Abstract
Nuclear receptors TR2 and TR4 (TR2/TR4) were previously shown to bind in vitro to direct repeat elements in the mouse and human embryonic and fetal β-type globin gene promoters and to play critical roles in the silencing of these genes. By chromatin immunoprecipitation (ChIP) we show that, in adult erythroid cells, TR2/TR4 bind to the embryonic β-type globin promoters but not to the adult β-globin promoter. We purified protein complexes containing biotin-tagged TR2/TR4 from adult erythroid cells and identified DNMT1, NuRD, and LSD1/CoREST repressor complexes, as well as HDAC3 and TIF1β, all known to confer epigenetic gene silencing, as potential corepressors of TR2/TR4. Coimmunoprecipitation assays of endogenous abundance proteins indicated that TR2/TR4 complexes consist of at least four distinct molecular species. In ChIP assays we found that, in undifferentiated murine adult erythroid cells, many of these corepressors associate with both the embryonic and the adult β-type globin promoters but, upon terminal differentiation, they specifically dissociate only from the adult β-globin promoter concomitant with its activation but remain bound to the silenced embryonic globin gene promoters. These data suggest that TR2/TR4 recruit an array of transcriptional corepressors to elicit adult stage-specific silencing of the embryonic β-type globin genes through coordinated epigenetic chromatin modifications.
INTRODUCTION
Regulatory pathways that control development through temporally specified gene activation and repression mechanisms have been recognized as “epigenetic” (i.e., heritable changes not involving alterations in the primary DNA code) for decades, although the molecules that elicit those developmental programs through epigenetic means have only been elucidated during the past several years. It is currently widely accepted that metazoan transcription factors (both activators and repressors) elicit their specific transcriptional responses through an enormous variety of cofactor molecules whose major purpose is to modulate chromatin structure (8, 31). Many such cofactors have been shown to chemically modify histones, transcription factors, and cofactors, as well as DNA, in order to elicit the required transcriptional responses.
The β-globin locus has been extensively studied as a paradigm for epigenetic regulation of lineage-specific and developmentally specific gene expression (29), as well as for its clinical relevance to β-globin disorders such as sickle cell disease and β-thalassemia. The human β-globin locus is composed of ε- (embryonic), Gγ- and Aγ- (fetal), and δ- and β-globin (adult) genes, which are spatially arranged from 5′ to 3′ and developmentally expressed in the same order (72). The elucidation of the molecular basis for γ-globin silencing in the adult stage in particular has been the focus of intense investigation, since it has been observed that coinheritance of genetic conditions that confer elevated γ-globin synthesis can significantly alleviate the symptoms of β-globin disorders (44, 56). Previously, several adult-stage γ-globin repressors have been identified, such as BCL11A and SOX6 that physically interact with each other to repress the γ-globin genes (67, 84, 86), as well as Ikaros (42) and GATA1 (20). In addition, KLF1 was recently shown to indirectly repress γ-globin synthesis through activation of the BCL11A gene (3, 92), whereas Myb (66), FOP (78), and COUP-TFII (1) also repress γ-globin expression by currently undefined mechanisms. While the precise mechanisms by which any of these factors repress γ-globin transcription is not yet fully understood, overall, the available evidence suggests that the collaborative action of multiple complex signaling pathways, which are still to be fully elucidated, are required for adult stage γ-globin gene silencing.
We previously identified DRED (direct repeat erythroid definitive) as a putative repressor complex that binds in vitro to the direct repeat (DR) elements, consensus binding sites for nonsteroidal nuclear receptors, in the ε- and γ-globin promoters (76). Subsequently, we purified and characterized DRED as a multiprotein complex with a molecular mass exceeding 500 kDa containing a heterodimer of the nuclear receptors TR2 and TR4 (TR2/TR4; in standardized nomenclature, NR2C1 and NR2C2, respectively) that can specifically bind in vitro to the DR elements of the human embryonic ε- and fetal γ-globin promoters, as well as to the murine embryonic εY- and βH1-globin promoters (73) (74). Another nuclear receptor COUP-TFII has also been reported to bind to the ε- and γ-globin DR sequences in vitro (10). However, the functional significance of COUP-TFII binding has not been clearly determined. Mutating the DR sequences in the ε- or γ-globin promoters borne on a YAC (for yeast artificial chromosome) transgene led to derepression of these genes in definitive erythroid cells of transgenic mice (55, 76). Further studies of mice in which wild-type or dominant-negative TR2 or TR4 was forcibly expressed, as well as analysis of mice bearing germ line null mutations in the TR2 and TR4 genes, have demonstrated their roles in repression of the human ε- and γ-globin genes, as well as the murine embryonic globin genes (74). We also found that the Gata1 gene is a direct and temporally selective target of TR2/TR4 repression (75). These studies, while underscoring the participation of TR2 and TR4 in erythroid cell physiology, did not provide insights into the mechanistic underpinnings by which TR2 and TR4 might elicit those temporal and lineage-specific transcriptional responses.
Nuclear receptors comprise a unique category of ligand-dependent activators and repressors since they have the ability, usually mediated through their association with individually unique small molecule ligands, to activate or repress gene transcription. TR2 and TR4 are both widely expressed and form homodimers and heterodimers that are capable of binding to DR sequences to either activate or repress target genes (38). Analyses of germ line mutant mice have shown that TR4 activates the genes for ApoE/C-I/C-II (30), PEPCK (41), LH receptor (6), and CD36 (83). Transcriptional repression by TR2 was shown to be mediated through direct association with RIP140 (34) and histone deacetylase 3 (HDAC3) (19). However, the molecular mechanisms that convert TR2 and TR4 from transcriptional activators to repressors and vice versa are not well understood, although it was recently shown that unsaturated fatty acids and their metabolites, as well as retinol and all-trans retinoic acid, can act as activating ligands for TR4, strongly implying that TR4 is no longer an “orphan” (77, 83) (93).
We provide here direct in vivo evidence for TR2/TR4 binding to the proximal promoters of embryonic β-type globin genes in adult erythroid cells, as would be predicted from their role as adult-stage repressors of the embryonic and fetal β-type globin genes. We also report that the most prominent TR2/TR4-interacting proteins in adult erythroid (erythroleukemia) cells are molecular complexes that are normally associated with epigenetic gene silencing. These TR2/TR4 interacting proteins include DNMT1 (DNA methyltransferase 1), HDAC3, and TIF1β (transcriptional intermediary factor 1β), as well as the NuRD (nucleosome remodeling and deacetylase) repressor complex containing the chromatin remodeling ATPase Mi2 and HDAC1/2, and the LSD1/CoREST repressor complex containing HDAC1/2 and LSD1 (lysine-specific histone demethylase 1) that demethylates mono- and dimethyl-histone H3 lysine 4 (H3K4), an activating histone epigenetic modification. Finally, by chromatin immunoprecipitation (ChIP) analysis, we found that, in undifferentiated murine adult erythroleukemia cells, these corepressor proteins associate with both embryonic and adult β-type globin promoters; however, upon terminal differentiation they specifically dissociate only from the adult β-globin promoter concomitant with its activation but remain bound to the silenced embryonic globin gene promoters. These data collectively suggest that TR2 and TR4 recruit and coordinate a well-defined array of distinct transcriptional corepressors to elicit developmental stage-specific gene silencing through a series of epigenetic chromatin modifications by nucleosome remodeling, histone deacetylation and H3K4 demethylation, and DNA CpG methylation.
MATERIALS AND METHODS
Real-time PCR analysis for murine β-type globin gene expression in MEL cells.
Mouse erythroleukemia (MEL) cells were cultured in RPMI medium 1640 (Gibco) supplemented with 10% fetal calf serum plus 20 mg of glutamine/liter. Total RNA was prepared from three independent cultures of MEL cells either before or 1, 3, or 5 days after differentiation induction with 2% dimethyl sulfoxide (DMSO) and then used as a template for cDNA synthesis as described previously (74). Real-time quantitative PCR was performed with 0.1 μl of the cDNA samples in triplicate in a 25-μl reaction using SYBR green Master Mix (Applied Biosystems) on an ABI Prism 7000 (Applied Biosystems). The abundance of each cDNA was calculated based on the threshold cycle (CT) value and the amplification efficiency, which was determined experimentally for each primer set, and then normalized to the abundance of 18S rRNA transcript as the internal control. All of the primer sets except for 18S were designed to span introns. PCR primers used to quantify the expression of murine embryonic εY- and βH1-globin, adult β-globin (coamplifying both βmajor and βminor) mRNAs and 18S rRNA are shown in Table 1.
Table 1.
Primers used for real-time quantitative PCR
| Method and target | Sequence (5′–3′) |
|
|---|---|---|
| Sense | Antisense | |
| RT-PCR | ||
| εY-globin | ACCCTCATCAATGGCCTGTGGA | CATGGGCTTTGACCCTTGGG |
| βH1-globin | ATCATGGGAAACCCCCGGA | GGGTGAATTCCTTGGCAAAATGAGT |
| Adult β-globin | TCTGCTATCATGGGTAATGCCAAA | GAAGGCAGCCTGTGCAGCG |
| 18S rRNA | GCTGCTGGCACCAGACTT | CGGCTACCACATCCAAGG |
| ChIP assay | ||
| εY promoter | GAAAGAATACCTCCATATCTAATGTGCAT | CTGCATTATTCTTTGAAGCTATTGGT |
| βH1 promoter | GGACCCCACCCCTGTCTT | TTACCCCTCCCCAGGACTCT |
| βmajor promoter | GAAGCCTGATTCCGTAGAGC | CAACTGATCCTACCTCACCTTATATGC |
| βH1-βmajor intergenic | CGGGATGGGCATTAAAGGTA | AACAACCTGTGTCAGAAGCAGATG |
| SCAP | CGCGGTCCGGTGTTTG | GGAAAGGTAGGAGTTGAGAGGTGAA |
| Human ε promoter | CACAAACTTAGTGTCCATCCATCAC | CCCTGTTCTCCATGGTACTTAAAAG |
| Human γ promoter | GCAAATATCTGTCTGAAACGG | GTGGAACTGCTGAAGGGTGCTT |
| Human β promoter | GAGGGTTTGAAGTCCAACTCCTAA | CAGGGTGAGGTCTAAGTGATGACA |
Antibodies.
Rabbit polyclonal antibodies against TR2 and TR4 generated and purified as described previously (74) were further purified with a Melon Gel IgG spin purification kit (Thermo Scientific). For coimmunoprecipitation and Western blotting, we used antibodies against DNMT1 (sc-20701), MTA1 (sc-10813), MTA2 (sc-9447), HDAC2 (sc-7899), Mi2β (sc-8774), RbAp46/48 (sc-8272), MBD2 (sc-12444), MBD3 (sc-9402), CoREST (sc-23448), LSD1 (sc-67272), and TIF1β (sc-33186) from Santa Cruz Biotechnology; antibodies against HDAC1 (ab7028), p66 (ab76924), HDAC3 (ab7030), and LSD1 (ab17721) from Abcam; and an antibody against TIF1β (K0075-04) from US Biologicals. For the ChIP assays, we used the same Abcam antibodies against HDAC1, HDAC3, and LSD1, as well as antibodies against CoREST (ab24166), TIF1β (ab10483), DNMT1 (ab16632), and Mi2β (ab72418) and normal rabbit IgG (ab46540) from Abcam, and an MTA1 antibody (sc-10813) from Santa Cruz Biotechnology.
ChIP assays.
