Abstract
Protein families are generated by successive rounds of gene duplication and subsequent diversification. However, the paths by which duplicated genes acquire distinct functions are not well characterized. We focused on a pair of duplicated deacetylases from Saccharomyces cerevisiae, Sir2 and Hst1, that subfunctionalized after duplication. As a proxy for the ancestral, nonduplicated deacetylase, we studied Sir2 from another yeast, Kluyveromyces lactis. We compared the interaction domains of these deacetylases for the Sir transcriptional silencing complex, which acts with ScSir2, and the Sum1 repressor, which acts with ScHst1, and found that these interaction domains have been retained over the course of evolution and can be disrupted by simple amino acid substitutions. Therefore, Sir2 and Hst1 subfunctionalized by acquiring complementary inactivating mutations in these interaction domains.
INTRODUCTION
Gene duplication is an important evolutionary mechanism to develop novel protein functions without losing ancestral functions, thereby generating genetic diversity. Several models describe how duplicated genes diverge after duplication, but few gene pairs have been examined experimentally to confirm or refute these models. Furthermore, the types of mutations that lead to functional divergence have been poorly characterized. As a case study, we examined the duplicated deacetylases Sir2 and Hst1 from yeast and found that they subfunctionalized by acquiring complementary inactivating mutations in distinct interaction domains.
Duplicated genes can acquire distinct functions in two ways (8, 16, 28). Neofunctionalization occurs when one copy continues to perform the ancestral function and the other copy acquires a novel function. In contrast, subfunctionalization results in each duplicated gene performing a subset of the ancestral functions. The majority of duplicate pairs that have been studied have subfunctionalized (11, 20, 43). Subfunctionalization can occur through a duplication, degeneration, and complementation (DDC) mechanism, in which the duplicates acquire complementary degenerative mutations (13). Alternatively, the escape from adaptive conflict, or specialization, model proposes that multifunctional proteins cannot be optimized for each of their functions (11, 20, 22). Hence, after duplication, mutations improve one function and worsen another.
Yeasts are ideal organisms in which to study the fates of duplicated genes, as many species are experimentally tractable. In addition, a whole-genome duplication that occurred about 100 million years ago (44) provides a number of candidate gene pairs, such as Sir2 and Hst1, that can be studied in duplicated and nonduplicated species.
Sir2 family deacetylases are ubiquitous in all kingdoms of life (15, 33). Unlike other deacetylases, Sir2 proteins require NAD+ for catalysis, tying their activity to the metabolic state of the cell and thus nutrient availability (26, 46). Some Sir2 enzymes have been linked to life span and life cycle progression, and the dependence on NAD+ may connect life span with nutrition (23, 26, 46). These deacetylases have a variety of targets and biological functions, and this diversity has been generated through gene duplications. Nevertheless, all Sir2 proteins retain a well-conserved catalytic core, including a zinc-binding module critical for the structural integrity of the protein. The conservation of this catalytic core despite the variety of targets makes these proteins an interesting family in which to study the evolution of specificity.
