Abstract
Cdc25A is a cell cycle-activating phosphatase, and its overexpression in breast cancers has been shown to correlate with poor prognosis. Most recent studies related to Cdc25A and tumor progression have focused on its role in regulating cell cycle progression. However, less is known about how Cdc25A modulates the metastasis of breast cancer cells. In this study, we revealed that Cdc25A enhances Foxo1 stability by dephosphorylating Cdk2, and Foxo1 was shown to directly regulate transcription of the metastatic factor MMP1. Further studies have shown that overexpression of Cdc25A in breast cancer cells enhances metastasis, whereas its downmodulation inhibits metastasis in mouse models, and the effects of Cdc25A on breast cancer cell metastasis are independent of cell proliferation and apoptosis. Furthermore, we have demonstrated that aberrant Cdc25A in breast cancer patient samples directly correlates with the metastatic phenotype. Further insights into this critical role of Cdc25A in the metastasis of breast cancer cells and the trial of an anti-Cdc25A strategy in mouse models may reveal its therapeutic potential in prevention and treatment of breast cancer cell dissemination.
INTRODUCTION
Breast cancer is one of the leading causes of morbidity and mortality in women worldwide (10, 30, 37). The majority of breast cancer deaths result from metastasis of breast cancer to other organs (14, 16, 36). Breast cancer metastasis has been shown to reduce the chances of long-term survival from 90% to around 5% (22). Therefore, insight into the molecular mechanisms underlying breast cancer invasion and metastasis would enable more effective treatment for breast cancer.
Cdc25 phosphatases (Cdc25A, Cdc25B, and Cdc25C) promote cell cycle progression by dephosphorylating and activating cyclin-dependent kinases (Cdks) (38). Cdc25A activates cyclin E (A)-Cdk2 during G1 through S and also seems to be involved in activation of Cdk1 at the G2/M boundary, and Cdc25A plays a nonredundant role in embryogenesis and oncogenesis (4, 12, 28). Cdc25B and Cdc25C, which collaborate for activation of cyclin B-Cdk1 at the G2/M boundary, are dispensable for checkpoint function (11, 18, 34). Among the Cdc25 family, Cdc25A is highlighted as an indispensable regulator of cell development and a driver of tumorigenesis (12, 28, 43, 55). While the known role of Cdc25A in cell cycle progression and proliferation has been studied with respect to breast cancer pathogenesis (12, 20, 28, 43), little is understood about whether and how Cdc25A affects the metastatic potential of breast cancer cells. Although epidemiologic studies have shown that most breast cancer patients (50 to 69%) have overexpressed Cdc25A and show poor prognosis (8, 25), not much is known about its association with breast cancer metastasis. In this study, we show that Cdc25A overexpression is correlated with the metastatic phenotype in breast cancer patient samples.
Foxo1, one of the mammalian Forkhead transcription factors of class O (FoxO), is involved in a wide range of biological processes (2, 24). The Foxo1 protein is tightly regulated by multiple posttranslational modifications, including phosphorylation, acetylation, ubiquitination, and methylation, and Foxo1 expression is highly variable in different tissues (47, 52). Interestingly, Cdk2, a downstream target of Cdc25A, directly phosphorylates Foxo1 and affects Foxo1 stability in cells (23, 54), which varies in different cell types (23, 47, 52, 54). However, less is known about whether and how Cdc25A modulates Foxo1 activity and stability in breast cancer cells.
In the present study, we elucidated the molecular mechanisms by which Cdc25A mediates Foxo1 function, which further regulates expression of matrix metalloprotease 1 (MMP1). Interestingly, MMP1 is a hallmark of human metastatic cancer, and its overexpression represents a high risk factor that adversely correlates with overall survival of patients with invasive breast carcinoma (14, 31, 33, 40, 53). Our findings explore a novel role of Cdc25A in breast cancer cell invasion and metastasis, which may provide novel therapeutic strategies to combat breast cancer metastasis by targeting Cdc25A.
MATERIALS AND METHODS
Cell culture.
Human breast cancer MDA-MB-231, MCF7, MDA-MB-453, MDA-MB-436, and MDA-MB-468 cells were obtained from the American Type Culture Collection. The metastatic Mary-X cell line was kindly provided by S. Barsky. The cells were cultured in Dulbecco's minimum essential medium (DMEM) (Gibco, Rockville, MD) supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin in a 5% CO2 atmosphere at 37°C.
Plasmid and pLenti-virus vector preparation.
The modified sequence of anti-CDC25A small interfering RNA (siRNA) was 5′-AGCAACCACTGGAGGTGAAGT-3′. pLenti-virus-siRNA preparation and infection of breast cancer cells were performed following the instructions of the Block-iT lentiviral RNA interference (RNAi) manual (Invitrogen). The full-length cDNA of CDC25A was amplified from MCF10A cells and cloned into pLent6/V5 Topo vector (Invitrogen) and pCDNA4 vector. The GFP-Foxo1 and GFP-Foxo1-S249A plasmids were kindly provided by A. Bonni. Cdk2 and Cdk2-AF (T14A and Y15A) were kindly provided by H. Huang. RNAi-resistant Foxo1-RE and Cdk2-RE constructs (see below), as well as Foxo1 and Cdk2 hpRNA, were generated by the method described previously (23, 54). All mutations were verified by sequencing. MMP1 short hairpin RNA (shRNA) (h) pLenti-virus particles were from Santa Cruz Biotechnology, Inc.
RNA preparation and real-time PCR.
Total RNA was prepared from the breast cancer cell lines by using TRIzol reagent (Invitrogen, Carlsbad, CA). First-strand cDNA was synthesized from total RNA with SuperScript (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Real-time quantitative reverse transcription (RT)-PCR (qRT-PCR) analysis was performed on the ABI Prism 7000 sequence detection system (Applied Biosystems, Foster City, CA). The specific primer sets were as follows: Cdc25A, 5′-CCTCCGAGTCAACAGATTCA-3′ and 5′-GGGTCGATGAGCTGAAAGAT-3′; GAPDH (glyceraldehyde-3-phosphate dehydrogenase), 5′-GAAGGTGAAGGTCGGAGTC-3′ and 5′-GAAGATGGTGATGGGATTTC-3′; MMP1, 5′-AAATGCAGGAATTCTTTGGG-3′ and 5′-ATGGTCCACATCTGCTCTTG-3′; Foxo1, 5′-GTGACTTGGATGGCATGTTC-3′ and 5′-CCCAGCTATGTGTCGTTGTC-3′; and Cdk2, 5′-CAGTACTGCCATCCGAGAGA-3′ and 5′-GAATGCCAGTGAGAGCAGAG-3′. The relative expression levels were calculated using the comparative cycle threshold (CT) method (2−ΔCT) (User Bulletin 2, ABI Prism 7700 Sequence Detection System; Applied Biosystems, Foster City, CA).
Luciferase reporter assay.
Cells were seeded in 24-well plates at a density of 1 × 105. After 24 h, the cells were transfected using Superfect (Qiagen). Briefly, luciferase reporter gene constructs (200 ng) and the pRL-SV40 Renilla luciferase construct (1 ng) (for normalization) were cotransfected into the wells. Cell extracts were prepared 48 h after transfection, and the luciferase activity was measured using the Dual-Luciferase reporter assay system (Promega).
Western blotting.
Lysates of the tested cells were generated in SDS sample buffer (60 mM Tris-HCl at pH 6.8, 10% glycerol, 2% SDS, and 5% 2-mercaptoethanol). Equal amounts of lysate protein (50 μg) were separated on 8 to 12% SDS-PAGE gels, transferred to polyvinylidene fluoride (PVDF) membranes (Amersham), and then incubated with primary antibodies specific for Cdc25A, Cdc25B, Cdc25C, Cdk2, and p-Cdk2 (Cell Signaling); CXCL1 and MMP1 (Santa Cruz Biotechnology); green fluorescent protein (GFP) (Molecular Probes); Foxo1, Foxo3, Foxo4, p-Foxo1-S249, p-Foxo1-T24, and p-Foxo1-S256 (Invitrogen); or β-actin (Sigma). The blots were then rinsed in Tris-buffered saline-Tween (TBST) and further incubated in peroxidase-conjugated anti-mouse or anti-rabbit secondary antibody (49, 56). Protein bands were visualized using the enhanced chemiluminescence (ECL) detection system (Pierce Biotech) and exposed to film. All experiments were repeated in triplicate.
ChIP.
Chromatin immunoprecipitation (ChIP) analysis was performed as described previously (39). In brief, breast cancer cells were fixed for 10 min at room temperature with 10% formaldehyde. After incubation, glycine was added to a final concentration of 0.125 M to “quench” the formaldehyde. The cells were pelleted, washed once with ice-cold phosphate-buffered saline (PBS), and then lysed. The lysates were pelleted, resuspended, and sonicated to reduce DNA length to between 500 and 1,000 bp. Chromatin was precleared with protein A agarose beads for 1 h and then incubated with 5 μg of Foxo1 antibody (Santa Cruz Biotechnology) or control rabbit IgG overnight. Immune complexes were precipitated with protein A agarose beads, washed, and eluted in 100 μl of Tris-EDTA (TE) with 0.5% SDS and 200 μg/ml proteinase K. The precipitated DNA was further purified by phenol-chloroform extraction and ethanol precipitation and analyzed by quantitative PCR (qPCR). The primers used in the analysis of Foxo1 potential binding sites in the MMP1 promoter were as follows: −687 to −456, 5′-TGCCTCGGCCTCCTGAAATT-3′ and 5′-CAGTGTTAGTAATTCCACCC-3′; −468 to −239, 5′-AGTGTTAGTAATTCCACCCT-3′ and 5′-TGTTTGAAGTTAATCATGAC-3′; −316 to −11, 5′-AATGAATTGGAGAAAACCAC-3′ and 5-CTTGCACTGAGAAAGAAGAC-3′.
Invasion and wound healing assays.