ChIP assays were performed essentially as described previously (13, 54) with minor modifications. MEL cells were harvested before or after differentiation induction (with 2% DMSO for 5 days). Differentiation was confirmed by benzidine staining of the cells (>90%). Human primary erythroid cells differentiated ex vivo from CD34+ cells were harvested on days 8, 11, and 14 of the differentiation culture (below). A total of 108 cells were washed twice with phosphate-buffered saline (PBS) and then incubated with 1% formaldehyde (Polysciences) in 50-ml PBS at room temperature for 10 min with gentle shaking, followed by the addition of 0.125 M glycine and continued incubation for 5 min. For ChIP assays with the TR2 or TR4 antibodies, the cells were treated with 2 mM ethylene glycol-bis(succinimidyl succinate) (EGS; Pierce) in 50 ml of PBS at room temperature for 30 min prior to the addition of formaldehyde as described previously (89). All procedures were carried out at 0 to 4°C unless specifically stated otherwise. After the cells were lysed with a Dounce homogenizer in 10 ml of cell lysis buffer (5 mM PIPES [pH 8.0], 85 mM KCl, 1% Igepal) with protease inhibitors (10 μg of aprotinin/ml, 10 μg of leupeptin/ml, and 1 mM phenylmethylsulfonyl fluoride [PMSF]), nuclei were collected by centrifugation at 13,000 × g for 5 min and then lysed by incubation in 2 ml of ChIP lysis buffer (50 mM Tris [pH 8.0], 10 mM EDTA, 1% sodium dodecyl sulfate [SDS]) containing the protease inhibitors for 30 min. After shearing DNA by sonication, the lysate was clarified by centrifugation (at 10,000 × g for 10 min) and then divided into 20-μl aliquots for immunoprecipitation (IP), to which 500 μl of IP dilution buffer (50 mM Tris [pH 7.4], 150 mM NaCl, 1% Igepal, 0.25% deoxycholic acid, 1 mM EDTA, and protease inhibitors) was added. After the addition of 1 to 10 μg of antibody to the 500 μl of lysate (equivalent to 106 cells) and incubation for 12 h, 15 μl of protein A-agarose beads (Millipore) was added to the lysate and mixed by rotation for an additional 2 h. The beads were washed with 1 ml of IP dilution buffer without protease inhibitors with rotation three times for 5 min and then once with PBS. Protein-DNA cross-linked material was then eluted by vortexing in 150 μl of elution buffer (50 mM NaHCO3, 1% SDS) for 30 min at room temperature. After the beads were removed by centrifugation (190 × g for 5 min at room temperature), 5 M NaCl was added to a final concentration of 0.54 M. After the samples were heated to 67°C for 2 h to reverse the cross-linking, the DNA was purified by using a QIAquick PCR purification kit (Qiagen) and then subjected to real-time quantitative PCR assay with SYBR green. PCR assays were performed in triplicate from at least two independent immunoprecipitations. The abundance of specifically immunoprecipitated DNA relative to input was calculated from the CT values for the samples, and the amplification efficiency was determined experimentally for each primer set. The PCR primers used to quantify murine genomic sequences in the embryonic εY- and βH1-globin and the adult βmajor-globin promoters, the intergenic region between βH1- and βmajor-globin genes, and the SCAP gene promoter, as well as human genomic sequences in the embryonic ε-, fetal γ-, and adult β-globin promoters, are shown in Table 1.
Ex vivo differentiation of purified human CD34+ cells.
Cryopreserved vials of purified human CD34+ cells, which were mobilized in and isolated from the peripheral blood of healthy donors, were purchased from the Fred Hutchinson Cancer Research Center. The cells were grown and differentiated ex vivo into the erythroid lineage by a two-phase culture method reported previously (16). Briefly, during the first phase (from days 0 to 8), the cells were cultured in IMDM basic medium (Biochrom) supplemented with 40 μg of inositol/ml, 10 μg of folic acid/ml, 160 μM monothioglycerol, 120 μg of iron-saturated human transferrin/ml, 10 μg of insulin/ml, 900 ng of ferrous sulfate/ml, 90 ng of ferric nitrate/ml, 1 μM hydrocortisone, and 5 ng of interleukin-3 (IL-3)/ml, which were all purchased from Sigma-Aldrich, and 200 mM l-glutamine (Invitrogen), 1% deionized bovine serum albumin (Stemcell Technologies), 100 ng of stem cell factor (Prospec)/ml, and 3 IU of erythropoietin/ml, which was purchased from Amgen. During the second phase (day 8 through day 14), the cells were cultured in the same medium without hydrocortisone or IL-3.
Plasmid construction and transfection.
The 23-amino-acid biotinylation tag sequence (7) was cloned into the NcoI site overlapping the translation initiation codon of the N-terminally FLAG-tagged TR2 and TR4 cDNAs previously cloned in pBluescript II SK(+) (73). After the introduction of an EagI linker into the XhoI site of the multiple cloning site at the C terminus of the biotin-FLAG-TR2 or biotin-FLAG-TR4 construct, the tagged cDNAs were released from the vector by EagI digestion and cloned into a unique NotI site in the erythroid-specific expression vector pEV3neo (52). The pEV3neo vectors expressing tagged TR2 or TR4 were transfected into MEL cells stably transformed with the BirA biotin ligase, as previously described (7). Clones were obtained by G418 and puromycin selection (the latter selects for BirA-expressing cells) and screened by immunoblotting for efficient expression and biotinylation of tagged TR2 and TR4.
Fractionation of nuclear extract by Superose 6 gel filtration.
Nuclear extracts were prepared from MEL cells expressing tagged TR2 and TR4, which were induced to differentiate, as previously described (7, 60). Size fractionation of MEL nuclear extracts using an analytical Superose 6 column was carried out as previously described (59, 60). Preparative size fractionation by gel filtration was done by injecting 40 mg of nuclear extract protein (in 5 ml) into a preparative-grade Superose 6 XK50/600 column connected to an AKTAFPLC system (GE Healthcare Life Sciences) equilibrated in running buffer (20 mM HEPES [pH 7.9], 0.5 mM EGTA, 1 mM MgCl2, 200 mM KCl, 10% glycerol). Gel filtration was carried out at 4°C, and 10-ml fractions were collected and concentrated by trichloroacetic acid precipitation for analysis by immunoblotting or pooled and bound directly to streptavidin beads, as described below. A detailed protocol for preparative Superose 6 gel filtration has already been described (60).
Binding to streptavidin beads.
Direct binding of nuclear extracts to streptavidin beads was carried out as described previously (7, 60). After pooling fractions 20 to 38 from the preparative gel filtration, the KCl and NP-40 concentrations were adjusted to 150 mM and 0.3%, respectively. Diluted, pooled fractions were then divided into 50-ml Falcon tubes, to which Dynabeads M-280–streptavidin (Invitrogen), preblocked as described previously (7), was added for overnight incubation at 4°C on a rotating wheel. Approximately 10 μl of resuspended beads were used per original 10-ml fraction. After overnight incubation, the beads were washed as described previously (7). Bound proteins were eluted with 1× Laemmli sample buffer (50 μl of sample buffer per 20 μl of beads), and then resolved by SDS-PAGE. Alternatively, bound proteins were treated with trypsin directly on the beads and processed for mass spectrometry as described below.
Mass spectrometry.
Proteins eluted from streptavidin beads were resolved by SDS-PAGE, and gel lanes were cut into slices by using an automatic gel slicer and subjected to in-gel trypsinization, essentially as described previously (69). Alternatively, bound proteins were treated with trypsin on the beads after resuspending in 50 mM ammonium bicarbonate and adding trypsin (sequencing grade; Promega) to approximately 60 ng/mg of total protein, followed by overnight incubation at 37°C (64). The supernatant containing the trypsin-treated peptides was then recovered by magnetically removing the beads. Peptides released by in-gel or on-bead trypsinization were analyzed by nano-LC-MS/MS performed on either a CapLC system (Waters, Manchester, United Kingdom) coupled to a Q-ToF Ultima mass spectrometer (Waters), operating in positive mode and equipped with a Z-spray source, or on a 1100 series capillary LC system (Agilent Technologies) coupled to an LTQ-Orbitrap or LTQ-FT-MS mass spectrometer (both from Thermo Scientific) operating in positive mode and equipped with a nanospray source. Peptides were trapped and separated on a Jupiter C18 reversed-phase column (Phenomenex) using a linear gradient from 0 to 80% medium B (where medium A = 0.1 M acetic acid and medium B = 80% [vol/vol] acetonitrile, 0.1 M acetic acid) using a splitter. The column eluate was directly sprayed into the electrospray ionization source of the mass spectrometer. Mass spectra were acquired in continuum mode; fragmentation of the peptides was performed in data-dependent mode.
Data analysis and protein identification.
Peak lists were automatically created from raw data files using ProteinLynx Global Server software (version 2.0; Waters, Manchester, United Kingdom) for Q-ToF spectra and Mascot Distiller software (version 2.0; MatrixScience, London, United Kingdom) for LTQ-Orbitrap and LTQ-FT-MS spectra. The Mascot search algorithm was used for searching the National Center for Biotechnology Information (NCBI) database (NCBI no. 20060106; taxonomy Mus musculus). The Mascot score cutoff value for a positive hit was set to 65. Individual peptide MS/MS spectra with Mowse scores below 40 were checked manually and either interpreted as valid identifications or discarded. Identified proteins listed as NCBI database entries were screened to identify proteins that were also identified in mass spectrometry experiments from control BirA-expressing cells (7). These were removed as background binding proteins. The remaining proteins were classified according to gene ontology criteria as listed for each protein in the Mouse Genome Informatics database (http://www.informatics.jax.org/) and were then grouped according to a highly representative identifier based on biological process or molecular function.
Streptavidin pulldown assay.
Nuclear extracts were prepared as described previously (76) from MEL cells expressing biotin-tagged TR2 and TR4. Preblocked Dynabeads M-270–streptavidin (Invitrogen) was incubated with the nuclear extracts containing biotin-tagged TR2 and TR4 proteins for 1 h at 4°C with rotation according to the manufacturer's instructions. Portions (150 μg) of nuclear extract protein were used for each assay. After the beads were washed three times with buffer (10 mM HEPES-KOH [pH 9.0], 250 mM KCl, 1.5 mM MgCl2, 0.25 mM EDTA, 20% glycerol, 0.3% NP-40, 1 mM PMSF), bound proteins were eluted from the beads by boiling in 1× Laemmli sample buffer and then subjected to SDS-PAGE, followed by immunoblotting.
Immunoprecipitation.
Antibodies were coupled to Dynabeads-protein G (Invitrogen) by incubation for 2 h at 4°C with rotation according to the manufacturer's instructions. The beads were then incubated with nuclear extracts (150 μg of protein) prepared from untransfected MEL cells for 2 h at 4°C. After the beads were washed with ice-cold PBS three times, immunoprecipitated proteins were eluted by boiling in 1× Laemmli sample buffer and then subjected to immunoblotting.
Immunoblotting.
After SDS-PAGE, the proteins were transferred to a nitrocellulose membrane (Li-Cor) and probed with specific primary antibodies (described above) and fluorescence-conjugated secondary antibodies (Li-Cor). The proteins were visualized on an Odyssey infrared imaging system (Li-Cor).
RESULTS
Binding of TR2 and TR4 to the embryonic β-type globin promoters in adult erythroid cells.
Our previous biochemical and genetic studies suggested that a TR2/TR4 heterodimer directly represses the embryonic and fetal β-type globin genes in adult erythroid cells through direct repeat (DR) elements in their promoters. In order to provide further evidence for this contention, we performed ChIP assays to demonstrate in vivo TR2/TR4 binding to the embryonic β-type globin promoters in adult erythroid cells. To do so, we used the MEL cell line, the only established cell line with a gene expression profile typical of adult erythroid cells, where the embryonic εY and βH1 genes, orthologues of human embryonic ε- and fetal γ-globin genes, respectively (which both possess DR sequences in their promoters), are silenced.