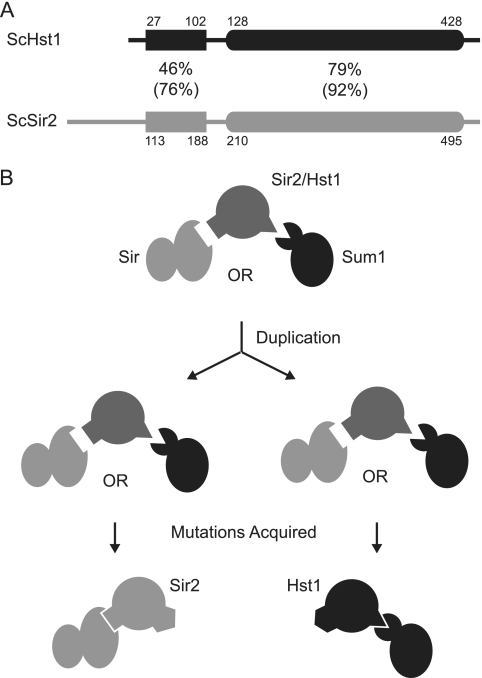
In Saccharomyces cerevisiae, Sir2 and Hst1 share two regions of sequence conservation (Fig. 1 A). The N-terminal domain is modestly conserved and functions in protein interactions. The C-terminal catalytic domain is highly conserved and contains the active site, as well as the structural zinc-binding module. Despite extensive regions of sequence conservation, ScSir2 and ScHst1 each perform a specific function due to the association with a distinct repressive complex. ScSir2 is involved in long-range silencing at the mating-type loci and telomeres as part of the Sir complex, in which ScSir2 interacts with ScSir4 (7, 9, 17, 36). ScSir2 also suppresses unequal sister chromatid exchange of the rRNA gene repeats as part of the RENT complex, in which ScSir2 interacts with ScNet1 (25, 42). In contrast, ScHst1 is a promoter-specific repressor of middle-sporulation, α-specific, and NAD+ biosynthetic genes (2, 29, 45, 47) as part of the Sum1 complex, in which it interacts with ScRfm1 (29). The Sum1 complex is recruited to target genes through the DNA-binding protein ScSum1 (45).
Fig. 1.
Model of the subfunctionalization of ScSir2 and ScHst1. (A) The conserved domains of ScSir2 and ScHst1 are shown with identity (similarity) percentages. ScHst1 is numbered using previously annotated M29 (Saccharomyces Genome Database [http://www.yeastgenome.org/]) as the start codon, based on observations that expression is unaffected by the mutagenesis of M1 and M6 but eliminated by the mutagenesis of M29 and M31. (B) A proposed model of the subfunctionalization of Sir2.
Although ScSir2 and ScHst1 have different functions, they can partially substitute for one another (3, 17). Moreover, a chimeric protein composed of the N terminus of ScSir2 and the C terminus of ScHst1 complements both sir2Δ and hst1Δ mutations in S. cerevisiae and interacts with ScSir4 and ScSum1 (17, 30). Therefore, different portions of the proteins confer specificity for the Sir or Sum1 complex.
Kluyveromyces lactis, another yeast species, diverged from S. cerevisiae prior to the whole-genome duplication. It has one ortholog of ScSir2 and ScHst1, which we have used as a proxy for the ancestral Sir2 protein (17, 18). KlSir2 has both ScSir2-like and ScHst1-like functions. Like ScSir2, KlSir2 interacts with Sir4 and is required for silencing of the cryptic mating-type loci and telomeres (18, 19). And like ScHst1, KlSir2 interacts with Rfm1 and is required for the repression of sporulation genes (18). Therefore, the common ancestor of KlSir2, ScSir2, and ScHst1 also had these functions and subfunctionalization occurred after duplication.
We propose that the ancestral Sir2 protein possessed two interaction domains, one of which interacted with Rfm1 and another of which interacted with Sir4 (Fig. 1B). After duplication, each deacetylase acquired mutations in one of the interaction domains, reducing or eliminating its interaction with one partner and making it specific for the other. If this model is correct, the interaction domain for Sir4 should be shared between a nonduplicated Sir2 protein and ScSir2 but not ScHst1, and the interaction domain for Rfm1 should be shared between a nonduplicated Sir2 protein and ScHst1 but not ScSir2. However, prior to this study, it was not known how nonduplicated Sir2 proteins interact with Sir4 or Rfm1.
To test our model (Fig. 1B), we first determined the minimal regions required for the specificity of ScSir2 and ScHst1 for their respective complexes and then identified mutations that disrupt those interactions. Then, using KlSir2 as a proxy for the nonduplicated, ancestral state, we tested whether homologous mutations in KlSir2 were also disruptive. We found that the interaction domain for Rfm1 is conserved between ScHst1 and KlSir2 and the interaction domain for Sir4 is conserved between ScSir2 and KlSir2. We also examined and rejected a variant of our model in which a single interaction domain enabled the ancestral Sir2 protein to interact with either Sir4 or Rfm1 and mutations acquired after duplication restricted interaction to either Sir4 or Rfm1. Thus, Sir2 and Hst1 represent a clear example of subfunctionalization through the acquisition of degenerative mutations in interaction domains.