In vitro invasion assays were conducted using Transwells (Costar, Cambridge, MA) with 8-μm-pore-size polycarbonate membrane filters in 24-well culture plates. The upper surface of the filter was coated with Matrigel (Becton Dickinson, Bedford, MA) in a volume of 12.5 μg per filter. The Matrigel was dried and reconstituted at 37°C into a solid gel on the filter surface. The lower surface of the filter was coated with fibronectin (20 μg/ml), vitronectin (10 μg/ml), collagen IV (50 μg/ml), or 1% bovine serum albumin (BSA), used as chemoattractants. After starving in BSA-free DMEM overnight, the cells were seeded in the upper chamber at a density of 2 × 104. The cells were allowed to invade for 24 h. Cells that invaded the lower surface of the filter were counted in 10 random fields under a light microscope at high magnification. Experiments were repeated at least in triplicate. Wound healing assays were conducted by seeding 2 × 105 cells onto 96-well plates. Confluent monolayers were wounded using a pipette tip.
Histopathology.
Paraffin-embedded (5 μm), 10% neutral-buffered-formalin-fixed tissue sections derived from tumor nodules or other tissues were stained with hematoxylin and eosin (H&E) to evaluate the morphology and invasiveness of breast cancer cells.
Metastasis assay of cells in a xenograft mouse model.
For the tumorigenesis and metastasis assays by intravenous (i.v.) injection, 5 × 106 breast cancer cells were injected into the mammary fat pads or lateral flanks of 6- to 8-week-old BALB/c female athymic mice. The palpable tumor diameters were measured once per week. Tumor length (L) and width (W) were measured with a caliper, and volume (V) was calculated by the following equation: V=(L × W2)/2 (41). For experimental metastasis assays by i.v. injection, 1 × 106 cancer cells were injected into the tail veins of the 6- to 8-week-old female BALB/c athymic mice. For indole-3-carbinol (I3C) assays, the mice were injected i.v. with cancer cells. Metastatic lung nodules >0.5 mm in diameter were counted. Statistical analyses were done by analysis of variance (ANOVA) tests. To ensure representative sampling of lung tumor nodules, four sections were made per lung at various depths along the coronal plane of the lung (53). The nodules per lung (four sections) were counted under a light microscope.
The injected mice were maintained under identical conditions and were monitored regularly. Endpoint assays were conducted at 10 weeks after injection unless significant morbidity required that the mouse be euthanized earlier. Survival curves were drawn and analyzed using Prism software (GraphPad Software, San Diego, CA). All animal experiments in this study were done under the animal use guidelines of the NIH (Bethesda, MD) and the Institutional Animal Care and Use Committees of Ohio State University and Beijing Medical University.
Statistical analysis.
Statistical analyses were performed with Student's t test for comparison of two samples.
RESULTS
Cdc25A positively correlates with expression of metastatic factor MMP1.
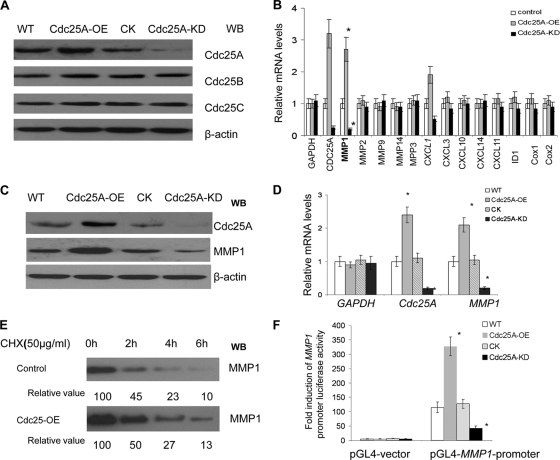
To gain insight into the potential interaction between Cdc25A and breast cancer cell metastasis, we first generated Cdc25A-overexpressing MDA-MB-231 cell lines (Cdc25A-OE) by transfecting Cdc25A using pLenti-expressing vector. As shown in Fig. 1A, second lane from the left, high expression of Cdc25A was observed in Cdc25A-OE compared to the wild type (WT). In addition, we also downregulated Cdc25A in MDA-MB-231 cells using pLenti-Cdc25A-siRNA (Cdc25A-KD [knockdown]). As shown in shown in Fig. 1A, fourth lane, significant downregulation of Cdc25A was observed in Cdc25A-KD- compared to nontargeted-siRNA-transfected cells (CK). Our experiments showed no difference between the cells with pLenti-scramble-siRNA and those with empty pLenti-expressing vector (Invitrogen) that harbor the same pLenti vector base. Thus, cells with empty pLenti-siRNA vector served as the control (CK) as a representative of the two empty vectors.
Fig. 1.
Cdc25A correlates with expression of metastasis factor MMP1. (A) Cdc25A was effectively and specifically overexpressed (Cdc25A-OE) or knocked down (Cdc25A-KD) in breast cancer cells. The controls Cdc25B and Cdc25C were intact in these cells. Our preexperiments showed no difference between the cells with pLenti-scramble-siRNA and those with empty pLenti-expressing vector (Invitrogen) that harbor the same pLenti vector base. Thus, either of them could serve as the control Cdc25A-CK. WB, Western blotting. (B) The relative mRNA expression levels of the metastasis-related factors were evaluated in the MDA-MD-231 breast cancer cell line and its derivatives Cdc25A-OE (overexpression) and Cdc25A-KD (knockdown) by quantitative real-time PCR; human GAPDH served as a loading control. These data represent three independent experiments (*, P < 0.01; means ± standard deviations [SD]; n=5). (C) Detection of MMP1 protein levels in breast cancer cells versus their derivatives Cdc25A-OE and Cdc25A-KD. (D) Evaluation of human Cdc25A and MMP1 mRNA levels in different breast cancer cells by qRT-PCR. Human GAPDH served as a loading control (*, P < 0.01 compared with the CK or WT; means ± SD; n=5). (E) Degradation rates of MMP1 protein in breast cancer cells. Exponentially proliferating cells were treated with 50 μg/ml cycloheximide (Chx) for the times indicated, and cellular MMP1 levels were determined by Western blotting. The data represent three independent experiments. (F) The pGL4-MMP1-promoter and the control pGL4 vector were transfected into the breast cancer cell line and its derivative Cdc25A-CK, Cdk25A-OE, or Cdc25A-KD, and fold induction of MMP1 promoter-induced luciferase activity was analyzed (*, P < 0.001 compared with the control or WT cells; mean ± SD; n=5).
We further analyzed the effects of Cdc25A overexpression and downregulation on a variety of metastasis-related factors in the breast cancer cells with modulation of Cdc25A levels. Our studies showed that MMP1, a strong cancer cell metastasis factor and a high risk for overall survival of patients with invasive breast carcinoma (13, 31, 46), was significantly positively correlated with Cdc25A levels in the breast cancer cells (Fig. 1B) (P < 0.01). Cdc25A overexpression led to an increase in MMP1 protein, whereas Cdc25A knockdown suppressed expression of MMP1 (Fig. 1C).
Subsequent experiments confirmed that Cdc25A expression positively correlates with MMP1 mRNA levels (Fig. 1D) but is not involved in the stability and degradation of MMP1 protein (Fig. 1E). To investigate whether the regulation of MMP1 takes place at the transcript level, we further cloned the MMP1 promoter into the PGL4 vector and measured its activity by the luciferase signal in the different cells. As shown in Fig. 1F, the MMP1 promoter activity was greatly increased in the Cdc25A-overexpressing cells and was significantly reduced in the Cdc25A knockdown cells compared to the WT or CK cells (P < 0.001).
Similar results were observed in the MDA-MB-453 breast cancer cells in response to Cdc25A up- or downregulation. The data suggest that Cdc25A mediates expression of the metastatic factor MMP1 gene at the transcriptional level.
Cdc25A mediates phosphorylation and cellular localization of Foxo1.
Next, we analyzed the mechanism through which Cdc25A regulates MMP1 expression at the transcriptional level. Since Cdc25A is a phosphatase and not a transcription factor, we hypothesized that Cdc25A may indirectly mediate MMP1 expression through modification of a specific transcription factor. Cdk2, a downstream target of Cdc25A (19, 55), directly phosphorylates the transcription factor Foxo1 (23, 54). We set out to determine whether Cdc25A can mediate Foxo1 stability and activity though regulation of Cdk2 in breast cancer cells.
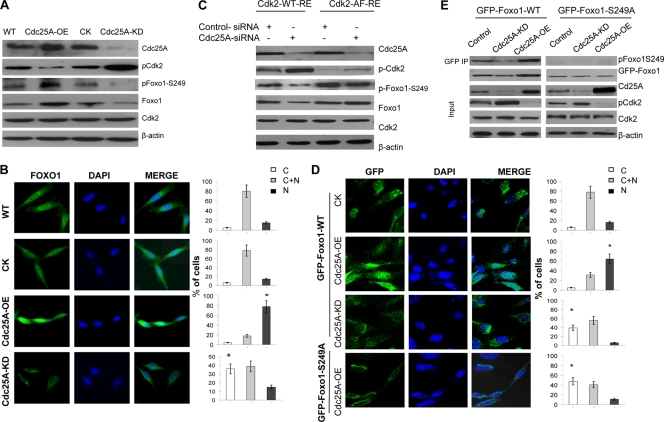
We first detected the status of Foxo1 phosphorylation at Ser249 in MDA-MB-231 breast cancer cells under conditions of Cdc25A overexpression or Cdc25A knockdown. As suspected, Cdc25A overexpression, which enhanced Cdk2 activity by dephosphorylating Cdk2 at Thr14 and Tyr15 (Fig. 2A) (19, 27, 57), increased the levels of phospho-Ser249 Foxo1 (Fig. 2A). Conversely, Cdc25A suppression attenuated Cdk2 activity and decreased the levels of phospho-Ser249 Foxo1 (Fig. 2A). Moreover, the elevated levels of phospho-Ser249 Foxo1 in the Cdc25A-OE cells led to an increase in the nuclear localization and stability of Foxo1; in contrast, the decreased levels of phospho-Ser249 Foxo1 mediated by Cdc25A knockdown reduced the nuclear localization and stability of Foxo1 (Fig. 2B).
