeric association on ligand binding derived from FRET-based monomer exchange studies in the absence of a cellular environment (27). Although the reason(s) for this discrepancy is not clear, it is probably due to the different analytical approaches employed. More specifically, we used living cells in our studies, while a cell-free system was utilized for thermodynamic and kinetic measurements in an earlier work (27). To further analyze the interaction of SP1 or C/EBPβ with ERα, we utilized a renilla luciferase gene (Rluc)-based protein fragment complementation approach. Because ERα could exist as a stable dimer in the absence of ligand binding, split Rluc fragments (aa 1 to 229 and aa 174 to 310 aa, with 55 aa overlapping) were placed in frame with ERα. Using this system, we established the presence of constitutive ERα dimer formation by luminescence activity. This led us to speculate that homodimers of ERα complex with SP1 or C/EBPβ. This aspect was conclusively demonstrated by BRET, where the observed ERα dimer (reconstituted Rluc due to ER dimerization) basal interaction with C/EBPβ-YFP or YFP-SP1 was highly increased by E2 (by 200 to 300%) (Fig. 7B). During the course of the study, we also demonstrated by BRET the presence of the homodimerization of SP1 with high affinity (Fig. 6E). The physical interaction between SP1 monomers has been described in an earlier study utilizing a cross-linking reagent (3). Also, an indication of the functional importance of SP1-SP1 interaction was derived from studies where transcription was superactivated when a DNA binding-deficient mutant of SP1 interacted with pro-moter-bound wild-type SP1. Thus, it is reasonable to assume that a constitutive SP1 dimer has a role in modulating promoter activity. Given that C/EBPβ is known to be present as a dimer (24) (Fig. 6F), it is conceivable that SP1-ERα-C/EBPβ complex formation results from the bridging of three paired homodimerized proteins (ERα/ERα, SP1/SP1, and C-EBPβ C/EBPβ) through the heterodimerization of individual monomers from each homodimer (Fig. 11).
Fig. 11.
Model for E2-ERα-SP1-C/EBPβ formation and binding to the hPIII promoter of the human prolactin receptor gene. Schematic presentation of the functional domain of ERα, SP1, and C/EBPβ and the cognate DNA binding site of SP1 and C/EBPβ in the hPIII promoter (bp −391 to −351). Each protein is constitutively present as a homodimer. Arrows indicate the interaction at the specific domain between the proteins. ERα, AF transactivation function domain; DBD, DNA binding domain; H, hinge region; LBD, ligand binding domain; SP1 TAD, transactivation domain; ZF, zinc finger motif; C/EBPβ BR, basic region; LZ, leucine zipper domain.
The identification of the protein domain(s) responsible for assembling multiple protein complexes is essential to gain insights into numerous cellular processes. In this study, we mapped domains important for E2-inducible ERα, SP1, and C/EBPβ complex formation, which is essential for E2-induced PRLR transcription through promoter III. GST pulldown analysis revealed that ERα-DBD (Fig. 2), SP1-ZFs (Fig. 3), and the C/EBPβ-bZIP region (Fig. 4) were involved, and that E2 increased the recruitment of ERα to SP1 or C/EBPβ. The DNA binding domain of ERα is known to be involved in tethering ERα to DNA-bound transcription factors, including SP1, AP1, NF-κB, and C/EBPβ, for the positive or negative regulation of transcription (1, 13, 14, 18, 25, 26). Thus, ERα-DBD appears to be a common domain that is responsible for the tethering of ERα to transcription factors bound to ERE-lacking gene promoters. The relevance of the DBD was further indicated by our transfection studies of ERα deletion constructs in cells that lack ERα, where the DNA binding domain of ERα was shown to be essential for the transcriptional activation of the PRLR through promoter III (4).