MATERIALS AND METHODS
S. cerevisiae strains.
The S. cerevisiae strains used in this study were derived from W303-1a (Table 1). The hst1Δ::KanMX (37), sir2Δ::TRP1, and pPES4–HIS3 (17) alleles were described previously. The sir4Δ::LEU2 and sir2Δ::LEU2 alleles were obtained from J. Rine (University of California, Berkeley). The RFM1-myc allele was constructed by integrating the 9×myc tag plus the entire open reading frame of KlTRP1 from pWZV87 (K. Nasmyth) at the end of the RFM1 open reading frame. The correct integration of the tag was confirmed by PCR using primers flanking the sites of recombination. Expression of the tagged protein was confirmed by immunoblotting. These alleles were moved into various genetic backgrounds, as described in Table 1, through standard genetic crosses.
Table 1.
Strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| S. cerevisiae | ||
| W303-1a | MATaade2–1 can1–100 his3–11 leu2–3,112 trp1–1 ura3–1 | R. Rothstein |
| LRY1007 | W303-1a MATα | |
| LRY1022 | MATα his4 | P. Schatz |
| LRY2083 | W303-1a hst1Δ::KanMX sir2Δ::TRP1 pPES4–HIS3 ADE2 lys2Δ | |
| LRY2507 | W303-1a hst1Δ::KanMX sir2Δ::LEU2 pPES4–HIS3 RFM1–9xmyc::TRP1 | |
| LRY2590 | W303-1a hst1Δ::KanMX sir2Δ::TRP1 sir4Δ::LEU2 pPES4–HIS3 ADE2 lys2Δ | |
| K. lactis | ||
| LRY2128 | MATα sir2Δ::KanMX SIR4–FLAG::KanMX nej1::LEU2 ade1 leu2 trp1 uraA1 metA1 hmlΔp | |
| LRY2333 | MATasir2Δ::KanMX myc–SUM1 nej1::LEU2 leu2 trp1 uraA1 metA1 | |
| LRY2388 | MATasir2Δ::NatMX SIR4–FLAG::KanMX myc–SUM1 nej1::LEU2 leu2 trp1 uraA1 metA1 | |
| LRY2654 | MATα sir2Δ::KanMX SIR4–FLAG::KanMX RFM1–myc::TRP1 nej1::LEU2 ade1 leu2 trp1 uraA1 metA1 hmlΔp | |
K. lactis strains.
The K. lactis strains used in this study were derived from SAY538 (Table 1) (1). The sir2Δ::KanMX, nej1::LEU2 (1), sir2Δ::NatMX, sum1Δ::NatMX, SIR4-Flag, and myc-SUM1 (18) alleles were described previously. The KlRFM1-myc allele was constructed by integrating the 9×myc tag plus the entire open reading frame of KlTRP1 from pWZV87 (K. Nasmyth) at the end of the KlRFM1 open reading frame. The correct integration of the tag was confirmed by PCR using primers flanking the sites of recombination. Expression of the tagged protein was confirmed by immunoblotting. Alleles were moved into various backgrounds by genetic crosses.
S. cerevisiae plasmids.
The S. cerevisiae plasmids used in this study are listed in Table 2. The plasmids containing hemagglutinin (HA)-tagged ScHST1 (ScHST1-HA), pLR30, and the S-H chimera (the N terminus of ScSir2 and the C terminus of ScHst1), pLR488, were described previously (17, 37). The plasmid containing HA-ScSIR2 (pRO298) was obtained from Rohinton Kamakaka (University of California, Santa Cruz). To construct the plasmid containing ScSIR2-HA (pLR753), the HST1 open reading frame in pLR30 was replaced with the SIR2 open reading frame. First, an MfeI site was incorporated before the start of ScHST1 to yield pLR493. Then, an AgeI site after the HA tag on pLR493 was removed to yield pLR496. The ScHST1 open reading frame in pLR496 was replaced with the ScSIR2 open reading frame from pRO298 flanked by EcoRI and AgeI sites and cloned into the MfeI and AgeI sites of pLR496 to generate pLR499*. A frameshift mutation was identified in the HA tag of pLR499* and corrected by site-directed mutagenesis (4) to generate pLR499. Finally, the HST1 promoter was replaced with a SIR2 promoter as described below to yield pLR753.