Fig. 2.
Cdc25A mediates Foxo1 phosphorylation and its cellular localization and stability by dephosphorylating Cdk2. (A) Lysates of the different cancer cells were immunoblotted with the indicated antibodies. (B) Increased or decreased nuclear Foxo1 in Cdc25A-OE (overexpression) and Cdc25A-KD (knockdown) breast cancer cells, respectively. (Left) Foxo1. (Middle) DAPI (4′,6-diamidino-2-phenylindole). (Right) Merge. Quantification of Foxo1 localization is shown on the right (bar graphs). C, cytoplasm; N, nucleus; C+N, cytoplasm and nucleus. n=200 for each cell type. *, P < 0.01 compared to the CK or WT cells. (C) The mutation Cdk2T14A Y15P interrupted the effect of Cdc25A on Foxo1. The endogenous Cdk2-depleted (by RNAi) breast cancer cells with Cdc25A-WT or Cdc25A knockdown were transfected with Cdk2-WT-RE or Cdk2-AF-RE plasmids. (D) Breast cancer cells engineered to express GFP-Foxo1 or GFP-Foxo1-S249A were assessed by localization of GFP-Foxo1 fusion proteins. (Left) GFP fluorescence. (Middle) DAPI. (Right) Merge. Quantification of GFP-Foxo1-WT or GFP-Foxo1-S329A (bottom) is shown on the right (bar graphs). n=200 for each cell type. *, P < 0.01 compared to the control cells. (E) The indicated breast cancer cells engineered to express exogenous GFP-Foxo1 were harvested, and the lysates were immunoprecipitated with GFP antibody. Similar results were observed using MDA-MB-231 and MDA-MB-453 breast cancer cells versus their derivatives.
Furthermore, to determine if Cdk2 links Cdc25A to Foxo1, we transfected Cdk2-WT-RE or Cdk2-AF-RE (23) (AF, mutation Cdk2T14A Y15P; RE, resistance to Cdk2-siRNA) into MDA-MB-231 or MDA-MB-453 breast cancer cells in which both endogenous Cdk2 and Cdc25A were already downregulated. As shown in Fig. 2C, Cdc25A knockdown displayed reduced phosphorylation of Foxo1 at Ser249 and total Foxo1 in the cells with the transfected Cdk2-WT (Fig. 2C, lane 2 versus lane 1). On the other hand, the Cdk2T14A Y15P (Cdk2-AF) mutant interrupted the effect of Cdc25A suppression on Foxo1, and the levels of phospho-S249 Foxo1 were not attenuated and total Foxo1 was not reduced compared to the control cells without Cdc25A downmodulation (Fig. 2C, lane 4 versus lane 3).
Next, we expressed a GFP-Foxo1-S249A mutant in the breast cancer cells while GFP-Foxo1-WT served as the control. Consistent with the results shown in Fig. 2B, Cdc25A overexpression resulted in nuclear localization and stabilization of exogenous GFP-Foxo1-WT (Fig. 2D, second row versus the top row), whereas silencing Cdc25A by RNAi reversed these effects (Fig. 2D, third row versus top row). However, in the mutated GFP-Foxo1-S249A cells, Cdc25A overexpression failed to enhance the nuclear localization and stability of GFP-Foxo1-S249A (Fig. 2D, bottom row). No significant difference was observed in the cellular localization and stability of GFP-Foxo1-S249A between Cdc25A-OE, Cdc25A-KD, and WT cells due to the S249A mutation of Foxo1. Additional experiments and analyses also showed that the S249A mutation in Foxo1-GFP inhibited the effect of Cdc25A on Foxo1-GFP (Fig. 2E), which further confirmed that Ser249 of Foxo1 is required for regulation of Cdc25A on Foxo1.
Foxo1 directly regulates MMP1 transcription and mediates the link between Cdc25A and MMP1.
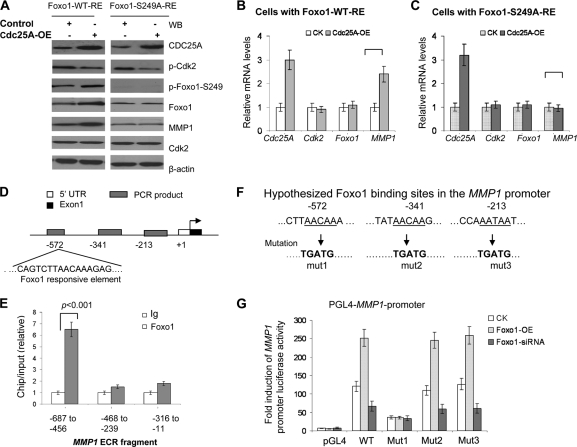
We sought to test the hypothesis that Foxo1 mediates the connection between Cdc25A and MMP1. We transfected the plasmid Foxo1-WT-RE or Foxo1-S249A-RE (RE, resistance to Foxo1-siRNA) into the breast cancer cells in which the endogenous Foxo1 was knocked down and Cdc25A was overexpressed. The experiments and analyses revealed that Cdc25A overexpression increased the levels of phospho-S249 Foxo1, total Foxo1 protein, and MMP1 expression in the cells transfected with Foxo1-WT-RE versus the control cells (Fig. 3A, lane 2 versus lane 1 [from left], and 3B). In contrast, the S249A mutation in Foxo1 interrupted this effect of Cdc25A overexpression on MMP1 (Fig. 3A, lane 4 versus lane 3, and C).
Fig. 3.
Foxo1 directly regulates MMP1 and mediates the connection between Cdc25A and MMP1. (A) Modulation of Foxo1 impacts MMP1 protein levels. The plasmid Foxo1-WT-RE or Foxo1-S249A-RE was transfected into the indicated cells with Foxo1 depletion via Foxo1-siRNA. The cell lysates were immunoblotted with the indicated antibodies. (B and C) The mutation Foxo1-S249A blocks the effects of Foxo1 on MMP1. The human Cdc25A and MMP1 mRNA levels were evaluated in the cells with Foxo1-WT-RE (B) and Foxo1-S249A-RE (C) by qRT-PCR (mean ± SD; n=5). The Cdk2 and Foxo1 mRNA levels served as loading controls. (D) The conserved putative Foxo1 binding sites in the MMP1 promoter (http://www.ifti.org/cgi-bin/ifti/Tfsitescan.pl). (E) ChIP analyses of Foxo1 binding in breast cancer cells established binding of Foxo1 to a conserved Foxo1 binding motif at 572 bp upstream of the MMP1 transcription start site. The results are presented as fold template enrichment in immunoprecipitates of Foxo1 antibody relative to those of a control antibody (mean ± SD; n=5). (F) Mutated derivatives of the pGL4-MMP1 promoter with three putative Foxo1 binding sites. (G) pGL4-MMP1-promoter or the mutated derivatives pGL4-MMP1-promoter-mut (Mut1, -2, or -3) was transfected into breast cancer cells with Foxo1-WT or Foxo1-siRNA and then assayed for its activity. The empty vectors served as controls (mean ± SD; n=5).
To determine whether Foxo1 directly regulates MMP1 gene transcription, we first conducted bioinformatics analysis of transcription factor binding sites and detected three highly conserved Foxo1 binding sequences within the MMP1 promoter, located in the regions around −572 bp, −341 bp, and −213 bp upstream of the start codon site (Fig. 3D). We further performed chromatin immunoprecipitation from the breast cancer cells, using Foxo1 or control antibody and primer sets designed to amplify sequences located in the noncoding promoter regions. The genomic fragment around −572 bp upstream of the start codon of MMP1 was selectively enriched in the Foxo1 antibody immunoprecipitation (Fig. 3D and E). These findings suggest that MMP1 is a direct Foxo1 target gene in breast cancer cells.
To further support this conclusion, we produced a mutation in each putative Foxo1 binding site of the MMP1 promoter (about 1 kb) (Fig. 3F) and measured the relative activity of each promoter mutation derivative by the luciferase signal. Consistent with the results from the chromatin immunoprecipitation assay (Fig. 3D and E), mutation of the putative Foxo1 binding site (mut1) of the MMP1 promoter significantly blocked MMP1 promoter activity induced by altered Foxo1 expression (Fig. 3G).
Similar results were observed in the assays done using MDA-MB-231 and MDA-MB-453 cells. These data suggest that Foxo1 directly regulates MMP1 transcription by its promoter and mediates the connection between Cdc25A and metastatic factor MMP1.
Effect of Cdc25A o the invasive phenotype of breast cancer cells in vitro.
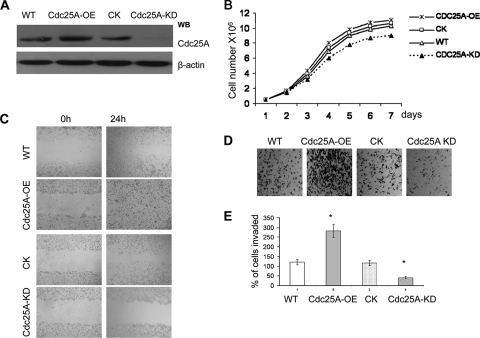
Cdc25A positively correlates with expression of MMP1 (Fig. 1 to 3), a strong cancer cell metastasis factor (13, 31, 46). This connection implies that altered Cdc25A may be related to breast cancer cell metastasis. To investigate the potential role of Cdc25A in breast cancer cell metastasis, we first analyzed the role of Cdc25A in regulating transendothelial migration and wound healing, which are important factors in regulating metastasis. As shown in Fig. 4A to C, Cdc25A overexpression (Cdc25A-OE) enhanced wound healing (Fig. 4C), although it did not significantly accelerate cell growth in vitro (Cdc25A-OE versus WT or CK; P > 0.5 [Fig. 4B]), whereas Cdc25A downregulation (Cdc25A-KD) inhibited wound healing compared to WT or vector control (CK) cells. Although Cdc25A-KD slightly reduced cell growth, the extended wound-healing assay for Cdc25A-KD cells (up to 48 or 72 h) showed the result was not significantly different from that of the 24-h assay (data not shown). These data suggest that the effect of Cdc25A on breast cancer cell migration in vitro is independent of cell proliferation.