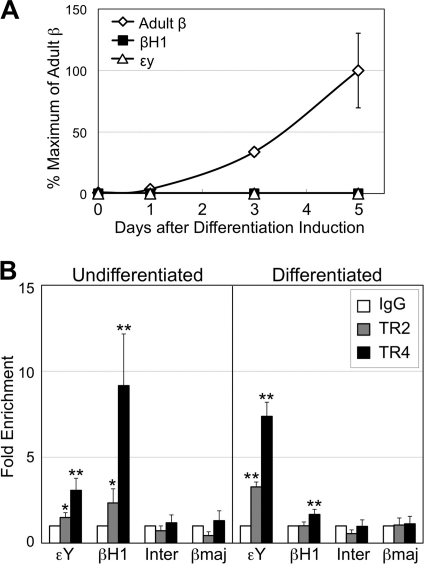
Differentiation of MEL cells was induced by exposure of cells to 2% DMSO for 5 days, which was confirmed by accumulation of adult β-globin mRNA (Fig. 1A), as well as by benzidine staining (>90% benzidine-positive cells). Cross-linked chromatin from MEL cells before and after differentiation induction with DMSO was prepared and analyzed for TR2 and TR4 binding to the globin gene promoters (Fig. 1B). In undifferentiated MEL cells, statistically significant TR4 enrichment at both εY and βH1 promoters compared to control IgG was detected. TR2 was also enriched at both promoters, albeit with slightly lower statistical significance, whereas no TR2 or TR4 binding was detectable to the adult βmajor-globin promoter (that does not have a DR consensus sequence) or to a random intergenic region between the βH1 and βmajor genes. In differentiated MEL cells, statistically significant binding of both TR2 and TR4 was detected at the εY promoter, whereas only TR4 was detected at the βH1 promoter. Interestingly, TR2/TR4 binding to the εY promoter increased after differentiation induction, whereas their binding to the βH1 promoter was diminished, which may suggest differential regulation and roles of TR2/TR4 in the silencing of the εY versus the βH1 gene, but its significance and underlying mechanisms remain unexplored (see Discussion). By ChIP assay, we also detected in vivo TR2/TR4 binding to the human embryonic ε- and fetal γ-globin promoters, which also have DR sequences (74), in primary human adult erythroid cells, which were differentiated ex vivo from purified CD34+ hematopoietic progenitor cells (data not shown). TR2/TR4 binding was not detectable on the adult β-globin promoter (that again, lacks a DR consensus sequence).
Fig. 1.
TR2/TR4 bind to the murine embryonic β-type globin gene promoters in adult erythroid (MEL) cells during differentiation. (A) Accumulation of murine adult β-globin transcripts (total of βmajor- and βminor-globin mRNAs), determined by reverse transcription and real-time quantitative PCR (RT-qPCR), in MEL cells during differentiation induced by 2% DMSO. (B) Binding of TR2 and TR4 to the proximal promoter regions, including the DR sequences, of the murine embryonic εY- and βH1-globin genes in undifferentiated or differentiated MEL cells was analyzed by ChIP assay. The statistical significance of TR2 or TR4 enrichment at the embryonic globin promoters compared to control IgG values is indicated with asterisks (*, P < 0.08; **, P < 0.04 [Student t test]). For negative controls, the proximal promoter of the adult βmajor-globin gene (βmaj; which has no DR sequence), as well as an intergenic DNA segment (Inter) lying between the βH1 and βmajor genes (5.9 kbp 5′ to the βmajor promoter), was also analyzed. Error bars represent standard errors of the mean (SEM).
Expression of biotin-tagged TR2 and TR4 in MEL cells.
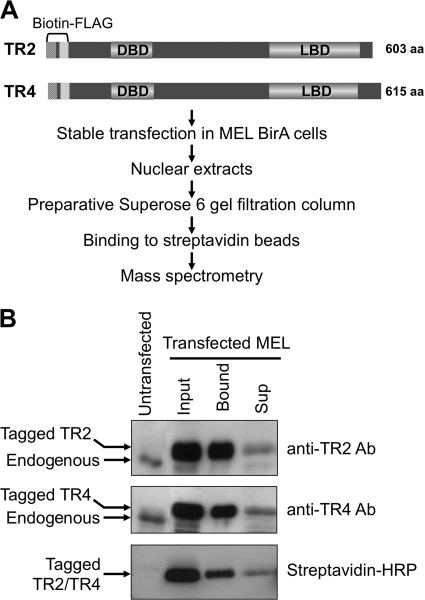
In order to investigate details of the molecular mechanisms underlying silencing by TR2/TR4 of the embryonic and fetal β-type globin genes in adult erythroid cells, we next set out to identify molecules that physically interact with TR2/TR4 and are thereby recruited to those genes to elicit stage-specific silencing. We transfected both biotin-FLAG-tagged TR2 and TR4 into MEL cells expressing the BirA biotin ligase to establish stably transformed cell lines (Fig. 2A) (7), since it has been previously shown that TR2 and TR4 preferentially heterodimerize (35) and that the TR2/TR4 heterodimer forms the core of the DRED complex that binds with high affinity to the DR sequences in the human ε- and γ-globin promoters (73, 74). We further reasoned that affinity tagging both proteins would help enrich for the DRED complex during protein complex purification. Immunoblot analysis of double transfectants using TR2 and TR4 antibodies showed that the tagged orphan receptors were expressed at higher levels than their endogenous counterparts, possibly as a consequence of using the heterologous human β-globin gene promoter of the pEV3neo vector to direct TR2 and TR4 expression (Fig. 2B). In fact, the forcibly expressed, tagged TR2 and TR4 proteins appeared to repress endogenous receptor levels (suggesting a potential negative transcriptional feedback loop, which was not explored further here). We tested the efficiency of TR2 and TR4 biotinylation by direct binding of nuclear extracts to streptavidin beads and found that 80 to 90% of the tagged protein was biotinylated by comparison of band intensities in the input and supernatant (Sup) lanes (Fig. 2B).
Fig. 2.
Experimental strategy for characterizing TR2/TR4-interacting proteins in MEL cells. (A) TR2 and TR4 cDNAs were tagged with biotinylation and FLAG sequences and then stably transfected into BirA-expressing MEL cells. DBD, DNA-binding domain; LBD, ligand binding domain. (B) Efficient biotinylation and recovery of tagged TR2 and TR4 from transformed MEL cells. Untransfected MEL and input lanes indicate the relative abundance of endogenous and biotin-tagged transfected TR2 and TR4 proteins. For input and supernatant (Sup) lanes, the same volumes of nuclear extracts before and after streptavidin-bead binding were loaded. Endogenous and transfected TR2 and TR4 proteins were detected by immunoblotting with anti-TR2 and anti-TR4 antibodies (upper and middle panels) (74). Biotin-tagged TR2 and TR4 were detected by using HRP-conjugated streptavidin (bottom panel).
Identification of TR2/TR4 interacting proteins by mass spectrometry.
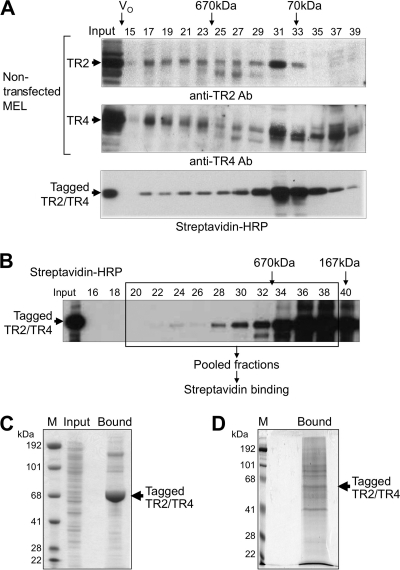
We next assessed, by size exclusion chromatography (with an analytical Superose 6 gel filtration column), the presence of TR2/TR4 in high-molecular-weight protein complexes in MEL nuclear extracts. The fractionation profiles of tagged TR2/TR4 proteins, as detected by streptavidin-horseradish peroxidase (HRP), were similar to those for the endogenous proteins in nuclear extracts from untransfected MEL cells (Fig. 3A). The majority of tagged TR2 and TR4 proteins elute in peaks equivalent to, or only slightly larger than, the molecular mass of a heterodimer (approximately 167 kDa, fraction 27 and higher), whereas less of the tagged protein elutes with massive (>670-kDa) molecular mass fractions. When we attempted larger-scale streptavidin binding of biotin-tagged TR2/TR4 from MEL nuclear extracts and then visualized the eluted material by Coomassie blue staining, we observed a strongly staining band migrating with a molecular mass consistent with that of the TR2 and TR4 monomers, with other co-eluting proteins staining more weakly (Fig. 3C). This was also reflected in the mass spectrometric analysis of this same gel lane, showing that the vast majority of peptides identified were derived from TR2 and TR4, with little information regarding the identity of other co-eluted proteins (data not shown).
Fig. 3.
Recovery of proteins that copurify with biotinylated TR2/TR4. (A) Size fractionation of nuclear extracts by analytical Superose 6 gel filtration. Fractions from induced (wild-type) MEL cells were analyzed by immunoblotting with anti-TR2 (top panel) or anti-TR4 (middle panel) antibodies. Fractions from induced MEL cells transformed with biotin-tagged TR2 and TR4 were analyzed by affinity blotting with HRP-conjugated streptavidin (bottom panel). The void volume (V0) and the elution of two molecular mass markers are indicated with arrows. (B) Fractionation of biotin-tagged TR2 and TR4 on a preparative Superose 6 column. Biotinylated TR2/TR4 proteins were detected by affinity blotting with HRP-streptavidin. Fractions that were pooled for streptavidin binding and mass spectrometric analysis are boxed. (C) Coomassie blue-stained SDS-polyacrylamide (4 to 12% gradient) gel of proteins bound to streptavidin beads from unfractionated MEL nuclear extract. Unfractionated nuclear extract prepared from induced MEL cells transfected with tagged TR2 and TR4 was incubated with streptavidin beads, from which bound proteins were eluted with 1× Laemmli sample buffer for SDS-PAGE. The arrow indicates the migration position of tagged TR2 and TR4 proteins. Input, MEL nuclear extract before binding to streptavidin beads; M, molecular mass markers. (D) Coomassie blue-stained SDS-polyacrylamide (8%) gel showing proteins bound to streptavidin beads from size-fractionated MEL nuclear extract. Proteins in size-fractionated nuclear extract from the transfected MEL cells were bound to and then eluted from streptavidin beads prior to SDS-PAGE.
On the basis of these initial observations and in order to enrich for the TR2/TR4 high-molecular-weight complexes, we used a preparative Superose 6 gel filtration column with a matrix bed volume of 550 ml, thus allowing us to fractionate more than 40 mg of protein prepared from the MEL nuclear extracts. Fractions with a molecular mass greater than that of the TR2/TR4 heterodimer were pooled in order to ensure that all high-molecular-weight TR2/TR4 complexes were captured and then bound to streptavidin beads (Fig. 3B). By Coomassie blue staining, numerous copurified protein bands were observed on SDS-polyacrylamide gels (Fig. 3D). The retained proteins were eluted by boiling and then separated by SDS-PAGE, followed by trypsinization of gel slices or treated with trypsin while still bound to the beads.