Structural and mutational studies defining regions of ZF responsible for protein-protein interaction have indicated the involvement of ZF residues in the α-helix and β-sheet, as well as in the linker region between ZFs depending on the interacting partner proteins (2). In this study, we demonstrated that the second and third ZFs of SP1 are important for its association with ERα and C/EBPβ (Fig. 3). This is consistent in part with earlier FRET studies (13) showing a relevant role of the C-terminal domain of SP1 for its interaction with ERα in breast cancer cells. Our pulldown assay further showed the leucine zipper as the major interaction domain of C/EBPβ for its association with ERα or SP1. The repression of interleukin-6 promoter activity involved the interaction of ERα with the C/EBPβ leucine zipper domain (26). In the case of SP1, the interaction domain of C/EBPβ further extends to the distal basic region, which is the DNA binding domain of the protein. This additional interaction between ZF of SP1 and the basic region of C/EBPβ might stabilize the C/EBPβ complex in the complex at the promoter.
The physiological relevance of the requirement of complex formation (ERα-SP1-C/EBPβ) for hPRLR hPIII-directed transcription derived from in vitro studies (Fig. 1 to 7) has been demonstrated clearly using ChIP and RNA interfering strategies applied to ERα-negative (MDA-MB-231) (Fig. 8) and -positive (MCF-7A2) (Fig. 9 and 10) breast cancer cells expressing the PRLR gene. For cells with SP1 or C/EBPβ knockdown, as expected the exogenous expression of SP1 or C/EBPβ restored the transcriptional activities induced by E2 in ERα-positive MCF-7A2 cells. In contrast to SP1 or C/EBPβ knockdown, the expression of mutant Sp1 (Sp1ΔZf-V5; deletion of the zinc finger domain) and C/EBPβ (C/EBPβΔBR&LP-V5; deletion of the basic region and leucine zipper) failed to restore the activity (Fig. 9B and C). The recovery effect of the C/EBPβ and SP1 transfactors on PIII promoter activity in the knockdown experiments, together with the requirement of ERα expression in MDA-MB-231 to restore the hPIII promoter activity, indicated the importance of protein-protein interactions among three transfactors on the promoter for the E2-induced transcriptional activation. Further ChIP/siRNA knockdown study (Fig. 10) in MCF7-A2 cells demonstrated SP1 constitutively binding to its cognate site on the promoter and the E2-induced recruitment of ERα and C/EBPβ proteins through the interaction between SP1, C/EBPβ, and ERα in WT cells (transfected with scramble siRNA). These recruitments were almost abolished or significantly reduced in cells that were individually knocked down by their respective specific siRNAs. The nearly complete loss of the transfactor associations at the promoter was observed in the SP1-knocked-down cells, further indicating that SP1 protein plays a major role in the assembly of the complex. SP1 recruitment still was robust in the C/EBPβ- or ERα-knocked-down cells, while the recruitment of ERα in the C/EBPβ knockdown or the recruitment of C/EBPβ in the ERα knockdown to the promoter was significantly reduced and/or abolished, respectively. It is noteworthy that ERα recruitment was detectable in the cells with the knockdown of C/EBPβ but not SP1, which resulted from its interaction with DNA-bound SP1 protein. These studies reaffirmed the major role of SP1 in the formation of the complex.
Since we have demonstrated the presence of the constitutive homodimer formation of ERα, SP1, and C/EBPβ, it is reasonable to assume that the complex formation of ERα-SP1-C/EBPβ relies on the specific binding domain of each monomer from the individual homodimerized protein (Fig. 11). We propose that the mechanism of ERE-independent ERα action on the PRLR gene is initiated by the binding of SP1 and C/EBPβ to its cognate sites located in close proximity to the promoter. The ERα-SP1-C/EBPβ complex was formed by the association of the DNA binding domain of ERα with the ZF motif of SP1, followed by recruiting C/EBPβ through its leucine zipper domain. The interaction between zinc fingers of SP1 and the basic region/leucine zipper domain of C/EBPβ stabilizes the ERα-SP1-C/EBPβ complex on the promoter to activate PRLR gene transcription. The stimulatory effect of E2 on the dimerization of ERα-ERα, ERα-SP1, and ERα-C/EBPβ and on ERα translocation to the nucleus leads to an increase in overall ERα-SP1-C/EBPβ complex formation and the subsequent activation of hPRLR gene expression. Overall, the results from in vivo experiments demonstrated that interactions between the three transfactors through ERα tethered to DNA-bound C/EBPβ and SP1 on the hPRLR PIII promoter are essential to mediate the E2-induced transcription of the hPRLR gene.