Fig. 4.
Cdc25A expression mediates breast cancer cell invasion phenotypes in vitro. (A) Cdc25A was effectively and specially overexpressed (Cdc25A-OE) or knocked down (Cdc25A-KD) in breast cancer cells. (B) Growth curve of MDA-MB-231 breast cancer cells and their derivatives Cdc25A-OE and Cdc25A-KD. The WT or cells with empty vector served as controls (mean ± SD; n=5). P > 0.5 for Cdc25A-OE compared with the control or WT cells; P < 0.05 for Cdc25A compared with the control or WT cells. (C) Wound-healing assays. (D) Cell invasion assay on Matrigel-coated Transwells. (E) Statistical analysis of cell invasive ability. The cells that invaded the lower surface of the filter were counted in 10 random fields under a light microscope at high magnification (×100 [D]). P < 0.01 compared with the control and WT cells. The images are representative of those for the assay of breast cancer MDA-MB-231 and MDA-MB-453 cells versus their derivatives.
We further analyzed the invasive ability of the cell lines by chemoinvasion assays using Matrigel-coated Transwell Boyden chambers. As shown in Fig. 4D and E, the breast cancer cells with Cdc25A overexpression showed enhanced invasive properties compared to WT or CK cells (P < 0.01). In contrast, Cdc25A knockdown significantly reduced breast cancer cell migration and invasion (Fig. 4B to E; P < 0.01). Similar data were observed using another breast cancer cell line, MDA-MB-453, in response to Cdc25A up- or downregulation. These results suggest that Cdc25A regulates breast cancer cell migration and invasion in vitro.
Cdc25A overexpression promotes metastasis in a xenograft model.
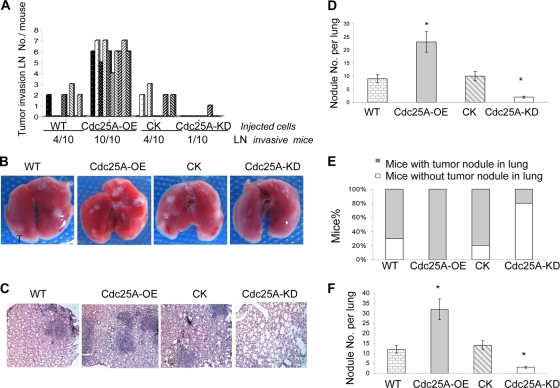
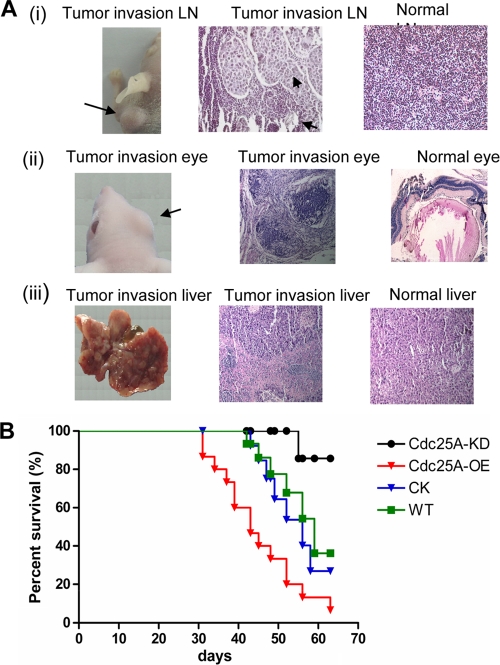
To further confirm the connection between Cdc25A and breast cancer metastasis observed in vitro, we assessed Cdc25A effects on metastasis using in vivo mouse model systems. The MDA-MB-231 (Cdc25A-WT), vector control (Cdc25A-CK), and Cdc25A overexpression (Cdc25A-OE) cell lines were injected into the mammary fat pads of 6- to 8-week-old athymic nude mice and monitored for tumor growth. Compared to those injected with either Cdc25A-WT or Cdc25A-CK, we found that all mice (10/10) injected with the Cdc25A-OE cells had massively enlarged lymph nodes with the invasive breast cancer cells (Fig. 5A and 6A, row i). In contrast, only approximately 40% of the mice (4/10) injected with the Cdc25A-WT or Cdc25A-CK cells had enlarged lymph nodes at termination of the study (Fig. 5A). Further histological examination showed that cancer cells also metastasize into the lung. To ensure representative sampling of the lungs, four sections were made per lung at various depths along the coronal plane of the lung (53). The nodules per lung (four sections) were counted under a light microscope (Fig. 5B and C). The results showed a remarkable increase in metastatic incidence in the lungs of mice injected with Cdc25A-OE cells versus WT or CK cells (Fig. 5D; P < 0.05). Interestingly, metastases were also observed in the eyes and livers in some mice implanted with Cdc25A-OE cells, but not in mice injected with the WT and CK cells (Fig. 6A). Similar results were observed with the implants injected via mouse flanks. These studies indicate that Cdc25A overexpression may enhance the breast cancer cell ability to metastasize to multiple tissues.
Fig. 5.
Cdc25A expression positively correlates with metastasis of breast cancer cells in vivo. (A) Lymph node (LN) invasion rate of breast cancer cells in nude mice (similar results were observed for cells injected via mammary fat pads or flanks). (B and C) Representative images of mouse lungs (B) and sections (C). The lung sections, which were from mice injected with different breast cancer cells via mammary fat pads, flanks, or tail veins, were stained with hematoxylin and eosin. (D) Assay and analysis for lung tumor nodules formed in nude mice (similar results were obtained for mice injected via the mammary fat pads and flanks). *, P < 0.05 compared with the control or WT cells; n=10 mice for each cell type. The error bars indicate SD. (E and F) Statistical analysis for the number of mice with metastasis (E) and tumor nodules in the mouse lung (F) (injected via the tail vein; mean ± SD; *, P < 0.01 compared with the control or WT cells). The nodules per lung for all 4 sections were counted under a light microscope (n=10 mice for each cell type). Similar results were also observed for the assay of MDA-MB-231 and MDA-MB-453 breast cancer cells versus their derivatives.
Fig. 6.
Modulation of Cdc25A expression impacts the survival of mice with implanted breast cancer cells. (A) Cdc25A overexpression leads to breast cancer cell invasion and metastasis into multiple mouse organs. In the mice injected with Cdc25A-OE cells, approximately 20% (4 of 20) had eye metastasis and 30% (6 of 20) had tumor nodules in their livers. In addition, almost all mice had aggressive lymph and lung metastasis. In contrast, no tumor nodules in the eyes or livers were observed and there was a marked reduction in lymph and lung metastasis for the control and Cdc25A-KD breast cancer cells. (B) Kaplan-Meier survival curves of the mice implanted with breast cancer cells with modulation of Cdc25A expression. The horizontal line indicates the time after the mice were injected with the cells via the tail vein. WT, parent MAD-MB-231 breast cancer cells; CK, breast cancer cells with the empty vector; Cdc25A-OE, breast cancer cells with Cdc25 overexpression; Cdc25A-KD, breast cancer cells with Cdc25 knockdown. The number of tested mice was 15 for each group.
We further analyzed the effect of Cdc25A overexpression on cancer cell metastasis by injecting these cells via the tail vein into female nude mice. The study also revealed a significant increase in metastatic incidence in the implants (via the tail vein) with Cdc25A-OE cells versus WT or CK cells (Fig. 5E). More tumor nodules were detected in the lungs of transplants with Cdc25A-OE cells (27 to 33 per lung) versus WT or CK cells (Fig. 5F; P < 0.01). The results also demonstrate that Cdc25A overexpression enhances breast cancer cell invasive and metastatic abilities in vivo.
Cdc25A suppression attenuates metastasis of breast cancer in vivo.
To further confirm that Cdc25A regulates metastasis in vivo, we injected the Cdc25A-KD cells into the mammary fat pads or flanks of athymic nude mice. As expected, it was shown that fewer instances of tumor invasion into lymph nodules were observed in the mice implanted with Cdc25A-KD cells (1 of 10 mice) versus the mice implanted with WT and CK cells (Fig. 5A). The number of tumor nodules in the lung was also significantly reduced in the implants with Cdc25A-KD versus WT and CK cells (Fig. 5D) (P < 0.05). Similarly, the xenograft assay by injection via the tail vein demonstrated that Cdc25A knockdown significantly attenuated breast cancer cell metastasis. Our results also showed that only 20% of mice injected with Cdc25A knockdown cells via the tail vein were found to have tumor nodules in the lung (Fig. 5E, bar 4 from the left). Moreover, in the Cdc25A-KD implants that showed metastasis, fewer tumor nodules (1 to 3 per lung) were observed (Fig. 5F, bar 4). In contrast, 70 to 80% of mice injected with WT or CK cells and 100% of implants with Cdc25A-OE cells exhibited metastases in the lung (Fig. 5F, bars 1 to 3). Further survival studies revealed that Cdc25A overexpression accelerated breast cancer cell invasion and mouse mortality, whereas Cdc25A knockdown attenuated the cell metastases and extended mouse survival times (Fig. 6B).
Aberrant Cdc25A expression correlates with human breast cancer metastasis.