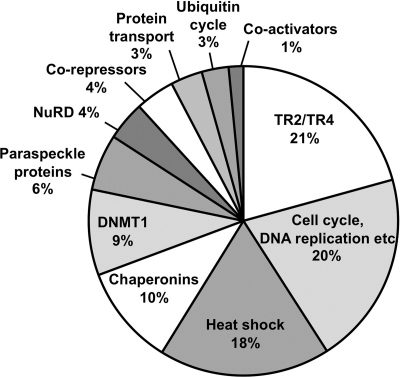
Peptides were identified by Q-TOF or LTQ-FT mass spectrometry in three separate experiments. After subtraction of the background protein binding (7), the remaining peptides were manually curated and the proteins were then grouped according to broadly representative gene ontology (GO) criteria. Identified proteins known to play direct roles in transcriptional regulation are summarized in Table 2, whereas a complete list of all peptides identified by three independent mass spectrometric analyses is available elsewhere (see Table S2 at http://www.fleming.gr/files/Supplementary_table_S2.doc). Since the total number of peptides identified per protein provides some indication of its relative abundance, we also depicted graphically the number of peptides corresponding to each GO class as a percentage of the total number of peptides (e.g., for experiment 1 in Fig. 4).
Table 2.
Mass spectrometric identification of TR2/TR4 interacting proteins
| Protein identity | Peptide content |
|||||
|---|---|---|---|---|---|---|
| Expt 1 |
Expt 2 |
Expt 3 |
||||
| Totala | % Coverage | Total | % Coverage | Total | % Coverage | |
| Orphan nuclear receptors | ||||||
| TR4 | 53 | 32.0 | 101 | 36.7 | 26 | 27.7 |
| TR2 | 28 | 9.7 | 22 | 22.5 | 9 | 17.3 |
| DNMT1 | 35 | 13.5 | 14 | 10.1 | 15 | 10.4 |
| NuRD complex | ||||||
| Mi2β | 1 | 1.1 | 4 | 5.3 | 6 | 4.4 |
| HDAC1 | 2 | 4.4 | 3 | 8.9 | ||
| HDAC2 | 2 | 4.9 | ||||
| RbAp48 | 2 | 2.0 | 3 | 6.3 | 1 | 1.9 |
| RbAp46 | 2 | 2.0 | 3 | 6.8 | ||
| MTA1 | 7 | 8.7 | 1 | 1.7 | 1 | 1.8 |
| MTA2 | 2 | 2.8 | ||||
| Corepressors | ||||||
| TIF1β | 16 | 12.7 | 6 | 9.0 | 1 | 1.8 |
| LSD1 | 1 | 3.4 | ||||
| NonO | 10 | 7.8 | 9 | 17.8 | 7 | 11.0 |
| SFPQ | 12 | 15.3 | 8 | 15.9 | 10 | 18.2 |
| PSPC1 | 1 | 3.3 | 3 | 7.8 | 2 | 4.6 |
| Sin3A | 1 | 1.1 | ||||
| Coactivators | ||||||
| HCF1 | 4 | 2.5 | 2 | 1.4 | 1 | 0.8 |
| CAPER | 2 | 4.9 | 1 | 2.6 | ||
| BRG1 | 2 | 1.4 | 1 | 0.8 | ||
| Total | 388 | 371 | 229 | |||
“Total” specifies the numbers of total peptide sequences for each protein identified in three independent mass spectrometry experiments. The percent coverage refers to the percentage of full-length sequence of each protein identified in these analyses.
Fig. 4.
Different classes of proteins identified by mass spectrometry as TR2/TR4-interacting proteins. The number of peptides corresponding to the different GO classes (see the text) is presented as a percentage of the total number of peptides shown in mass spectrometry experiment 1 in Table 2 and in Table S2 at http://www.fleming.gr/files/Supplementary_table_S2.doc.
While, as anticipated, we found that TR2 and TR4 were the most abundant proteins identified, a number of enzymes and accessory factors related to transcriptional regulation and chromatin remodeling/modification were copurified with TR2 and TR4. Intriguingly, we found DNMT1 to be among the most abundant of these copurifying proteins in all three experiments. A number of subunits of the NuRD repressor complex (85) were also consistently identified in all experiments. In addition, we identified a number of corepressor proteins that have been previously reported to interact with other nuclear receptors. These include TIF1β (33), LSD1 (70), and Sin3A (50). Other coregulatory proteins for nuclear receptors, NonO (p54nrb), SFPQ (PSF), and PSPC1 (PSP1) (45) (25), which are less well characterized and also known as paraspeckle-associated proteins, were also identified in all mass spectrometric analyses. Coactivators HCF1 (host cell factor 1) (82), CAPER (27), and BRG1 (28) were also identified in multiple experiments.
Other classes of identified proteins included those involved in cellular functions related to cell cycle, cell division, DNA replication and recombination (broadly classified as cell cycle related), heat shock proteins, chaperonins, proteins involved in intracellular protein transport and in the ubiquitin cycle (Fig. 4; see also Table S2 at http://www.fleming.gr/files/Supplementary_table_S2.doc). Interactions of nuclear receptors with heat shock proteins acting as chaperones are well characterized, and these chaperones have been implicated in refolding and nuclear translocation of nuclear receptors (49). It is possible that copurification of chaperonins, intracellular protein transporters, and ubiquitin cycle proteins is related to proper folding, intracellular localization, and regulated degradation of TR2 and TR4.
Interactions of putative corepressors with biotin-tagged TR2 and TR4.
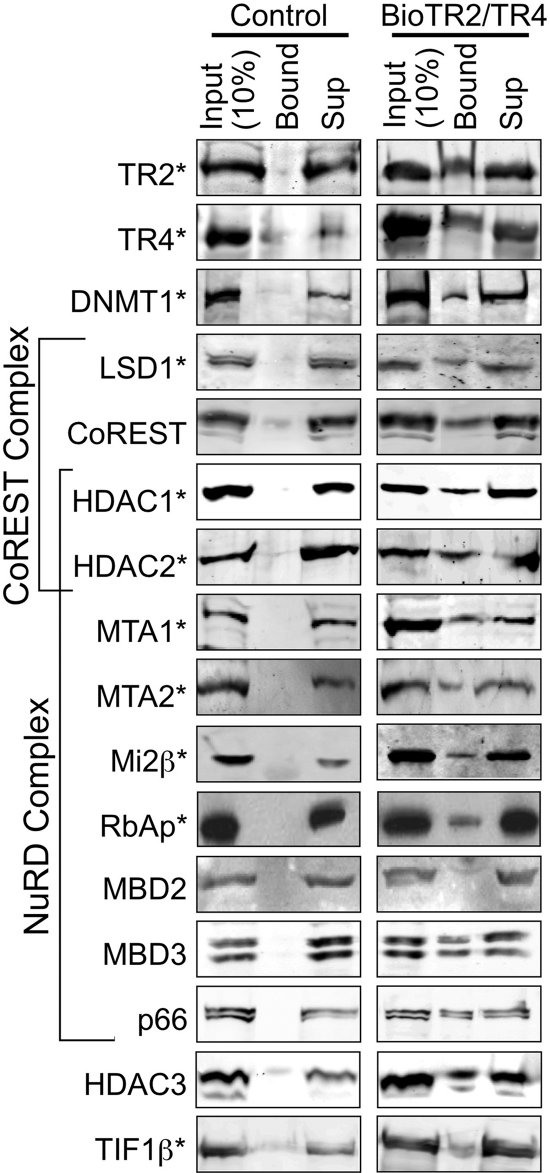
The association of TR2/TR4 with the NuRD repressor complex subunits (HDAC1/2, MTA1/2, Mi2β, and RbAp46/48), as initially indicated by mass spectrometry, was confirmed by immunoblotting of proteins recovered using streptavidin beads from nuclear extracts of MEL cells expressing biotin-tagged TR2 and TR4 (Fig. 5). In addition, other NuRD subunits that were not detected by mass spectrometry (e.g., MBD3 and p66) were also confirmed by immunoblot analysis, whereas MBD2 (39) was never detected by either mass spectrometry or immunoblotting.
Fig. 5.
Immunoblotting analysis of proteins precipitated with streptavidin beads from MEL cells expressing biotin-tagged TR2 and TR4. Nuclear extracts were prepared from induced MEL cells expressing the biotin ligase gene (birA) without (control) or with biotin-tagged TR2 and TR4 and then incubated with streptavidin beads. A total of 150 μg of nuclear extract protein was used in each binding reaction. Proteins precipitated with the beads (Bound), 10% of the input (15 μg of protein), and 10% of supernatants (Sup) were subjected to SDS-PAGE, followed by immunoblotting with antibodies that recognize the proteins shown to the left of the figure. TR2/TR4-interacting proteins identified by mass spectrometry are indicated by asterisks.
The interactions between TR2/TR4 and DNMT1, LSD1, and TIF1β, as originally identified in the mass spectrometry experiments, were also verified by streptavidin pulldown and immunoblotting (Fig. 5). In addition, CoREST, a component of the LSD1/CoREST repressor complex together with HDAC1/2 (37), was also found to interact with biotinylated TR2/TR4, even though it was not originally detected by mass spectrometry. Finally, a previously reported interaction of HDAC3 with TR2 (19) was also confirmed by immunoblotting, although it was not detected by mass spectrometry.
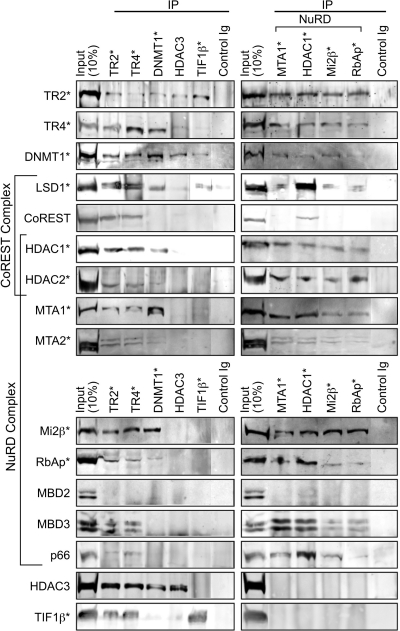
Interactions between natural abundance proteins.
In order to verify that the protein-protein interactions detected by mass spectrometry and immunoblotting following streptavidin-bead purification of tagged TR2/TR4 were genuine and to specifically rule out potentially artifactual interactions that might result from the (aberrantly abundant) expression of tagged TR2/TR4, we performed coimmunoprecipitation assays using nuclear extracts prepared from untransfected MEL cells. TR2, TR4, DNMT1, HDAC3, TIF1β, LSD1, and NuRD subunits (MTA1, HDAC1, Mi2β, and RbAp46/48) (Fig. 6 and 7) were individually immunoprecipitated from nuclear extracts of untransfected MEL cells and then probed on immunoblots to detect potentially interacting proteins. In immune complexes precipitated with either the anti-TR2 or anti-TR4 antibodies, all of the NuRD subunits that were detected by streptavidin pull-down, namely, HDAC1/2, MTA1/2, Mi2β, RbAp46/48, MBD3, and p66, were also detected, whereas MBD2, once again, was not (Fig. 6, left). In the reverse experiments, where antibodies against MTA1, HDAC1, Mi2β, and RbAp46/48 were used for immunoprecipitation, both TR2 and TR4 were detected in immune complexes (Fig. 6, right). These reciprocally reinforced data confirmed that a bona fide physical interaction takes place between TR2/TR4 and the NuRD complex that contains the MTA1/2, HDAC1/2, Mi2β, RbAp46/48, MBD3, and p66 subunits.
Fig. 6.