Table 2.
Plasmids used in this studya
| Plasmid | Alias | Description | Source |
|---|---|---|---|
| S. cerevisiae | |||
| pRS416 | Vector | R. S. Sikorski and P. Hieter, 1989 | |
| pRO298 | HA-ScSir2 | HA-ScSIR2 | R. Kamakaka |
| pLR30 | ScHst1 | ScHST1-HA | 37 |
| pLR488 | S-H | HA-ScSIR21-255-ScHST1174-475 | 17 |
| pLR733 | A | ScHST11-26-ScSIR294-198-ScHST1112-475-HA | |
| pLR753 | ScSir2 | ScSIR2-HA | |
| pLR754 | ScSir2-2H | ScSIR2N378Q L379I-HA | |
| pLR755 | ScSir2-2H′ | ScSIR2N386E K387N-HA | |
| pLR756 | ScSir2-4H | ScSIR2N378Q L379I N386E K387N-HA | |
| pLR757 | H-S | ScHST11-173-ScSIR2256-562-HA | |
| pLR770 | B1 | ScHST11-26-ScSIR294-149-ScHST164-475-HA | |
| pLR771 | B2 | ScHST11-63-ScSIR2150-198-ScHST1112-475-HA | |
| pLR788 | ScHst1-2S | ScHST1Q296N I297L-HA | |
| pLR789 | ScHst1-4S | ScHST1Q296N I297L E304N N305K-HA | |
| pLR801 | C1 | ScHST11-26-ScSIR2106-149-ScHST164-475-HA | |
| pLR802 | C2 | ScHST11-26-ScSIR294-136-ScHST151-475-HA | |
| pLR814 | A* | ScHST11-26-ScSIR294-198Y145N G146D-ScHST1112-475-HA | |
| pLR830 | ScSir2* | ScSIR2Y145N G146D-HA | |
| K. lactis | |||
| pLR849 | Vector | ||
| pLR850 | KlSir2 | HA-KlSIR2 | |
| pLR852 | KlSir2-2S | HA-KlSIR2K434N I435L | |
| pLR853 | KlSir2-4S | HA-KlSIR2K434N I435L P442N N443K | |
| pLR854 | KlSir2* | HA-KlSIR2F188N G189D | |
| pLR876 | KlSir2-F188N | HA-KlSIR2F188N | |
| pLR877 | KlSir2-G189D | HA-KlSIR2G189D | |
S. cerevisiae plasmids use the pRS416 backbone (ScCEN/ARS URA3). K. lactis plasmids use the pLR849 backbone (KlCEN/ARS hphR).
Mutations in ScSir2 (pLR754-756) were made by site-directed mutagenesis and confirmed by sequencing. On our original plasmids, pLR499 and derivatives, the ScSIR2 gene was under the control of a truncated ScHST1 promoter rather than the native ScSIR2 promoter. Consequently, ScSIR2 was poorly expressed. Therefore, the promoters were replaced with the ScSIR2 promoter, which was amplified by PCR from pLR196 using primers with flanking homology to the target plasmids. The target plasmids were linearized with MfeI, and the PCR product containing the ScSIR2 promoter was incorporated into each plasmid by homologous recombination in S. cerevisiae. Correct plasmids were confirmed by sequencing.