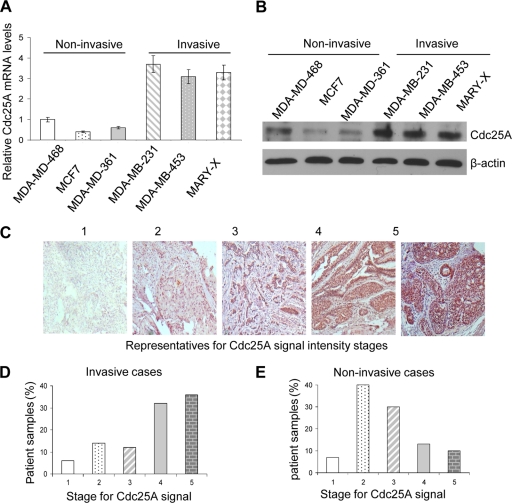
Although high expression of Cdc25A (approximately 50 to 69%) has been observed in breast cancer patients (8, 25), less is known about its expression in different stages of breast cancers, especially invasive types. Initially, we determined expression of Cdc25A in various human invasive and noninvasive breast cancer cell lines by qRT-PCR and Western blotting. As shown in Fig. 7A and B, higher Cdc25A expression was observed in highly invasive metastatic cell lines, such as MDA-MB-231, MDA-MB-453, and Mary-X cells, than in noninvasive MDA-MB-468, MCF7, and MDA-MB-361 cells.
Fig. 7.
Aberrant Cdc25A expression correlates with human breast cancer metastasis. (A and B) The relative Cdc25A mRNA (A) and protein (B) levels in human invasive cells versus noninvasive cells were evaluated by qRT-PCR and Western blotting. The error bars indicate SD. (C) Representatives of the Cdc25A signal intensity stages. The samples were graded on a scale of 1 (lowest) to 5 (maximum staining) scores or stages according to the Cdc25A signal intensity in the patient breast cancer samples. (D and E) Distribution of the invasive breast cancer patient cases (D) and noninvasive breast cancer patient cases (E) among the different Cdc25A signal intensity stages.
Next, to determine whether aberrant Cdc25A expression correlates with human breast cancer metastasis, we randomly collected microarray samples from 175 invasive/metastatic and 138 noninvasive breast cancer patients from the Ohio State University Department of Pathology and U.S. Biomax Inc. and then stained the samples with a specific Cdc25A antibody. In general, little to no Cdc25A expression was detected in normal mammary cells, but most breast cancer cells showed higher levels of Cdc25A protein expression. According to the Cdc25A signal intensity, the samples were graded on a scale of staining scores from 1 (lowest) to 5 (maximum) (Fig. 7C) and used for subsequent statistical analysis.
Among all invasive samples, approximately 67% (118 of 175) showed a broad range of staining and higher Cdc25A signal intensity (stages 4 and 5) (Fig. 7D). In contrast, only approximately 17% of noninvasive cases (24 of 138) showed stronger Cdc25A signal intensity (stage 4 or 5) (Fig. 7E). These data establish a significant correlation between aberrant Cdc25A expression and the human breast cancer metastatic cases (P < 0.01).
Taken together, these results suggest that Cdc25A is highly expressed in primary metastatic breast cancer patient tissues, as well as in invasive breast cancer cell lines.
MMP1 downmodulation suppresses the effect of Cdc25A on metastasis of breast cancer cells.
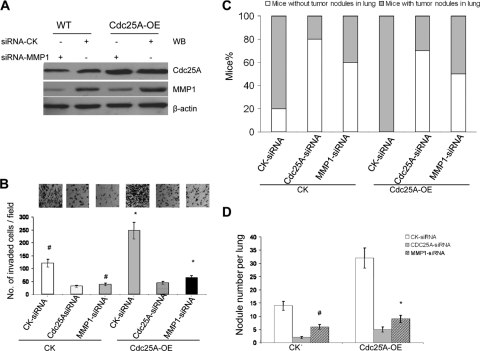
To further confirm that Cdc25A mediates breast cancer cell metastasis through regulating MMP1 (at least in part), we knocked down MMP1 in the cells (Fig. 8A) and analyzed the effect on cell invasion. As shown in Fig. 8B, MMP1 suppression significantly attenuated the impact of Cdc25A overexpression on the invasion of breast cancer cells in vitro (P < 0.01).
Fig. 8.
MMP1 blockage inhibits the effects of Cdc25A on metastasis of breast cancer cells in vitro and in vivo. (A) Detection of MMP1 levels in breast cancer cells versus their Cdc25A overexpression derivative. (B) Cell invasion assay on Matrigel-coated Transwells. The migrated cells were counted per high-power field (×40). The bars and error bars represent the means ± SD; n=10 random fields. #, P < 0.01, and *, P < 0.001 compared with the CK-siRNA control. (C and D) Statistical analysis of the numbers of mice with metastasis (C) and tumor nodules in lungs (D) of the mice injected with the different breast cancer cells via the tail vein. The lungs and the H&E staining of metastatic tumors in the lungs are similar to those in shown in Fig. 5B and C. The nodules per lung for all 4 sections were counted under a light microscope. #, P < 0.05, and *, P < 0.01 compared with the CK-siRNA control (n=10 mice per cell type). Similar results were observed in MDA-MB-231 and MDA-MB-453 breast cancer cells versus their derivatives.
We further assayed the metastatic ability of these cells in mouse models in vivo. The cells were injected into nude mice, and lung metastasis was analyzed as previously described. MMP1 downregulation showed reduced metastasis, similar to the results observed with Cdc25A downregulation. The results showed that the numbers of mice with metastasis (Fig. 8C) and tumor nodules in the lung were markedly reduced in the breast cancer cells with siRNA-MMP1 versus siRNA-CK (Fig. 8D) (P < 0.05). Similar effects of MMP1 knockdown were observed in the MDA-MB-231 and MDA-MB-453 cells and their Cdc25-OE derivatives. The results demonstrated that MMP1 downmodulation significantly inhibits the effect of Cdc25A on breast cancer cell metastasis in vivo.
Effects of Cdc25A on breast cancer cell metastasis are independent of cell proliferation and apoptosis.
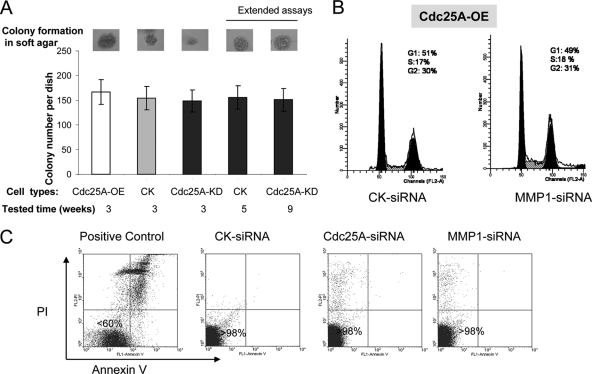
Cell proliferation and metastasis are two different properties of cancer cells, and it was reported that cancer cell cycle progression did not affect their invasion and metastasis (32). In order to determine if the metastatic effects observed were due to enhanced growth of these breast cancer cells in our models, we first performed a soft-agar colony formation assay and showed that the number of colonies formed in soft agar (the number of tumors formed in vitro in a certain site or location) was independent of the growth rate of the cells (Fig. 9A). Our further assays showed that the length of the study (10 weeks) was sufficient to distinguish the metastatic difference between the breast cancer cells in our tested models and that the extended cell growth did not significantly affect the metastatic phenotypes of these cells (Table 1). Moreover, we also showed that MMP1 suppression did not change the cell cycle and proliferation of breast cancer cells (Fig. 9B) but markedly blocked the effect of Cdc25A on breast cancer cell invasion and metastasis (Fig. 8). In addition, we revealed that Cdc25C, a member of the Cdc25 phosphatase family, does not affect the invasion and metastasis of breast cancer cells (Fig. 10), although altered expression of Cdc25C impacts cell proliferation through G2/M transition (11, 18). Furthermore, no apoptosis under the normal culture conditions was observed in these cell lines with modulation of Cdc25A or MMP1 (Fig. 9C). These findings suggest that effects of Cdc25A on breast cancer cell metastasis are independent of cell proliferation and apoptosis.
Fig. 9.
Effects of Cdc25A on breast cancer cell metastasis are independent of cell proliferation and apoptosis. (A) Soft-agar colony formation assays were performed using control MDA-MB-231 breast cancer cells with empty vector, Cdc25A-OE cells, and Cdc25A-KD cells, following the procedure previously described (56). The numbers of colonies in soft agar per plate were scored on day 21 after plating the cells (first three bars from left) (P > 0.5 for Cdc25A-OE or Cdc25A-KD versus control cells; means ± SD; n=5). To match the colony size formed by the Cdc25A-OE cells (recorded at 3 weeks), we further extended soft-agar colony formation assays for the control cells (to 5 weeks) and for the Cdc25A-KD cells (to 9 weeks). The results showed no significant change in the number of colonies formed for the extended groups (P > 0.5) (last two bars versus first three bars). The results showed that the colony formation numbers in soft agar (tumorigenesis in vitro) were independent of the proliferation of these breast cancer cells. (B) MMP1 knockdown does not affect the cell cycle of breast cancer cells driven by Cdc25A overexpression. The MDA-MB-231 breast cancer cell derivatives (overexpressing Cdc25A [CdC25A-OE]) with the CK-siRNA control and MMP1-siRNA were harvested in PBS and stained with a hypotonic solution containing propidium iodide (PI). The stained nuclei were subjected to flow cytometry analysis. The results are representative of five independent sets of experiments. Similar results were also observed for the assay of MDA-MB-453 breast cancer cell derivatives (Cdc25A-OE) with CK-siRNA and MMP1-siRNA. (C) The MDA-MB-231 breast cancer cell derivatives with CK-siRNA, MMP1-siRNA, and Cdc25A-siRNA were harvested and stained with PI-annexin V and further subjected to flow cytometry and apoptosis analysis. Apoptosis of the original breast cancer cells treated with UV served as a control (1). The results are representative of five independent sets of experiments. The results showed that under normal culture conditions, MMP1 knockdown or Cdc25A knockdown alone did not cause breast cancer cell apoptosis.
Table 1.