Immunoblotting analysis of protein interactions in untransformed MEL cells. Nuclear extracts were prepared from untransfected, induced MEL cells and incubated with protein G beads coupled with antibodies against proteins shown at the top of the figures or with a control IgG. A total of 150 μg of nuclear extract protein was used for each lane. Immunoprecipitates with the antibody-coupled beads, and 10% of the input (15 μg of protein) was subjected to SDS-PAGE, followed by immunoblotting with antibodies against proteins indicated to the left of the figures. For immunoprecipitation, antibodies against TR2, TR4, DNMT1, HDAC3, and TIF1β were used in the left panels. Antibodies against NuRD subunits (MTA1, HDAC1, Mi2β, and RbAp46/48) were used in the right panels. TR2/TR4-interacting proteins identified by mass spectrometry are indicated by asterisks.
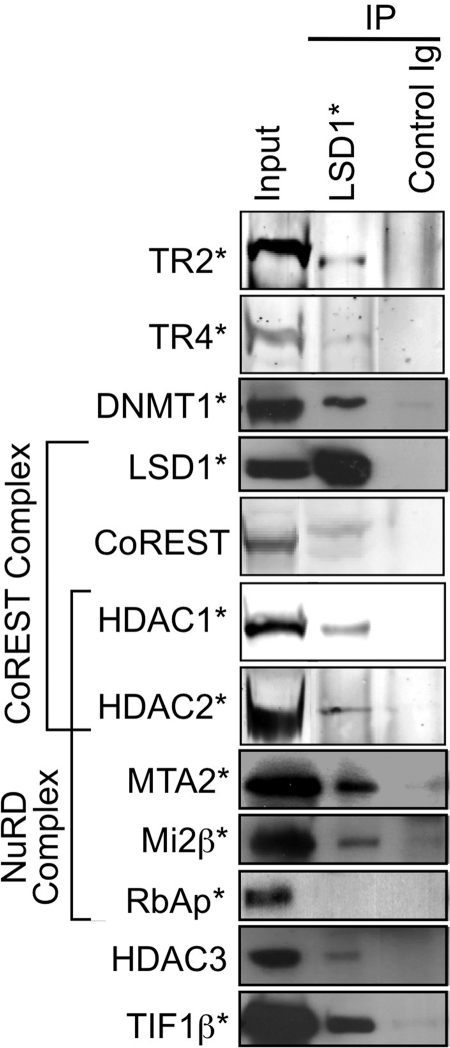
Fig. 7.
Proteins that coimmunoprecipitate with LSD1 in MEL cells. Nuclear extracts were prepared from normal, induced MEL cells and incubated with beads coupled with an anti-LSD1 antibody or control IgG. Immunoprecipitates with the anti-LSD1 or control antibody and 10% of the input were subjected to SDS-PAGE, followed by immunoblotting with antibodies against proteins indicated on the left. TR2/TR4-interacting proteins identified by mass spectrometry are indicated by asterisks.
DNMT1 and HDAC3 were also coprecipitated in immune complexes formed with either anti-TR2 or TR4 antibodies (Fig. 6, left). In the reverse experiment with anti-DNMT1 or HDAC3 antibody for primary immunoprecipitation, both TR2 and TR4 were detected (Fig. 6, left). These results confirmed the in vivo interaction of TR2/TR4 with DNMT1, as well as with HDAC3. TIF1β was also detected in the complex precipitated with either the anti-TR2 or anti-TR4 antibodies. When the reverse experiment was performed, TR2 was detected in the TIF1β immunoprecipitate, although TR4 was barely detectable. These results confirmed the in vivo interaction of TR2 with TIF1β, despite the lack of clear detection of coprecipitated TR4, possibly due to the lower abundance of TR4 compared to TR2 in MEL cells (data not shown). LSD1/CoREST complex subunits that were detected in the streptavidin purifications were also detected in the TR2 or TR4 immunoprecipitates. In the reverse experiments with an antibody recognizing LSD1 in the immunoprecipitation step, both TR2 and TR4 were detected (Fig. 7). These final coprecipitation results showed that TR2 and TR4 also interact with the LSD1/CoREST complex in vivo.
Interactions between the corepressor proteins.
In addition to the interactions of TR2 and TR4 with potential accessory cofactors, novel as well as known interactions between these coregulatory proteins were further investigated by coimmunoprecipitation assays. When several NuRD components (MTA1, HDAC1, Mi2β, or RbAp46/48) were immunoprecipitated with unique antibodies, most of the other NuRD components could clearly be detected in the immune complexes (Fig. 6, right), a finding consistent with the reported subunit structure of the NuRD complex (85). In the immune complex that was precipitated using an anti-HDAC1 antibody, the CoREST complex subunits, LSD1 and CoREST, were detected (Fig. 6, right), whereas in the immune complex precipitated with an anti-LSD1 antibody, HDAC1, HDAC2, and CoREST were detected (Fig. 7). These data are consistent with the reported subunit structure of the LSD1/CoREST complex (37).
Among the immune complexes precipitated with the anti-DNMT1 antibody, most of the NuRD subunits (MTA1/2, HDAC1/2, Mi2β, and RbAp46/48) were coprecipitated and detected by immunoblotting (Fig. 6, left). In the reverse experiments, DNMT1 was also detected in immune complexes that were first precipitated with antibodies against the NuRD subunits MTA1, HDAC1, Mi2β, or RbAp46/48 (Fig. 6, right). These data clearly demonstrated a novel in vivo interaction between DNMT1 and the NuRD complex. In the DNMT1 immune precipitates, LSD1 was also detected by immunoblotting (Fig. 6, left). In the reverse experiment, DNMT1 was similarly detected in LSD1 immunoprecipitates (Fig. 7), which is consistent with a previously reported interaction between DNMT1 and LSD1 (79). DNMT1 was also detected in both HDAC3 and TIF1β immunoprecipitates, whereas in the DNMT1 immunoprecipitate HDAC3 was detected, but TIF1β was not (Fig. 6, left). These data also demonstrated novel DNMT1 interactions with TIF1β and with HDAC3, despite the failure to detect coprecipitated TIF1β, possibly due to steric hindrance of a DNMT1 epitope by bound TIF1β.
In immune complexes precipitated with the anti-LSD1 antibody, most of the NuRD subunits (specifically MTA2, HDAC1/2, and Mi2β) were detectable by immunoblotting (Fig. 7). In the reverse experiments, LSD1 could be detected among immune complexes precipitated with antibodies recognizing individual NuRD components (MTA1, HDAC1, Mi2β, and RbAp46/48; Fig. 6, right). These data are consistent with the previously reported interaction between LSD1 and the NuRD complex (81). In LSD1 immunoprecipitates, TIF1β and HDAC3 were also detected (Fig. 7). These data demonstrate the existence of novel interactions between LSD1 and TIF1β and with HDAC3.
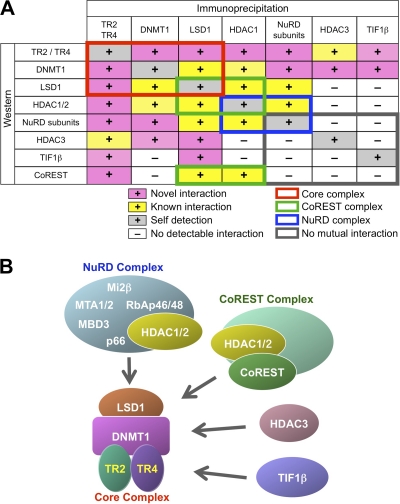
Hypothetical models for the TR2/TR4 repressor complexes.
The results of the coimmunoprecipitation analyses are summarized in Fig. 8A. Based on these, we postulate the existence of multiple TR2 and TR4 repressor complexes. TR2, TR4, DNMT1, and LSD1 were shown to mutually interact with one another in the coimmunoprecipitation assays, suggesting that these four proteins comprise a core complex (Fig. 8B). Most of these four core complex components commonly interact with HDAC1/2, other NuRD components (such as MTA1/2, Mi2β, or RbAp46/48), CoREST, HDAC3, and TIF1β, indicating that these proteins can interact with this core complex to form even larger complexes that share the core proteins. Interestingly, no physical interactions among the NuRD signature components (except HDAC1/2), HDAC3, TIF1β, or CoREST were detectable in multiple, independent coimmunoprecipitation experiments (Fig. 8A), suggesting that the interactions of TR2/TR4 with the NuRD complex, with HDAC3, with TIF1β, or with CoREST are each mutually exclusive and independent associations and that these proteins form at least four distinct larger complexes that share the core protein group (Fig. 8B).
Fig. 8.
Multiple TR2/TR4 repressor complexes in adult erythroid (MEL) cells. (A) Summary of coimmunoprecipitation experiments using normal MEL cells. Novel interactions identified in the present study are shown as plus signs on a pink background, whereas previously reported interactions are shown on a yellow background. Detection of a protein immunoprecipitated with an antibody against it is indicated on a gray background as a positive control (self-detection). Known protein interactions between components of the NuRD and CoREST repressor complexes are shown in blue and green rectangles, respectively. TR2, TR4, DNMT1, and LSD1 interactions are indicated by a red rectangle and comprise a core complex (shown in panel B). These four core constituents commonly interact with additional proteins, such as HDAC1/2, NuRD components (MTA1/2, Mi2β, or RbAp46/48), HDAC3, TIF1β, or CoREST, thus forming larger complexes. However, no mutual interactions between NuRD components (except HDAC1/2), HDAC3, TIF1β or CoREST were detected by coimmunoprecipitation (indicated by the gray rectangle), suggesting that NuRD, HDAC3, TIF1β, and the CoREST complex bind to the core complex in a mutually exclusive manner, thus forming at least four distinct larger complexes that share the common (TR2+TR4+DNMT1+LSD1) core. (B) Structural models for multiple TR2/TR4 repressor complexes. Based on the results of the coimmunoprecipitation assays, we propose a structural model that contains multiple and diverse activities as TR2/TR4 repressor complexes. TR2, TR4, DNMT1, and LSD1 comprise the core negative regulatory silencing complex, to which the NuRD or CoREST complex, HDAC3, or TIF1β binds in a mutually exclusive manner to form at least four distinct larger complexes that share the core complex.
Size fractionation of components of TR2/TR4 repressor complexes.
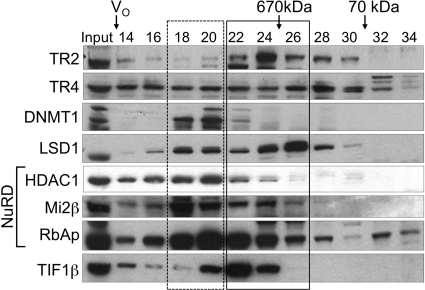
In order to assess how different TR2/TR4-interacting proteins might be differentially partitioned into distinct protein complexes, we determined the fractionation profiles of the proteins by gel filtration on a Superose 6 column (Fig. 9). The elution profile of TR4 is broadly spread almost equally from fractions 14 through 30, whereas TR2 elution shows a peak in fraction 24 but otherwise largely overlaps the profile of TR4. The differences in elution profiles between TR2 and TR4 may be due to the presence of either (or both) TR2 or TR4 homodimers that have a distinct elution profile from the TR2/TR4 heterodimer. For the TR2/TR4 interacting proteins, we observed two patterns in their elution profiles: LSD1 and TIF1β eluted with peaks in fractions 22 to 26 (solid box in Fig. 9), whereas DNMT1 and NuRD components (HDAC1, Mi2β, and RbAp46/48) peaked in the higher-molecular-mass range around fractions 18 to 20 (dotted box). Overall, these results are consistent with the hypothetical model that TR2, TR4, DNMT1, and LSD1 comprise a core complex, which in turn interacts with other proteins (including NuRD components and TIF1β) to form even larger protein complexes.
Fig. 9.