To generate pLR733 (A), pLR770 (B1), pLR801 (C1), and pLR802 (C2), in which portions of ScHST1 were replaced by the homologous region of ScSIR2, homologous recombination in S. cerevisiae was used. The desired ScSIR2 sequence was amplified by PCR from pLR499 using primers specific for ScSIR2 with flanking homology for ScHST1. The resulting PCR product was then incorporated by homologous recombination into ScHST1-HA on pLR30, which had been linearized with EcoNI. Using a similar strategy, pLR771 (B2) was generated by amplifying the sequence for ScHst1 amino acids (aa) 27 to 63 to replace ScSir2 aa 94 to 149 in pLR733, which had been cut with BsiWI. pLR757 (H-S) was generated by amplifying the sequence for ScSir2 aa 256 to 562 to replace ScHst1 aa 174 to 475 in pLR30, which had been cut with BglII and BsaBI. Amino acid substitutions to generate pLR788 and pLR789 (from pLR30), pLR814 (from pLR733), and pLR830 (from pLR753) were made by site-directed mutagenesis and confirmed by sequencing.
K. lactis plasmids.
K. lactis plasmids used in this study are listed in Table 2. To construct the plasmid with HA-KlSIR2 (pLR850), KlSIR2 was amplified by PCR from genomic DNA using primers 5′-GCTTACGTCGACCTTGGGGTCTATGATGCACTAC and 5′-CGAAATGCGGCCGCCGTGATACTATCACAACTGACGAAG, with the underlined sequences annealing to genomic DNA and the bold sequences creating NotI and SalI restriction sites. The KlSIR2 gene was cloned into the NotI and SalI sites of S. cerevisiae vector pRS316 to yield pLR666. Next, KlSIR2 was transferred from pLR666 into K. lactis plasmid pCXJ18 (6) using the SacI and SalI restriction sites, yielding pLR730. Finally, an epitope tag was added by cloning an AgeI-SacI fragment containing HA-KlSIR2 from pLR490 (17) into the same sites of pLR730, yielding pLR734. Because Sir2-mediated repression was less robust in the minimal medium required to select for these URA3-expressing plasmids, the hphMX marker, which confers hygromycin resistance, was cloned into HaeII and HindIII sites on pCXJ18 and pLR734 using a HaeII and HindIII fragment from pAG32 (14) to yield pLR849 and pLR850. Amino acid substitutions were made by site-directed mutagenesis (4) and confirmed by sequencing.
Co-IPs.
Coimmunoprecipitations (co-IPs) were performed using modifications of a previously described protocol (42). For S. cerevisiae co-IPs, cells were grown in complete synthetic medium lacking uracil to select for plasmids and harvested at an optical density at 600 nm (OD600) of 1.0. K. lactis cells were grown in yeast extract-peptone-dextrose (YPD) with 300 μg/ml hygromycin B to ensure retention of the plasmid and harvested at an OD600 of 1.4. Twenty-five OD equivalents of cells was resuspended in 400 μl lysis buffer (50 mM HEPES-KOH [pH 7.5], 0.5 M NaCl, 10% glycerol, 0.5% NP-40, 1 mM EDTA, 10 mM dithiothreitol, 1× Complete protease inhibitor [Roche], 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml pepstatin A, 2 mM benzamidine), and approximately 0.5 ml 0.5-mm zirconia-silica beads (Biospec) was added to each sample. Cells were lysed by vortexing for 30 min at 4°C. Samples were then spun for 10 min at 13,200 rpm at 4°C. The cleared supernatants were incubated with 5 μl anti-HA (Sigma H-6908), anti-myc (Millipore 06-549), anti-Flag (Sigma F-7425), or anti-ScSir4 (serum from rabbit 2913.7/27.3, gift from J. Rine) antibody for 8 h at 4°C. A 60-μl volume of a 50% slurry of protein A agarose or Sepharose beads (washed three times with phosphate-buffered saline [PBS]) was added, and samples were rotated overnight at 4°C. The beads were collected by centrifugation for 2 min at 2,000 rpm and washed three times with 1 ml of lysis buffer. The beads were then resuspended in 30 μl 1.5× protein sample buffer and heated at 95°C for 5 min. Fifteen-microliter volumes of IP samples were electrophoretically fractionated on either 7.5% or 10% polyacrylamide-SDS gels, transferred to nitrocellulose membranes, and probed with mouse monoclonal anti-HA antibody (Sigma H-3663), mouse polyclonal anti-myc antibody (Calbiochem OP10), mouse monoclonal anti-myc antibody (Millipore 05-724), or mouse monoclonal anti-Flag antibody (Sigma F-3165) and detected by chemiluminescence (GE RPN2135).