Metastatic phenotypes of breast cancer cells are independent of cell growth
| Parameter | Valuea |
||||
|---|---|---|---|---|---|
| Cdc25A-OE | CK | Cdc25A-KD | CK | Cdc25A-KD | |
| Tested time (mo) | 9 | 9 | 9 | 15 | 23 |
| Tumor size (mm3)b | 1,471 | 810 | 390 | 1,459 | 1,467 |
| No. of mice with LN-invasive tumors/total | 10/10 | 5/10 | 2/10 | 6/10 | 2/10 |
| No. of mice with lung-invasive tumors/total | 10/10 | 6/10 | 2/10 | 6/10 | 3/10 |
| No. of tumor nodules per lung | 32 | 12 | 3 | 13 | 4 |
CK, MDA-MB-231 breast cancer cells with the empty vector control; Cdc25A-OE, MDA-MB-231 breast cancer cells with Cdc25A overexpression; Cdc25A-KD, MDA-MB-231 breast cancer cells with Cdc25A knockdown.
Mean ± SD (n=10).
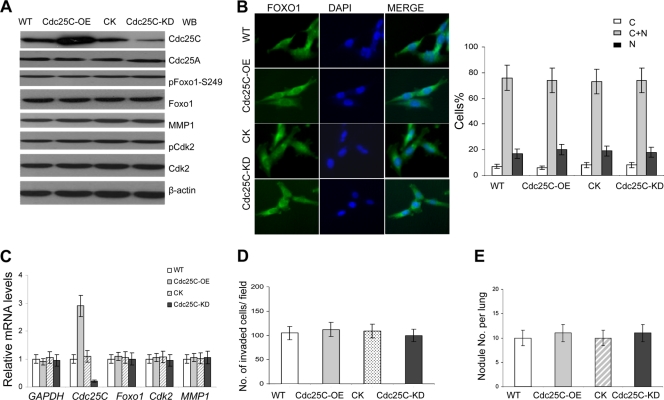
Fig. 10.
Altered Cdc25C expression does not affect the Fox1-MMP1 pathway and the metastatic phenotype of breast cancer cells. (A) Cdc25C was effectively and specifically overexpressed (Cdc25C-OE) or knocked down (Cdc25C-KD) in MDA-MB-231 breast cancer cells. The control Cdc25A levels were intact in the breast cancer cell lines. Lysates of the cancer cells were immunoblotted with the indicated antibodies. Total Foxo1 and MMP1 showed no significant changes in the cells with modulation of Cdc25C. (B) Increased or decreased Cdc25C expression (Cdc25C-OE or Cdc25C-KD) did not affect the cellular location and stability of Foxo1 in breast cancer cells. (Left) Foxo1. (Middle) DAPI. (Right) Merge. Quantification of Foxo1 localization in cells is shown in the bar graphs. n=200 for each cell type. (C) Foxo1 and MMP1 mRNA levels were evaluated in breast cancer cells with modulation of Cdc25C by quantitative real-time PCR; human GAPDH and Cdk2 served as loading controls (means ± SD; n=5). (D) Cell invasion assay on Matrigel-coated Transwells and statistical analysis of cell invasive ability in vitro. The cells that invaded the lower surface of the filter were counted in 10 random fields under a light microscope at high magnification (×100 [B]). The error bars represent the means ± SD; n=10. (E) Metastasis assay in vivo. The nodules per mouse lung for all 4 sections were counted under a light microscope (injected via the tail vein; 10 mice for each cell type). Similar results were also observed for the assay of MDA-MB-231 and MDA-MB-453 breast cancer cells versus their derivatives. The experimental evidence showed that Cdc25C did not regulate Foxo1-MMP1 and metastasis of these cancer cells, although altered Cdc25C expression affected the cell cycle through the G2/M checkpoint (11, 18).
IC3 targets Cdc25A degradation and metastasis of breast cancer cells in vivo.
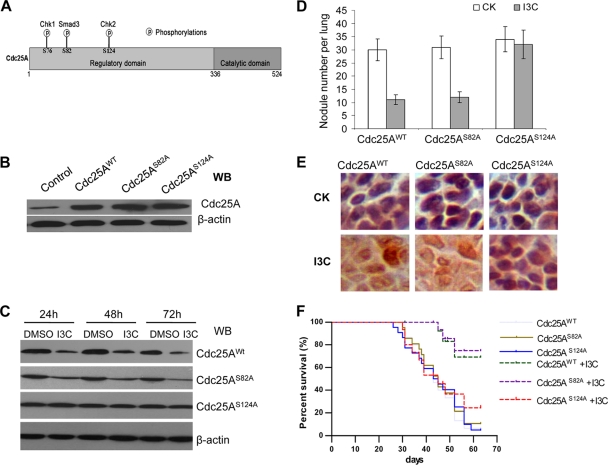
Our studies described above identify Cdc25A as an attractive target in the treatment of metastatic breast cancer. We further investigated whether the natural dietary phytochemical I3C, which induces Cdc25A degradation through the Chk2-Cdc25A pathway (51), also can attenuate breast cancer metastasis. We overexpressed Cdc25AS124A, Cdc25AS82A, Cdc25AWT, or their control empty vector using pLenti-virus into MDA-MB-231 cells (Fig. 11A to C). These special cells were injected into female BALB/c athymic mice at 6 to 8 weeks of age via the tail vein. After 2 days, the implanted mice were administered I3C (1.5 mg/day/mouse) daily by oral gavage as described in Materials and Methods. The mice were divided into eight groups based on the injected cell types and I3C treatment status: groups 1 and 2, vector with or without I3C treatment, i.e., control, I3C+ or I3C−; groups 3 and 4, Cdc25AWT, I3C+ or I3C−; groups 5 and 6, Cdc25AS82A, I3C+ or I3C−; and groups 7 and 8, Cdc25AS124A, I3C+ or I3C− (Table 2).
Fig. 11.
I3C targets Cdc25A degradation and metastasis of breast cancer cells in vivo. (A) Diagram of the pathways related to Cdc25A degradation (7, 42). (B) Expression of exogenous Cdc25A and its derivatives by pLenti-expressing vectors. (C) Response of the mutation Cdc25AS124A to I3C treatment in metastatic breast cancer cells. Cdc25AWT and Cdc25AS82A served as controls. DMSO, dimethyl sulfoxide. (D) Assay and analysis of tumor nodules in mouse lungs. The mice were injected with different breast cancer cells. The tumor nodules per lung (4 sections) were counted under a light microscope (means ± SD; 10 mice per cell type). CK, without I3C treatment. (E) The mutation Cdc25AS124A failed to respond to I3C treatment (in the tumor nodules formed in the mouse lungs). The immunohistochemical staining (brown) of the breast cancer cells in the sections with anti-Cdc25A monoclonal antibody is presented at higher magnification. (F) Kaplan-Meier survival curves of the implanted mice with/without I3C treatment (15 mice for each group). The horizontal line indicates the time after the mice were injected with the breast cancer cells via the tail vein. Similar results were observed for the assay of MDA-MB-231 and MDA-MB-453 breast cancer cell lines versus their derivatives.
Table 2.
Effect of I3C on metastasis of breast cancer cells through targeting Cdc25A degradation in a xenograft mouse model
| Group | Cancer cells injecteda | No. of mice | I3Cb | % of mice with lung metastasis | No. of dead micec | No. of tumor nodules per lung (meand ± SD) | % reduction in tumor nodule no.e | P value |
|---|---|---|---|---|---|---|---|---|
| 1 | Control | 15 | − | 73 | 7 | 15 ± 3 | ||
| 2 | Control | 15 | + | 20 | 2 | 6 ± 1 | 60 | <0.01 |
| 3 | Cdc25AWT | 15 | − | 100 | 14 | 30 ± 4 | ||
| 4 | Cdc25AWT | 15 | + | 27 | 3 | 11 ± 2 | 63 | <0.01 |
| 5 | Cdc25AS82A | 15 | − | 100 | 14 | 31 ± 4 | ||
| 6 | Cdc25AS82A | 15 | + | 33 | 4 | 12 ± 3 | 61 | <0.01 |
| 7 | Cdc25AS124A | 15 | − | 100 | 14 | 34 ± 4 | ||
| 8 | Cdc25AS124A | 15 | + | 93 | 13 | 32 ± 3 | <6 | >0.5 |
Control, MDA-MB-231 breast cancer cells without exogenous Cdc25A; Cdc25AWT, cells with exogenous wild-type Cdc25A; Cdc25AS82A, cells with exogenous mutation Cdc25AS82A; Cdc25AS124A, cells with exogenous mutation Cdc25AS124A.
The concentration of I3C was 1.5 mg/day/mouse. +, treated; −, untreated.
The number of dead mice during the study period (10 weeks).
Mean for the mice with lung metastasis and counting tumor nodule numbers per lung in four sections as described in Materials and Methods.
Relative percentage of the group with I3C treatment versus that without I3C treatment among the mice for each breast cancer cell type.
As expected, more than 90% (14/15) of implants without I3C treatment (groups 3, 5, and 7) progressed to mortality during the 10 weeks following injection of cells with exogenous overexpression of Cdc25AWT, Cdc25AS82A, or Cdc25AS124A due to aggressive lung metastases and tumor nodules in the lung (Fig. 11D and F; Table 2). However, the mice treated with I3C (groups 2, 4, and 6) showed significant extension of survival time; 60 to 70% of the mice survived with I3C treatment over the course of the 10-week study. Although I3C treatment significantly attenuated lung metastasis and extended survival in mice injected with exogenous Cdc25AWT and Cdc25AS82A (Fig. 11D, bar 3 or 4 versus 1 or 3; Table 2, group 4 or 6 versus 3 or 5), the mutated Cdc25AS124A failed to respond to I3C treatment (Fig. 11C to E; Table 2, group 8 versus 7). Furthermore, immunohistochemical staining revealed that mutant Cdc25AS124A protein levels remained higher in the tumor nodules formed by the breast cancer cells with exogenous Cdc25AS124A, regardless of the presence or absence of I3C treatment; in contrast, levels of the control Cdc25AWT and Cdc25AS82A proteins decreased in response to I3C treatment (Fig. 11E).
These data indicate that I3C, which induces Cdc25A degradation, inhibits metastasis of breast cancer cells. These studies imply the therapeutic potential of targeting Cdc25A in inhibiting breast cancer cell metastasis.