ACKNOWLEDGMENTS
This work was supported by the Intramural Research program of the NICHD, National Institutes of Health.
Footnotes
Published ahead of print on 13 June 2011.
REFERENCES
- 1. Björnström L., Sjoberg M. 2005. Mechanisms of estrogen receptor signaling: convergence of genomic and nongenomic actions on target genes. Mol. Endocrinol. 19:833–842 [DOI] [PubMed] [Google Scholar]
- 2. Brayer K. J., Segal D. J. 2008. Keep your fingers off my DNA: protein-protein interactions mediated by C2H2 zinc finger domains. Cell Biochem. Biophys. 50:111–131 [DOI] [PubMed] [Google Scholar]
- 3. Courey A. J., Holtzman D. A., Jackson S. P., Tjian R. 1989. Synergistic activation by the glutamine-rich domains of human transcription factor Sp1. Cell 59:827–836 [DOI] [PubMed] [Google Scholar]
- 4. Dong J., Tsai-Morris C. H., Dufau M. L. 2006. A novel estradiol/estrogen receptor alpha-dependent transcriptional mechanism controls expression of the human prolactin receptor. J. Biol. Chem. 281:18825–18836 [DOI] [PubMed] [Google Scholar]
- 5. Harvey P. W. 2005. Human relevance of rodent prolactin-induced non-genotoxic mammary carcinogenesis: prolactin involvement in human breast cancer and significance for toxicology risk assessments. J. Appl. Toxicol. 25:179–183 [DOI] [PubMed] [Google Scholar]
- 6. Héroux M., Hogue M., Lemieux S., Bouvier M. 2007. Functional calcitonin gene-related peptide receptors are formed by the asymmetric assembly of a calcitonin receptor-like receptor homo-oligomer and a monomer of receptor activity-modifying protein-1. J. Biol. Chem. 282:31610–31620 [DOI] [PubMed] [Google Scholar]
- 7. Hu Z., Zhuang L., Dufau M. L. 1996. Multiple and tissue-specific promoter control of gonadal and non-gonadal prolactin receptor gene expression. J. Biol. Chem. 271:10242–10246 [DOI] [PubMed] [Google Scholar]
- 8. Hu Z. Z., Meng J., Dufau M. L. 2001. Isolation and characterization of two novel forms of the human prolactin receptor generated by alternative splicing of a newly identified exon 11. J. Biol. Chem. 276:41086–41094 [DOI] [PubMed] [Google Scholar]
- 9. Hu Z. Z., Zhuang L., Dufau M. L. 1998. Prolactin receptor gene diversity: structure and regulation. Trends Endocrinol. Metab. 9:94–102 [DOI] [PubMed] [Google Scholar]
- 10. Hu Z. Z., Zhuang L., Meng J., Dufau M. L. 1998. Transcriptional regulation of the generic promoter III of the rat prolactin receptor gene by C/EBPbeta and Sp1. J. Biol. Chem. 273:26225–26235 [DOI] [PubMed] [Google Scholar]
- 11. Hu Z. Z., Zhuang L., Meng J., Leondires M., Dufau M. L. 1999. The human prolactin receptor gene structure and alternative promoter utilization: the generic promoter hPIII and a novel human promoter hP(N). J. Clin. Endocrinol. Metab. 84:1153–1156 [DOI] [PubMed] [Google Scholar]
- 12. Hu Z. Z., Zhuang L., Meng J., Tsai-Morris C. H., Dufau M. L. 2002. Complex 5′ genomic structure of the human prolactin receptor: multiple alternative exons 1 and promoter utilization. Endocrinology 143:2139–2142 [DOI] [PubMed] [Google Scholar]
- 13. Kim K., Barhoumi R., Burghardt R., Safe S. 2005. Analysis of estrogen receptor alpha-Sp1 interactions in breast cancer cells by fluorescence resonance energy transfer. Mol. Endocrinol. 19:843–854 [DOI] [PubMed] [Google Scholar]
- 14. Kushner P. J., et al. 2000. Estrogen receptor pathways to AP-1. J. Steroid Biochem. Mol. Biol. 74:311–317 [DOI] [PubMed] [Google Scholar]