Size fractionation of TR2, TR4, and interacting proteins in nuclear extracts from untransfected, induced MEL cells. Fractions from a Superose 6 gel filtration column were subjected to immunoblotting with antibodies against the proteins shown to the left. Elution positions of molecular mass markers and the void volume (V0) are indicated at the top. Elution peaks of TR2, LSD1, and TIF1β were positioned within fractions 22 through 26 (solid box), while DNMT1 and NuRD components (HDAC1, Mi2β, and RbAp46/48) peaked in fractions 18 through 20 (dotted box) in a higher-molecular-mass range.
Binding of TR2/TR4 corepressors to the β-type globin promoters in adult erythroid cells.
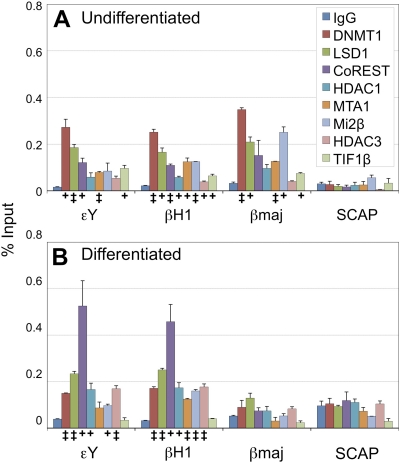
To gain further insight into the functional significance of the TR2/TR4-associated corepressors in β-type globin gene regulation, we investigated their localization to their promoters in adult erythroid (MEL) cells by ChIP assays. MEL cells were grown under conditions that would lead to either continued proliferation (Fig. 10A) or differentiation (with DMSO; Fig. 10B). The data clearly demonstrate that, in undifferentiated MEL cells, DNMT1, subunits of the CoREST complex (LSD1, CoREST, and HDAC1), subunits of the NuRD complex (HDAC1, MTA1, and Mi2β), and HDAC3, as well as TIF1β, are all recruited to the embryonic globin promoters (εY and βH1), although binding of HDAC1, Mi2β, and HDAC3 to the εY promoter is statistically less significant compared to other proteins, as shown in Fig. 10A (where statistically significant enrichment compared to control IgG is indicated with symbols). Many of the TR2/TR4-interacting proteins were also clearly shown to bind to the adult βmajor-globin promoter, whereas localization of these corepressors to the promoters of an active gene (SCAP) that is highly transcribed in MEL cells is essentially undetectable. These data imply that, in undifferentiated cells, the adult β-globin gene is, in fact, actively repressed even in cells poised to transcribe the gene, although the nature of DNA binding factors other than TR2/TR4 which recruit these multiple corepressors to the inactive, but transcriptionally poised, adult β-globin gene remain to be determined (see the Discussion).
Fig. 10.
Binding of TR2/TR4-interacting corepressors to the murine β-type globin gene promoters in undifferentiated (A) and differentiated (B) MEL cells. The binding of TR2/TR4-interacting proteins to the proximal promoter regions of the murine embryonic εY-, βH1-, and adult βmajor-globin genes in both undifferentiated and differentiated MEL cells was analyzed using ChIP assays. As a negative control, binding of the same corepressors to the promoter of a gene (SCAP) that is transcriptionally active in MEL cells was also examined. Statistically significant enrichment of the corepressors at the promoters compared to control IgG values is indicated with symbols (+, P < 0.05; ‡, P < 0.01 [Student t test]). Error bars represent the SEM.
Interestingly, upon terminal differentiation, these corepressors dissociate specifically from the adult βmajor-globin promoter (Fig. 10B), whereas all of the corepressors except TIF1β remain bound to the embryonic globin promoters. These data suggest that the specific binding of TR2/TR4 to the conserved DR sequences in the embryonic globin promoters cause persistent association of the TR2/TR4-interacting corepressors to the embryonic globin promoters through terminal differentiation, thereby fulfilling the TR2/TR4-induced silencing of the embryonic globin genes. Dissociation of TIF1β from both the embryonic and adult β-type globin promoters upon terminal differentiation implies a distinct regulatory role for TIF1β in β-type globin gene regulation during differentiation.
Previously, it was shown in undifferentiated murine adult erythroid cells that active histone marks (histone H3 and H4 acetylation and H3K4 methylation) are only modestly enriched even on the adult β-globin promoter and that, after differentiation, these active marks become highly enriched specifically on the adult, but not on the embryonic, β-type globin promoters (11, 24). It was also reported that in human adult hematopoietic progenitor cells the promoter DNA of both the fetal γ- and adult β-globin genes are highly methylated and that during differentiation the adult β-globin promoter DNA specifically undergoes extensive demethylation, another hallmark of gene activation, but the fetal γ-globin promoter does not (43). TR2/TR4-interacting corepressors include enzymes that can remove or cancel all of these epigenetic activation marks, namely, HDAC1/2/3, methyl-H3K4 demethylase LSD1, and DNMT1. The differentiation-dependent specific dissociation of these enzymes from the adult β-globin promoter shown here may constitute a key underlying mechanism for generation of the active chromatin structures exclusively on the adult β-globin genes in adult erythroid cells during the terminal differentiation events leading to definitive erythroid cell maturation, causing the high-level transcription of the adult genes, as well as the silencing of embryonic or fetal β-type globin genes.
DISCUSSION
TR2/TR4 bind to the promoters of the murine embryonic β-type globin genes and forms the scaffold for multiple, distinct epigenetic transcriptional repressor complexes.
We showed here that TR2 and TR4 bind in vivo to the murine embryonic β-type globin promoters in adult erythroid cells, which suggests that TR2 and TR4 not only temporarily repress the embryonic globin genes during the transition from embryonic to adult globin synthesis but are actively silencing the embryonic genes even in adults after the completion of switching, and hence could be viable targets for molecular intervention in attempting to induce embryonic or fetal β-type globin genes for therapeutic application. TR2 and TR4 were also shown to physically interact with a number of corepressors which the data here would argue play multiple roles in the silencing of the embryonic β-type globin genes: DNMT1, the NuRD complex, the LSD1/CoREST complex, HDAC3, and TIF1β. Furthermore, these TR2/TR4-interacting corepressors are recruited to the embryonic β-type globin promoters, strongly suggesting the functional significance of these interactions in silencing of the embryonic globin genes in adult erythroid cells.
TR2/TR4 binding to the embryonic εY-globin promoter was enhanced upon differentiation of MEL cells, whereas their binding to the embryonic βH1-globin promoter was diminished after differentiation (Fig. 1B), indicating differential regulation of TR2/TR4 binding to the εY-globin versus βH1-globin DR elements. One potential mechanism for this phenomenon may involve the presence of a TR2/TR4 regulator that positively or negatively influences TR2/TR4 binding to the DR elements in a gene-specific and differentiation stage-specific manner. In this regard, persistent binding, or even seemingly enhanced binding of the corepressors to the βH1 promoter upon differentiation in spite of diminution of TR2/TR4 binding, may appear puzzling (Fig. 10). A possible explanation for this curious phenomenon may be that TR2/TR4 binding is only required for initial corepressor recruitment to the embryonic genes, while the persistent presence of TR2/TR4 on the promoters is not required for the continued association of the corepressors or in fact for the gene silencing elicited by those enzymes. Continuous association of the corepressors to the genes may be autonomously maintained, or even reinforced, by chromatin modifications such as histone posttranslational modifications and DNA methylation, as discussed below, that are introduced by the corepressors themselves during the initial phase of gene silencing.
The association of the corepressors with the adult βmajor-globin promoter in the undifferentiated state is also noteworthy and implies that the adult globin gene is actively repressed even in cells poised to transcribe the gene. In contrast to the embryonic genes, the corepressors dissociate from the adult gene upon differentiation, suggesting that different regulatory mechanisms/molecules are controlling corepressor association with the embryonic and adult globin genes. However, the regulatory mechanisms or the identity of DNA-binding factors that recruit the corepressors to the adult β-globin gene is unknown but warrants further investigation.
Unique characteristics of TR2/TR4 in corepressor interaction.
Many nuclear receptors in the unliganded or antagonist-bound state repress transcription by recruiting N-CoR/SMRT that nucleates a corepressor complex containing HDAC3 (17, 61). In addition, a Sin3 complex containing HDAC1/2 was shown to physically interact with N-CoR/SMRT to mediate repression by unliganded nuclear receptors (21, 50). In contrast to these notions, the present study has revealed unconventional characteristics of TR2/TR4 with regard to nuclear receptor corepressor interactions in the following ways. First, there was no evidence for interaction with N-CoR/SMRT, which was undetectable either by repeated mass spectrometric or immunoblotting analysis of proteins that copurified with biotin-tagged TR2/TR4 or by coimmunoprecipitation assays (data not shown), although HDAC3 was identified as a TR2/TR4-interacting protein, possibly through direct interaction as previously reported (12). Second, we found that the nuclear receptor complex interacts with DNMT1. Previously, only two other sequence-specific DNA-binding proteins, E2F1 and STAT3, were shown to interact with DNMT1 (58, 91), and this is the first report to demonstrate a physical link between any nuclear receptor and DNMT1, raising the intriguing possibility that DNMT1 might play a direct role in developmental gene silencing triggered by nuclear receptors. Third, these data demonstrate interactions with a wide variety of corepressors known to possess activities and roles in different aspects of gene silencing, thus placing TR2/TR4 in a unique position even in the realm of transcriptional repressors in general. Whereas the NuRD repressor complex that contains both histone deacetylase (HDAC1/2) and chromatin remodeling ATPase (Mi2) activities (85) has been shown to interact with a number of transcription factors such as the GATA-1/FOG-1 complex (59) (22), the estrogen receptor is the only nuclear receptor other than TR2/TR4 that was previously shown to interact with NuRD (46). TR2/TR4 also interact with the LSD1/CoREST complex that contains HDAC1/2 and H3K4 demethylase LSD1 and acts as a corepressor for a number of transcription factors such as Gfi-1/Gfi-1b (65). However, TLX is the only nuclear receptor previously shown to recruit LSD1/CoREST for gene silencing (88). Recently, it was shown that LSD1 can also act as a coactivator for androgen or estrogen receptor-regulated transcription, possibly by stimulating the demethylation of histone H3K9, a repressive histone mark (15, 47). In this regard, a potential alternate role for LSD1, as well as TR2/TR4, in differentiation stage-specific transcriptional activation of the fetal γ-globin gene warrants further investigation in light of our previous observation that forced TR2/TR4 expression can paradoxically stimulate γ-globin gene expression in mice (74). TIF1β, also known as KAP-1, is another TR2/TR4-interacting protein, which is known to directly interact with HP1 (heterochromatin protein 1), implicating its role in transcriptional repression through heterochromatin formation (53, 63). Although TIF1β is known to interact with a number of transcription factors, only three other nuclear receptors (DAX-1, Nur77, and COUP-TFI) were previously shown to interact with TIF1β (57, 80, 90). Overall, interaction with such an unexpectedly wide variety of distinct epigenetic transcriptional corepressors may indicate unique characteristics and functions of TR2/TR4 to coordinate multiple corepressors to elicit developmental stage-specific gene silencing through a series of epigenetic chromatin modifications by nucleosome remodeling (through NuRD), histone deacetylation (through NuRD, LSD1/CoREST, and HDAC3), histone H3K4 demethylation (through LSD1/CoREST), and CpG DNA methylation (through DNMT1) (Fig. 8B). It is also possible that such complexity of corepressor interactions is shared by other nuclear receptors but was not previously detected.
Functional interactions among distinct epigenetic transcriptional repressors.