DISCUSSION
Despite compelling evidence for the correlation of Cdc25A and breast cancer outcomes (8, 25), almost all previous studies of Cdc25A pathogenesis focus on cell cycle progression and proliferation (20, 27, 28, 43, 55). In the present studies, we initially revealed that Cdc25A impacted breast cancer cell migration and invasion in vitro. Subsequent studies on xenograft mouse models and patient samples confirmed that aberrant accumulation of Cdc25A was strongly associated with metastatic phenotypes of Cdc25A. Interestingly, we observed abnormal and aggressive eye and liver metastasis in some implants with Cdc25A-overexpressing cells. Thus, Cdc25A overexpression may also expand breast cancer cell invasive and metastatic ability into multiple tissues. Our further findings support the idea that the effects of Cdc25A on breast cancer cell metastasis are independent of cell proliferation and apoptosis.
The results led us to the novel notion that the poor prognosis and survival associated with breast cancers overexpressing Cdc25A may be attributed to its role in cell metastasis. This relates to the larger observation that the majority of breast cancer deaths result from metastases rather than from direct effects of the primary tumor itself (14, 16, 35). Thus, our findings present a more plausible model to explain previous clinical observations and also underscore the therapeutic potential of anti-Cdc25A strategies that may target breast cancer cell metastasis.
Our studies uncovered a previously unrecognized molecular pathway, Cdc25A-Foxo1-MMP1, mediating the connection between Cdc25A and breast cancer cell metastasis. Foxo1, which is tightly regulated by multiple posttranslational modifications, including phosphorylation, acetylation, ubiquitination, and methylation, is involved in a wide range of biological processes (2, 24). The statuses of Foxo1's modifications and functions are altered under different environmental conditions and in different cell types (23, 47, 52, 54). Thus, it is possible that the Cdc25A-mediated phosphorylation and stability of Foxo1 may vary in different cell types or under different environmental conditions. Notably, the effects of Cdc25A on Foxo1 may be selective in the breast cancer cells, as the modulation of Cdc25A expression did not seem to impact Foxo3 and Foxo4 (data not shown).
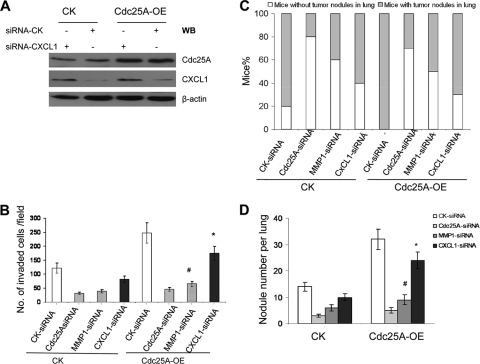
Interestingly, blocking MMP1 significantly attenuated Cdc25A-mediated metastasis of breast cancer cells (Fig. 8). This suggested that MMP1 is an important factor for mediation of the connection between Cdc25A and breast cancer metastasis. Notably, CXCL1, another cancer cell metastatic factor (3, 31), seems to be modulated by Cdc25A as well, but the modulated effects are weaker than that of Cdc25A on MMP1 (Fig. 1B and 12). Interestingly, the modulation of Cdc25A on CXCL1 is associated with the CXCL1 promoter activity through its NF-κB binding site (reference 17 and data not shown). Whether and how Cdc25A regulates NF-κB directly or indirectly, as well as downstream CXCL1, will be investigated separately in the future. In the present study, we focused on how Cdc25A modulates MMP1 and regulates breast cancer cell metastasis, and we revealed that MMP1 is an important and major factor for the mediation of the connection between Cdc25A and breast cancer metastasis.
Fig. 12.
Comparison of the effects of CXCL1 knockdown or MMP1 knockdown on invasion and metastasis of breast cancer cells driven by Cdc25A overexpression. The relative mRNA data (Fig. 1B) seem to indicate that CXCL1 is regulated by modulation of Cdc25A, as well, but these effects are weaker than that of Cdc25A on MMP1. To confirm and measure the contributions of MMP1 and CXCL1 in breast cancer cell metastatic phenotypes mediated by Cdc25A, we performed further assays. (A) CXCL1 protein levels in the indicated breast cancer cell line and its derivative with Cdc25A-OE were detected by Western blotting. (B) Cell invasion assay on Matrigel-coated Transwells and statistical analysis of cell invasive ability. The migrated cells were counted per high-power field (×200; 10 random fields). #, P < 0.01, and *, P < 0.1 compared to the CK-siRNA control in the Cdc25A-OE cells. The error bars indicate standard deviations. (C and D) The number of mice with metastasis (C) and tumor nodules in the lung (D) of the mice injected with the different breast cancer cells via the tail vein. The lungs and the H&E staining of metastatic tumors in the lungs were similar to those in shown in Fig. 5B and C. The nodules per lung for all 4 sections were counted under a light microscope. #, P=0.015, and *, P=0.21, compared to the CK-siRNA control cells and Cdc25A-OE cells; n=10. Similar results were observed in MDA-MB-231 and MDA-MB-453 breast cancer cells versus their derivatives. The results showed that CXCL1 may play a small role in mediating the link between Cdc25A and the metastatic phenotypes. In contrast, MMP1 is a major factor in mediating the connection between Cdc25A and breast cancer metastasis.
Notably, Cdc25A overexpression is associated with poor prognosis of breast, colorectal, lung, prostate, and esophageal cancers (5, 8, 15, 21, 50); similarly, MMP1 overexpression was also observed in these different types of cancer (6, 9, 26, 40, 45). The potential molecular connection between Cdc25A and MMP1 may reflect their clinical similarity. Although it was reported that Cdc25A associates with hepatocellular carcinoma metastasis (48) and that the 263C/T polymorphism of Cdc25A was significantly associated with breast cancer and risk of metastasis (29), details regarding the possible molecular mechanism(s) have so far been absent. Thus, it would be of great interest to investigate the possible connection between Cdc25A, Foxo1, MMP1, and metastasis in other cancer cells, in addition to breast cancer.
In relating Cdc25A to breast cancer cell metastasis, our studies indicate that Cdc25A would be an attractive pharmacological target in treatment of metastatic breast cancer. Recent advances in the Cdc25A field have led to the identification of anti-Cdc25A reagents, but it has been difficult to predict therapeutic success due to uncertainties surrounding safety and efficacy (44). We found that the natural dietary I3C, which induced Cdc25A degradation through its Ser124 site (Fig. 11A to C) (51), blocked breast cancer cell metastasis. This may represent an effective and safe strategy for prevention and treatment of human metastatic breast cancers.
In summary, our studies provide the novel notion that the poor prognosis and survival associated with breast cancers overexpressing Cdc25A may be attributed to its role in cell metastasis and a mechanism by which this occurs. These studies are important, as the majority of breast cancer deaths result from metastases rather than from direct effects of the primary tumor itself (14, 16, 35). Our findings for interaction of aberrant Cdc25A and an invasive phenotype present a plausible model to explain previous clinical observations and also underscore the therapeutic potential of anti-Cdc25A strategies that may target breast cancer cell metastasis. Our further findings of the therapeutic potential of an anti-Cdc25A strategy show that Cdc25A may be a novel chemopreventive tool for the effective treatment of breast cancer metastasis.
ACKNOWLEDGMENTS
We thank H. Huang for Cdk2-related plasmids, A. Bonni and S. Guo for kindly providing Foxo1 and Foxo-1-S249A-GFP plasmids, S. H. Barsky for the Mary-X cell line, G. L. Wang, and Y. Jin for help in plasmid construction, bioinformatics, and promoter analysis, and Michelle Smith, Weiwei Zhang, Yuanzhi Lu, and Legong Li for helpful suggestions.
This research was supported by grants NIH R01 CA113579 (X.Z.), NSFC 30571006 and 30671026 (X.Z.), KM200810028011 (X.Z.), and NIH R01 CA109527 (R.K.G.) in part.
Footnotes
Published ahead of print on 13 June 2011.