- 15. Lai J. S., Herr W. 1992. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc. Natl. Acad. Sci. U. S. A. 89:6958–6962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Leondires M. P., Hu Z. Z., Dong J., Tsai-Morris C. H., Dufau M. L. 2002. Estradiol stimulates expression of two human prolactin receptor isoforms with alternative exons-1 in T47D breast cancer cells. J. Steroid Biochem. Mol. Biol. 82:263–268 [DOI] [PubMed] [Google Scholar]
- 17. Meng J., Tsai-Morris C. H., Dufau M. L. 2004. Human prolactin receptor variants in breast cancer: low ratio of short forms to the long-form human prolactin receptor associated with mammary carcinoma. Cancer Res. 64:5677–5682 [DOI] [PubMed] [Google Scholar]
- 18. Paech K., et al. 1997. Differential ligand activation of estrogen receptors ERalpha and ERbeta at AP1 sites. Science 277:1508–1510 [DOI] [PubMed] [Google Scholar]
- 19. Peirce S. K., Chen W. Y. 2001. Quantification of prolactin receptor mRNA in multiple human tissues and cancer cell lines by real time RT-PCR. J. Endocrinol. 171:R1–R4 [DOI] [PubMed] [Google Scholar]
- 20. Pfleger K. D., Seeber R. M., Eidne K. A. 2006. Bioluminescence resonance energy transfer (BRET) for the real-time detection of protein-protein interactions. Nat. Protoc. 1:337–345 [DOI] [PubMed] [Google Scholar]
- 21. Porter W., Saville B., Hoivik D., Safe S. 1997. Functional synergy between the transcription factor Sp1 and the estrogen receptor. Mol. Endocrinol. 11:1569–1580 [DOI] [PubMed] [Google Scholar]
- 22. Powell E., Xu W. 2008. Intermolecular interactions identify ligand-selective activity of estrogen receptor alpha/beta dimers. Proc. Natl. Acad. Sci. U. S. A. 105:19012–19017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Qazi A. M., Tsai-Morris C. H., Dufau M. L. 2006. Ligand-independent homo- and heterodimerization of human prolactin receptor variants: inhibitory action of the short forms by heterodimerization. Mol. Endocrinol. 20:1912–1923 [DOI] [PubMed] [Google Scholar]
- 24. Ramji D. P., Foka P. 2002. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem. J. 365:561–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ray A., Prefontaine K. E., Ray P. 1994. Down-modulation of interleukin-6 gene expression by 17 beta-estradiol in the absence of high affinity DNA binding by the estrogen receptor. J. Biol. Chem. 269:12940–12946 [PubMed] [Google Scholar]
- 26. Stein B., Yang M. X. 1995. Repression of the interleukin-6 promoter by estrogen receptor is mediated by NF-kappa B and C/EBP beta. Mol. Cell. Biol. 15:4971–4979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tamrazi A., Carlson K. E., Daniels J. R., Hurth K. M., Katzenellenbogen J. A. 2002. Estrogen receptor dimerization: ligand binding regulates dimer affinity and dimer dissociation rate. Mol. Endocrinol. 16:2706–2719 [DOI] [PubMed] [Google Scholar]
- 28. Ujházy P., et al. 1994. Ecto-5′-nucleotidase (CD73) in multidrug-resistant cell lines generated by doxorubicin. Int. J. Cancer 59:83–93 [DOI] [PubMed] [Google Scholar]
- 29. Xie Y. L., Hassan S. A., Qazi A. M., Tsai-Morris C. H., Dufau M. L. 2009. Intramolecular disulfide bonds of the prolactin receptor short form are required for its inhibitory action on the function of the long form of the receptor. Mol. Cell. Biol. 29:2546–2555 [DOI] [PMC free article] [PubMed] [Google Scholar]