Recent studies have revealed functional and regulatory interplay between histone modifications (acetylation and methylation) and DNA methylation, all of which are regarded as heritable epigenetic marks to transmit gene expression propensities over multiple cell generations. It has been reported that deacetylation of nucleosomal histone H3 renders it a better substrate for H3K4 demethylation by LSD1, whereas LSD1 activity will conversely enhance the deacetylase activity of HDAC1, thus suggesting that HDACs and LSD1 synergistically generate repressive chromatin structures (36, 71). Physical association between LSD1 and the NuRD complex has been also reported, further supporting this notion (81). Conversely, coordination between histone modifications and DNA methylation in the regulation of chromatin structure and gene expression has been also reported, but the hierarchical order of events in this interplay is currently under active debate. Some studies have shown that gene silencing by DNA methylation is mediated by methyl-CpG-binding proteins that recruit HDAC activity to methylated DNA, resulting in a deacetylated repressive chromatin structure (26, 51). In contrast, other studies have shown that DNA methylation is guided by histone modification, suggesting that DNA methylation is secondary to repressive chromatin formation (40). These seemingly contradictory findings may indicate that the coordination between histone modifications and DNA methylation can proceed hierarchically in either direction or that genes differ in their relative susceptibility to different epigenetic signaling events. Physical associations between DNMT1 and HDAC1/2 (14, 58, 62), as well as between DNMT1 and LSD1 (79), have been defined and may substantiate these notions. Taken together, the data presented here suggest that TR2 and TR4 are unique in that these transcription factors elicit developmental stage-specific gene silencing through both histone modifications and DNA methylation by recruiting and coordinating multiple distinct corepressors, although the hierarchical order or possible regulatory interplay among these events in embryonic and fetal globin gene silencing is not currently understood.
Roles for TR2/TR4 corepressors in globin gene switching.
The β-globin locus has been extensively studied as a model for molecular mechanisms of lineage- and developmental stage-specific gene expression through epigenetic chromatin modifications. The mammalian embryonic and fetal β-type globin genes have been shown to acquire histone modifications associated with gene activation, namely, H3 and H4 acetylation and H3K4 methylation, when highly expressed, and afterward lose those activation marks as the genes become silenced during development (4, 11, 32, 48, 87). In addition to histone modifications, a regulatory role for local DNA methylation in the silencing of murine embryonic and human fetal β-type globin genes has been suggested by the inverse correlation between their expression and CpG methylation in their promoters during development (23) (43). Furthermore, using a transgenic mouse system to allow controlled Aγ-globin gene methylation, it was shown that DNA methylation works as a dominant repressor in Aγ gene expression (18). Regulatory roles of these epigenetic chromatin modifications are also supported by the effects of the pharmacological inhibitors of several chromatin modifying enzymes.
DNA methyltransferase inhibitors 5-azacytidine and 5-aza-2′-deoxycytidine have been shown to induce fetal γ-globin gene expression in primary erythroid cells in culture with concomitant demethylation of the γ-globin promoter and have proven to be effective in the treatment of β-globin disorders such as sickle cell disease and β-thalassemia for the induction of γ-globin synthesis (68). More specifically, depletion of DNMT1 protein by transfection of small interfering RNAs that degrade DNMT1 mRNA in primary cultures of erythroid progenitor cells derived from CD34+ baboon bone marrow cells was shown to depress DNA methylation of the embryonic ε- and fetal γ-globin promoters and increase the expression of both genes (2). These notions strongly suggest a functional significance of the TR2/TR4-DNMT1 interaction and recruitment of this complex to the embryonic globin promoters in gene silencing.
HDAC inhibitors such as butyrates have also been shown to increase fetal γ-globin synthesis with a concomitant increase in histone H3 and H4 acetylation at the γ-globin promoters in primary human erythroid cells in culture and are sometimes effective in the induction of γ-globin synthesis in patients with β-globin disorders (9) (5). These notions also indicate a critical role for HDACs in γ-globin silencing. However, butyrates and other HDAC inhibitors are known to inhibit all of the class I HDACs (HDAC1, HDAC2, HDAC3, and HDAC8), as well as some of the class II HDACs (5). Therefore, the identity of specific HDAC molecules, or the identity of repressor complexes containing the HDACs that elicit chromatin modification leading to γ-globin silencing, and hence those responsible for γ-globin induction in response to the HDAC inhibitors, have not been well defined. The direct physical association of TR2/TR4 with the NuRD and LSD1/CoREST complexes, both containing HDAC1/2, and independently with HDAC3, and their recruitment to the mouse embryonic β-type globin promoters, strongly suggest roles for these deacetylases in epigenetic chromatin modification leading to developmental stage-specific silencing of the embryonic and fetal β-type globin genes. The protein interfaces between TR2/TR4 and their epigenetic interacting cofactors (namely, DNMT1, the NuRD and LSD1/CoREST complexes, HDAC3, and TIF1β), as well as the enzymatic activities themselves, might serve as molecular targets for development of specific γ-globin inducing agents that can be effectively and safely applied to the treatment of sickle cell disease and β-thalassemia.
ACKNOWLEDGMENTS
This study was supported by NIH grants RC1 DK086956 (O.T. and J.D.E.), R01 HL24415 (J.D.E.), Cooley's Anemia Foundation Postdoctoral Fellowships (O.T. and N.O.), and an American Heart Association Postdoctoral Fellowship (L.S.).
We are grateful to our laboratory coworkers for many helpful discussions and to Gerd Blobel for initially suggesting the use of EGS in the ChIP assays. We thank Shoko Kobayashi and Carmen Yu for technical assistance.
Footnotes
Published ahead of print on 13 June 2011.
REFERENCES
- 1. Aerbajinai W., Zhu J., Kumkhaek C., Chin K., Rodgers G. P. 2009. SCF induces γ-globin gene expression by regulating downstream transcription factor COUP-TFII. Blood 114:187–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Banzon V., et al. 2011. siDNMT1 increases γ-globin expression in chemical inducer of dimerization (CID)-dependent mouse βYAC bone marrow cells and in baboon erythroid progenitor cell cultures. Exp. Hematol. 39:26–36 e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Borg J., et al. 2010. Haploinsufficiency for the erythroid transcription factor KLF1 causes hereditary persistence of fetal hemoglobin. Nat. Genet. 42:801–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bulger M., et al. 2003. A complex chromatin landscape revealed by patterns of nuclease sensitivity and histone modification within the mouse β-globin locus. Mol. Cell. Biol. 23:5234–5244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cao H. 2004. Pharmacological induction of fetal hemoglobin synthesis using histone deacetylase inhibitors. Hematology 9:223–233 [DOI] [PubMed] [Google Scholar]
- 6. Chen L. M., et al. 2008. Subfertility with defective folliculogenesis in female mice lacking testicular orphan nuclear receptor 4. Mol. Endocrinol. 22:858–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. de Boer E., et al. 2003. Efficient biotinylation and single-step purification of tagged transcription factors in mammalian cells and transgenic mice. Proc. Natl. Acad. Sci. U. S. A. 100:7480–7485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Delcuve G. P., Rastegar M., Davie J. R. 2009. Epigenetic control. J. Cell Physiol. 219:243–250 [DOI] [PubMed] [Google Scholar]
- 9. Fathallah H., Weinberg R. S., Galperin Y., Sutton M., Atweh G. F. 2007. Role of epigenetic modifications in normal globin gene regulation and butyrate-mediated induction of fetal hemoglobin. Blood 110:3391–3397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Filipe A., et al. 1999. Regulation of embryonic/fetal globin genes by nuclear hormone receptors: a novel perspective on hemoglobin switching. EMBO J. 18:687–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Forsberg E. C., et al. 2000. Developmentally dynamic histone acetylation pattern of a tissue-specific chromatin domain. Proc. Natl. Acad. Sci. U. S. A. 97:14494–14499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Franco P. J., Farooqui M., Seto E., Wei L. N. 2001. The orphan nuclear receptor TR2 interacts directly with both class I and class II histone deacetylases. Mol. Endocrinol. 15:1318–1328 [DOI] [PubMed] [Google Scholar]
- 13. Frietze S., O'Geen H., Blahnik K. R., Jin V. X., Farnham P. J. 2010. ZNF274 recruits the histone methyltransferase SETDB1 to the 3′ ends of ZNF genes. PLoS One 5:e15082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fuks F., Burgers W. A., Brehm A., Hughes-Davies L., Kouzarides T. 2000. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat. Genet. 24:88–91 [DOI] [PubMed] [Google Scholar]
- 15. Garcia-Bassets I., et al. 2007. Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell 128:505–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Giarratana M. C., et al. 2005. Ex vivo generation of fully mature human red blood cells from hematopoietic stem cells. Nat. Biotechnol. 23:69–74 [DOI] [PubMed] [Google Scholar]
- 17. Glass C. K., Rosenfeld M. G. 2000. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 14:121–141 [PubMed] [Google Scholar]
- 18. Goren A., et al. 2006. Fine tuning of globin gene expression by DNA methylation. PLoS One 1:e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gupta P., Ho P. C., Ha S. G., Lin Y. W., Wei L. N. 2009. HDAC3 as a molecular chaperone for shuttling phosphorylated TR2 to PML: a novel deacetylase activity-independent function of HDAC3. PLoS One 4:e4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Harju-Baker S., Costa F. C., Fedosyuk H., Neades R., Peterson K. R. 2008. Silencing of Aγ-globin gene expression during adult definitive erythropoiesis mediated by GATA-1-FOG-1-Mi2 complex binding at the −566 GATA site. Mol. Cell. Biol. 28:3101–3113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Heinzel T., et al. 1997. A complex containing N-CoR, mSin3, and histone deacetylase mediates transcriptional repression. Nature 387:43–48 [DOI] [PubMed] [Google Scholar]
- 22. Hong W., et al. 2005. FOG-1 recruits the NuRD repressor complex to mediate transcriptional repression by GATA-1. EMBO J. 24:2367–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hsu M., Mabaera R., Lowrey C. H., Martin D. I., Fiering S. 2007. CpG hypomethylation in a large domain encompassing the embryonic β-like globin genes in primitive erythrocytes. Mol. Cell. Biol. 27:5047–5054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Im H., et al. 2003. Dynamic regulation of histone H3 methylated at lysine 79 within a tissue-specific chromatin domain. J. Biol. Chem. 278:18346–18352 [DOI] [PubMed] [Google Scholar]
- 25. Ishitani K., et al. 2003. p54nrb acts as a transcriptional coactivator for activation function 1 of the human androgen receptor. Biochem. Biophys. Res. Commun. 306:660–665 [DOI] [PubMed] [Google Scholar]
- 26. Jones P. L., et al. 1998. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 19:187–191 [DOI] [PubMed] [Google Scholar]
- 27. Jung D. J., Na S. Y., Na D. S., Lee J. W. 2002. Molecular cloning and characterization of CAPER, a novel coactivator of activating protein-1 and estrogen receptors. J. Biol. Chem. 277:1229–1234 [DOI] [PubMed] [Google Scholar]
- 28. Khavari P. A., Peterson C. L., Tamkun J. W., Mendel D. B., Crabtree G. R. 1993. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature 366:170–174 [DOI] [PubMed] [Google Scholar]