REFERENCES
- 1. Abedin M. J., Wang D., McDonnell M. A., Lehmann U., Kelekar A. 2007. Autophagy delays apoptotic death in breast cancer cells following DNA damage. Cell Death Differ. 14:500–510 [DOI] [PubMed] [Google Scholar]
- 2. Arden K. C. 2007. FoxOs in tumor suppression and stem cell maintenance. Cell 128:235–237 [DOI] [PubMed] [Google Scholar]
- 3. Bachmeier B. E., et al. 2008. Curcumin downregulates the inflammatory cytokines CXCL1 and -2 in breast cancer cells via NFkappaB. Carcinogenesis 29:779–789 [DOI] [PubMed] [Google Scholar]
- 4. Blomberg I., Hoffmann I. 1999. Ectopic expression of Cdc25A accelerates the G(1)/S transition and leads to premature activation of cyclin E- and cyclin A-dependent kinases. Mol. Cell. Biol. 19:6183–6194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boutros R., Lobjois V., Ducommun B. 2007. CDC25 phosphatases in cancer cells: key players? Good targets? Nat. Rev. Cancer 7:495–507 [DOI] [PubMed] [Google Scholar]
- 6. Bradbury P. A., et al. 2009. Matrix metalloproteinase 1, 3 and 12 polymorphisms and esophageal adenocarcinoma risk and prognosis. Carcinogenesis 30:793–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Busino L., et al. 2003. Degradation of Cdc25A by beta-TrCP during S phase and in response to DNA damage. Nature 426:87–91 [DOI] [PubMed] [Google Scholar]
- 8. Cangi M. G., et al. 2000. Role of the Cdc25A phosphatase in human breast cancer. J. Clin. Invest. 106:753–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cao J., et al. 2008. Membrane type 1 matrix metalloproteinase induces epithelial-to-mesenchymal transition in prostate cancer. J. Biol. Chem. 283:6232–6240 [DOI] [PubMed] [Google Scholar]
- 10. Castrellon A. B., Gluck S. 2008. Chemoprevention of breast cancer. Expert Rev. Anticancer Ther. 8:443–452 [DOI] [PubMed] [Google Scholar]
- 11. Chen M. S., Hurov J., White L. S., Woodford-Thomas T., Piwnica-Worms H. 2001. Absence of apparent phenotype in mice lacking Cdc25C protein phosphatase. Mol. Cell. Biol. 21:3853–3861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen M. S., Ryan C. E., Piwnica-Worms H. 2003. Chk1 kinase negatively regulates mitotic function of Cdc25A phosphatase through 14-3-3 binding. Mol. Cell. Biol. 23:7488–7497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cheng S., et al. 2008. High MMP-1 mRNA expression is a risk factor for disease-free and overall survivals in patients with invasive breast carcinoma. J. Surg. Res. 146:104–109 [DOI] [PubMed] [Google Scholar]
- 14. Chiang A. C., Massague J. 2008. Molecular basis of metastasis. N. Engl. J. Med. 359:2814–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chiu Y. T., et al. 2009. CDC25A functions as a novel Ar corepressor in prostate cancer cells. J. Mol. Biol. 385:446–456 [DOI] [PubMed] [Google Scholar]
- 16. Culhane A. C., Quackenbush J. 2009. Confounding effects in “a six-gene signature predicting breast cancer lung metastasis”. Cancer Res. 69:7480–7485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dhawan P., Richmond A. 2002. A novel NF-kappa B-inducing kinase-MAPK signaling pathway up-regulates NF-kappa B activity in melanoma cells. J. Biol. Chem. 277:7920–7928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ferguson A. M., White L. S., Donovan P. J., Piwnica-Worms H. 2005. Normal cell cycle and checkpoint responses in mice and cells lacking Cdc25B and Cdc25C protein phosphatases. Mol. Cell. Biol. 25:2853–2860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Galaktionov K., Beach D. 1991. Specific activation of cdc25 tyrosine phosphatases by B-type cyclins: evidence for multiple roles of mitotic cyclins. Cell 67:1181–1194 [DOI] [PubMed] [Google Scholar]
- 20. Galaktionov K., et al. 1995. CDC25 phosphatases as potential human oncogenes. Science 269:1575–1577 [DOI] [PubMed] [Google Scholar]
- 21. Gasparotto D., et al. 1997. Overexpression of CDC25A and CDC25B in head and neck cancers. Cancer Res. 57:2366–2368 [PubMed] [Google Scholar]
- 22. Greenberg P. A., et al. 1996. Long-term follow-up of patients with complete remission following combination chemotherapy for metastatic breast cancer. J. Clin. Oncol. 14:2197–2205 [DOI] [PubMed] [Google Scholar]
- 23. Huang H., Regan K. M., Lou Z., Chen J., Tindall D. J. 2006. CDK2-dependent phosphorylation of FOXO1 as an apoptotic response to DNA damage. Science 314:294–297 [DOI] [PubMed] [Google Scholar]
- 24. Huang H., Tindall D. J. 2007. Dynamic FoxO transcription factors. J. Cell Sci. 120:2479–2487 [DOI] [PubMed] [Google Scholar]
- 25. Ito Y., et al. 2004. Expression of cdc25A and cdc25B phosphatase in breast carcinoma. Breast Cancer 11:295–300 [DOI] [PubMed] [Google Scholar]
- 26. Jeffery N., McLean M. H., El-Omar E. M., Murray G. I. 2009. The matrix metalloproteinase/tissue inhibitor of matrix metalloproteinase profile in colorectal polyp cancers. Histopathology 54:820–828 [DOI] [PubMed] [Google Scholar]
- 27. Jinno S., et al. 1994. Cdc25A is a novel phosphatase functioning early in the cell cycle. EMBO J. 13:1549–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kang T., et al. 2008. GSK-3 beta targets Cdc25A for ubiquitin-mediated proteolysis, and GSK-3 beta inactivation correlates with Cdc25A overproduction in human cancers. Cancer Cell 13:36–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Karagoz I. D., et al. 2010. CDC 25A gene 263C/T, -350C/T, and -51C/G polymorphisms in breast carcinoma. Tumour Biol. 31:597–604 [DOI] [PubMed] [Google Scholar]
- 30. Kennedy D. A., Lee T., Seely D. 2009. A comparative review of thermography as a breast cancer screening technique. Integr. Cancer Ther. 8:9–16 [DOI] [PubMed] [Google Scholar]
- 31. Kim M. Y., et al. 2009. Tumor self-seeding by circulating cancer cells. Cell 139:1315–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Krtolica A., Ludlow J. W. 1996. Hypoxia arrests ovarian carcinoma cell cycle progression, but invasion is unaffected. Cancer Res. 56:1168–1173 [PubMed] [Google Scholar]
- 33. Langley R. R., Fidler I. J. 2007. Tumor cell-organ microenvironment interactions in the pathogenesis of cancer metastasis. Endocr. Rev. 28:297–321 [DOI] [PubMed] [Google Scholar]
- 34. Lincoln A. J., et al. 2002. Cdc25b phosphatase is required for resumption of meiosis during oocyte maturation. Nat. Genet. 30:446–449 [DOI] [PubMed] [Google Scholar]
- 35. Ma L., Teruya-Feldstein J., Weinberg R. A. 2007. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 449:682–688 [DOI] [PubMed] [Google Scholar]
- 36. Minn A. J., et al. 2005. Distinct organ-specific metastatic potential of individual breast cancer cells and primary tumors. J. Clin. Invest. 115:44–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nelson M. H., Dolder C. R. 2007. A review of lapatinib ditosylate in the treatment of refractory or advanced breast cancer. Ther. Clin. Risk Manag. 3:665–673 [PMC free article] [PubMed] [Google Scholar]
- 38. Nilsson I., Hoffmann I. 2000. Cell cycle regulation by the Cdc25 phosphatase family. Prog. Cell Cycle Res. 4:107–114 [DOI] [PubMed] [Google Scholar]
- 39. Ouyang W., Beckett O., Flavell R. A., Li M. O. 2009. An essential role of the Forkhead-box transcription factor Foxo1 in control of T cell homeostasis and tolerance. Immunity 30:358–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Poola I., et al. 2005. Identification of MMP-1 as a putative breast cancer predictive marker by global gene expression analysis. Nat. Med. 11:481–483 [DOI] [PubMed] [Google Scholar]
- 41. Rahman K. M., et al. 2006. Therapeutic intervention of experimental breast cancer bone metastasis by indole-3-carbinol in SCID-human mouse model. Mol. Cancer Ther. 5:2747–2756 [DOI] [PubMed] [Google Scholar]
- 42. Ray D., et al. 2005. Transforming growth factor beta facilitates beta-TrCP-mediated degradation of Cdc25A in a Smad3-dependent manner. Mol. Cell. Biol. 25:3338–3347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ray D., et al. 2007. Hemizygous disruption of Cdc25A inhibits cellular transformation and mammary tumorigenesis in mice. Cancer Res. 67:6605–6611 [DOI] [PubMed] [Google Scholar]
- 44. Rudolph J. 2007. Inhibiting transient protein-protein interactions: lessons from the Cdc25 protein tyrosine phosphatases. Nat. Rev. Cancer 7:202–211 [DOI] [PubMed] [Google Scholar]
- 45. Sauter W., et al. 2008. Matrix metalloproteinase 1 (MMP1) is associated with early-onset lung cancer. Cancer Epidemiol. Biomarkers Prev. 17:1127–1135 [DOI] [PubMed] [Google Scholar]
- 46. Trivedi V., et al. 2009. Platelet matrix metalloprotease-1 mediates thrombogenesis by activating PAR1 at a cryptic ligand site. Cell 137:332–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. van der Horst A., Burgering B. M. 2007. Stressing the role of FoxO proteins in lifespan and disease. Nat. Rev. Mol. Cell Biol. 8:440–450 [DOI] [PubMed] [Google Scholar]
- 48. Wang X. Q., et al. 2008. Aberrant Polo-like kinase 1-Cdc25A pathway in metastatic hepatocellular carcinoma. Clin. Cancer Res. 14:6813–6820 [DOI] [PubMed] [Google Scholar]
- 49. Wen R., et al. 2006. Essential role of phospholipase C gamma 2 in early B-cell development and Myc-mediated lymphomagenesis. Mol. Cell. Biol. 26:9364–9376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wu W., Fan Y. H., Kemp B. L., Walsh G., Mao L. 1998. Overexpression of cdc25A and cdc25B is frequent in primary non-small cell lung cancer but is not associated with overexpression of c-myc. Cancer Res. 58:4082–4085 [PubMed] [Google Scholar]
- 51. Wu Y., et al. 2010. A novel mechanism of indole-3-carbinol effects on breast carcinogenesis involves induction of Cdc25A degradation. Cancer Prev. Res. 3:818–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yamagata K., et al. 2008. Arginine methylation of FOXO transcription factors inhibits their phosphorylation by Akt. Mol. Cell 32:221–231 [DOI] [PubMed] [Google Scholar]
- 53. Yang E., et al. 2009. Blockade of PAR1 signaling with cell-penetrating pepducins inhibits Akt survival pathways in breast cancer cells and suppresses tumor survival and metastasis. Cancer Res. 69:6223–6231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yuan Z., et al. 2008. Activation of FOXO1 by Cdk1 in cycling cells and postmitotic neurons. Science 319:1665–1668 [DOI] [PubMed] [Google Scholar]
- 55. Zhao H., Watkins J. L., Piwnica-Worms H. 2002. Disruption of the checkpoint kinase 1/cell division cycle 25A pathway abrogates ionizing radiation-induced S and G2 checkpoints. Proc. Natl. Acad. Sci. U. S. A. 99:14795–14800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zou X., et al. 2002. Cdk4 disruption renders primary mouse cells resistant to oncogenic transformation, leading to Arf/p53-independent senescence. Genes Dev. 16:2923–2934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zou X., et al. 2001. The cell cycle-regulatory CDC25A phosphatase inhibits apoptosis signal-regulating kinase 1. Mol. Cell. Biol. 21:4818–4828 [DOI] [PMC free article] [PubMed] [Google Scholar]