- 29. Kiefer C. M., Hou C., Little J. A., Dean A. 2008. Epigenetics of β-globin gene regulation. Mutat. Res. 647:68–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim E., et al. 2003. Disruption of TR4 orphan nuclear receptor reduces the expression of liver apolipoprotein E/C-I/C-II gene cluster. J. Biol. Chem. 278:46919–46926 [DOI] [PubMed] [Google Scholar]
- 31. Kim J. K., Samaranayake M., Pradhan S. 2009. Epigenetic mechanisms in mammals. Cell. Mol. Life Sci. 66:596–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kingsley P. D., et al. 2006. “Maturational” globin switching in primary primitive erythroid cells. Blood 107:1665–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Le Douarin B., et al. 1996. A possible involvement of TIF1α and TIF1β in the epigenetic control of transcription by nuclear receptors. EMBO J. 15:6701–6715 [PMC free article] [PubMed] [Google Scholar]
- 34. Lee C. H., Chinpaisal C., Wei L. N. 1998. Cloning and characterization of mouse RIP140, a corepressor for nuclear orphan receptor TR2. Mol. Cell. Biol. 18:6745–6755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee C. H., Chinpaisal C., Wei L. N. 1998. A novel nuclear receptor heterodimerization pathway mediated by orphan receptors TR2 and TR4. J. Biol. Chem. 273:25209–25215 [DOI] [PubMed] [Google Scholar]
- 36. Lee M. G., et al. 2006. Functional interplay between histone demethylase and deacetylase enzymes. Mol. Cell. Biol. 26:6395–6402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lee M. G., Wynder C., Cooch N., Shiekhattar R. 2005. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature 437:432–435 [DOI] [PubMed] [Google Scholar]
- 38. Lee Y. F., Lee H. J., Chang C. 2002. Recent advances in the TR2 and TR4 orphan receptors of the nuclear receptor superfamily. J. Steroid Biochem. Mol. Biol. 81:291–308 [DOI] [PubMed] [Google Scholar]
- 39. Le Guezennec X., et al. 2006. MBD2/NuRD and MBD3/NuRD, two distinct complexes with different biochemical and functional properties. Mol. Cell. Biol. 26:843–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lehnertz B., et al. 2003. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 13:1192–1200 [DOI] [PubMed] [Google Scholar]
- 41. Liu N. C., et al. 2007. Loss of TR4 orphan nuclear receptor reduces phosphoenolpyruvate carboxykinase-mediated gluconeogenesis. Diabetes 56:2901–2909 [DOI] [PubMed] [Google Scholar]
- 42. Lopez R. A., Schoetz S., DeAngelis K., O'Neill D., Bank A. 2002. Multiple hematopoietic defects and delayed globin switching in Ikaros null mice. Proc. Natl. Acad. Sci. U. S. A. 99:602–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mabaera R., et al. 2007. Developmental- and differentiation-specific patterns of human γ- and β-globin promoter DNA methylation. Blood 110:1343–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Marcus S. J., Kinney T. R., Schultz W. H., O'Branski E. E., Ware R. E. 1997. Quantitative analysis of erythrocytes containing fetal hemoglobin (F cells) in children with sickle cell disease. Am. J. Hematol. 54:40–46 [DOI] [PubMed] [Google Scholar]
- 45. Mathur M., Tucker P. W., Samuels H. H. 2001. PSF is a novel corepressor that mediates its effect through Sin3A and the DNA binding domain of nuclear hormone receptors. Mol. Cell. Biol. 21:2298–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mazumdar A., et al. 2001. Transcriptional repression of oestrogen receptor by metastasis-associated protein 1 corepressor. Nat. Cell Biol. 3:30–37 [DOI] [PubMed] [Google Scholar]
- 47. Metzger E., et al. 2005. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 437:436–439 [DOI] [PubMed] [Google Scholar]
- 48. Miles J., et al. 2007. Intergenic transcription, cell-cycle and the developmentally regulated epigenetic profile of the human β-globin locus. PLoS One 2:e630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Morimoto R. I. 2002. Dynamic remodeling of transcription complexes by molecular chaperones. Cell 110:281–284 [DOI] [PubMed] [Google Scholar]
- 50. Nagy L., et al. 1997. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell 89:373–380 [DOI] [PubMed] [Google Scholar]
- 51. Nan X., et al. 1998. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393:386–389 [DOI] [PubMed] [Google Scholar]
- 52. Needham M., et al. 1992. LCR/MEL: a versatile system for high-level expression of heterologous proteins in erythroid cells. Nucleic Acids Res. 20:997–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nielsen A. L., et al. 1999. Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. EMBO J. 18:6385–6395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. O'Geen H., et al. 2010. Genome-wide binding of the orphan nuclear receptor TR4 suggests its general role in fundamental biological processes. BMC Genomics 11:689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Omori A., Tanabe O., Engel J. D., Fukamizu A., Tanimoto K. 2005. Adult stage γ-globin silencing is mediated by a promoter direct repeat element. Mol. Cell. Biol. 25:3443–3451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Papadakis M. N., Patrinos G. P., Tsaftaridis P., Loutradi-Anagnostou A. 2002. A comparative study of Greek nondeletional hereditary persistence of fetal hemoglobin and β-thalassemia compound heterozygotes. J. Mol. Med. 80:243–247 [DOI] [PubMed] [Google Scholar]
- 57. Rambaud J., Desroches J., Balsalobre A., Drouin J. 2009. TIF1β/KAP-1 is a coactivator of the orphan nuclear receptor NGFI-B/Nur77. J. Biol. Chem. 284:14147–14156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Robertson K. D., et al. 2000. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat. Genet. 25:338–342 [DOI] [PubMed] [Google Scholar]
- 59. Rodriguez P., et al. 2005. GATA-1 forms distinct activating and repressive complexes in erythroid cells. EMBO J. 24:2354–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rodriguez P., et al. 2006. Isolation of transcription factor complexes by in vivo biotinylation tagging and direct binding to streptavidin beads. Methods Mol. Biol. 338:305–323 [DOI] [PubMed] [Google Scholar]
- 61. Rosenfeld M. G., Lunyak V. V., Glass C. K. 2006. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 20:1405–1428 [DOI] [PubMed] [Google Scholar]
- 62. Rountree M. R., Bachman K. E., Baylin S. B. 2000. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat. Genet. 25:269–277 [DOI] [PubMed] [Google Scholar]
- 63. Ryan R. F., et al. 1999. KAP-1 corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: a potential role for Kruppel-associated box-zinc finger proteins in heterochromatin-mediated gene silencing. Mol. Cell. Biol. 19:4366–4378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rybak J. N., et al. 2005. In vivo protein biotinylation for identification of organ-specific antigens accessible from the vasculature. Nat. Methods 2:291–298 [DOI] [PubMed] [Google Scholar]
- 65. Saleque S., Kim J., Rooke H. M., Orkin S. H. 2007. Epigenetic regulation of hematopoietic differentiation by Gfi-1 and Gfi-1b is mediated by the cofactors CoREST and LSD1. Mol. Cell 27:562–572 [DOI] [PubMed] [Google Scholar]
- 66. Sankaran V. G., et al. 2011. MicroRNA-15a and -16-1 act via MYB to elevate fetal hemoglobin expression in human trisomy 13. Proc. Natl. Acad. Sci. U. S. A. 108:1519–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sankaran V. G., et al. 2008. Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science 322:1839–1842 [DOI] [PubMed] [Google Scholar]
- 68. Saunthararajah Y., Lavelle D., DeSimone J. 2004. DNA hypo-methylating agents and sickle cell disease. Br. J. Haematol. 126:629–636 [DOI] [PubMed] [Google Scholar]
- 69. Shevchenko A., Wilm M., Vorm O., Mann M. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850–858 [DOI] [PubMed] [Google Scholar]
- 70. Shi Y., et al. 2004. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119:941–953 [DOI] [PubMed] [Google Scholar]
- 71. Shi Y. J., et al. 2005. Regulation of LSD1 histone demethylase activity by its associated factors. Mol. Cell 19:857–864 [DOI] [PubMed] [Google Scholar]
- 72. Stamatoyannopoulos G., Grosveld F. 2001. Hemoglobin switching, p. 135–182 In Stamatoyannopoulos G., et al. (ed.), Molecular basis of blood diseases, 3rd ed. W. B. Saunders Publishing Company, Philadelphia, PA [Google Scholar]
- 73. Tanabe O., et al. 2002. An embryonic/fetal β-type globin gene repressor contains a nuclear receptor TR2/TR4 heterodimer. EMBO J. 21:3434–3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Tanabe O., et al. 2007. Embryonic and fetal β-globin gene repression by the orphan nuclear receptors, TR2 and TR4. EMBO J. 26:2295–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Tanabe O., et al. 2007. The TR2 and TR4 orphan nuclear receptors repress Gata1 transcription. Genes Dev. 21:2832–2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Tanimoto K., Liu Q., Grosveld F., Bungert J., Engel J. D. 2000. Context-dependent EKLF responsiveness defines the developmental specificity of the human ε-globin gene in erythroid cells of YAC transgenic mice. Genes Dev. 14:2778–2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Tsai N. P., et al. 2009. Activation of testicular orphan receptor 4 by fatty acids. Biochim. Biophys. Acta 1789:734–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. van Dijk T. B., et al. 2010. Fetal globin expression is regulated by Friend of Prmt1. Blood 116:4349–4352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wang J., et al. 2009. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat. Genet. 41:125–129 [DOI] [PubMed] [Google Scholar]
- 80. Wang J., et al. 2006. A protein interaction network for pluripotency of embryonic stem cells. Nature 444:364–368 [DOI] [PubMed] [Google Scholar]
- 81. Wang Y., et al. 2009. LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell 138:660–672 [DOI] [PubMed] [Google Scholar]
- 82. Wilson A. C., LaMarco K., Peterson M. G., Herr W. 1993. The VP16 accessory protein HCF is a family of polypeptides processed from a large precursor protein. Cell 74:115–125 [DOI] [PubMed] [Google Scholar]
- 83. Xie S., et al. 2009. TR4 nuclear receptor functions as a fatty acid sensor to modulate CD36 expression and foam cell formation. Proc. Natl. Acad. Sci. U. S. A. 106:13353–13358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Xu J., et al. 2010. Transcriptional silencing of γ-globin by BCL11A involves long-range interactions and cooperation with SOX6. Genes Dev. 24:783–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Yang X. J., Seto E. 2008. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat. Rev. Mol. Cell. Biol. 9:206–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Yi Z., et al. 2006. Sox6 directly silences epsilon globin expression in definitive erythropoiesis. PLoS Genet. 2:e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Yin W., et al. 2007. Histone acetylation at the human β-globin locus changes with developmental age. Blood 110:4101–4107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Yokoyama A., Takezawa S., Schule R., Kitagawa H., Kato S. 2008. Transrepressive function of TLX requires the histone demethylase LSD1. Mol. Cell. Biol. 28:3995–4003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Zeng P. Y., Vakoc C. R., Chen Z. C., Blobel G. A., Berger S. L. 2006. In vivo dual cross-linking for identification of indirect DNA-associated proteins by chromatin immunoprecipitation. Biotechniques 41:694–698 [DOI] [PubMed] [Google Scholar]
- 90. Zhang L. J., Liu X., Gafken P. R., Kioussi C., Leid M. 2009. A chicken ovalbumin upstream promoter transcription factor I (COUP-TFI) complex represses expression of the gene encoding tumor necrosis factor α-induced protein 8 (TNFAIP8). J. Biol. Chem. 284:6156–6168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Zhang Q., et al. 2005. STAT3- and DNA methyltransferase 1-mediated epigenetic silencing of SHP-1 tyrosine phosphatase tumor suppressor gene in malignant T lymphocytes. Proc. Natl. Acad. Sci. U. S. A. 102:6948–6953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Zhou D., Liu K., Sun C. W., Pawlik K. M., Townes T. M. 2010. KLF1 regulates BCL11A expression and γ- to β-globin gene switching. Nat. Genet. 42:742–744 [DOI] [PubMed] [Google Scholar]
- 93. Zhou X. E., et al. 2010. The orphan nuclear receptor TR4 is a vitamin A-activated nuclear receptor. J. Biol. Chem. 286:2877–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]