Gene expression analysis.
RNA was isolated from logarithmically growing cultures (39) and treated with DNase I as previously described (17). To confirm that the DNase treatment was complete, 1 μl of DNase-treated RNA was used in a PCR mixture containing primers for the ScACT1 or KlACT1 transcript. One microgram of DNA-free RNA was used for cDNA synthesis as previously described (17). To quantify the relative amount of mRNA transcripts, cDNA was analyzed by real-time PCR in the presence of SYBR green using a Bio-Rad iCycler. Oligonucleotide sequences are provided in Table 3. A standard curve was generated with genomic DNA isolated from LRY1007, LRY2128, or LRY2388. Transcript levels of queried genes were first normalized to ScACT1 or KlACT1 for each sample. The n-fold induction was then calculated by normalizing to the strain expressing wild-type ScHst1-HA or HA-KlSir2. Results represent the average fold induction of two independent cultures. The standard error of the mean (SEM) was calculated from the differences in n-fold induction of two independent cultures from the mean.
Table 3.
Oligonucleotides used in this study for RT-PCR
| Target | Oligonucleotide sequence |
|---|---|
| S. cerevisiae | |
| ACT1 | GCCTTCTACGTTTCCATCCA |
| GGCCAAATCGATTCTCAAA | |
| DTR1 | GGTGGGCACCTCTCAGATTATC |
| CATACCAAAGGCAGTGAGAGCG | |
| SPS1 | AAGGTCCCTTTTCGGATGCAG |
| TTTCATCGTCGCGCGCAC | |
| K. lactis | |
| ACT1 | GTGGTACCACCGGACATGAC |
| CGTCGCTTTGGACTTCGA ACAA | |
| CDA2 | CGGATCTTAGGAAAGGATTAGAG |
| GTACACATACTTGGTCACATCC | |
| SPS4 | CCTCCTGGTTGTCCAAATTTACG |
| GAGGTTCGTTGGATCCACTTG | |
| KLLA0B14927g | AGCTCTAGTGTTGTTGTTGGCTC |
| CTTCTGGGGTATTAATGCTGCTG | |
ChIPs.
Chromatin IPs (ChIPs) were performed by harvesting approximately 50 OD units of logarithmically growing cells at an OD600 of around 1.0 for S. cerevisiae or 1.4 for K. lactis. Cells were collected, washed twice with PBS, resuspended in DMA (10 mM dimethyl adipimidate, 0.1% dimethyl sulfoxide, 1× PBS), and rocked at room temperature for 45 min for cross-linking. Cells were washed once with PBS, resuspended in PBS-1% formaldehyde, and rocked for 45 min. The preparation of soluble chromatin and IP were performed as previously described (37), using 5 μl of the same antibodies used for co-IPs. Chromatin IP samples were analyzed by real-time PCR using a standard curve prepared from input DNA and oligonucleotides listed in Table 4. The amounts of the immunoprecipitated DNA at experimental loci and a control locus, ScATS1 or KlRRP7, were determined relative to the standard curve, and then the relative enrichment of the experimental loci compared to that of the control locus was calculated. Results represent IP from two or more independent cultures of each transformed strain, and the SEM was calculated from the differences of the relative enrichment from the mean.
Table 4.
Oligonucleotides used in this study for chromatin IP
| Target | Oligonucleotide sequence |
|---|---|
| S. cerevisiae | |
| ATS1 | GGTAACGCAGCCGTTTGAGC |
| CCTCATCGTGCCCCAGTCC | |
| DTR1 | GTAGCCAAAGCTGCCTGTTG |
| CTTACTACCATCCTTCTAGCC | |
| PES4 | CATTGTACATTCTCCAAATGTGGTG |
| TCTAGTACCTACTGTGCCGAATAATGTG | |
| HML-E | CAGACTTCAACACAATCAGAATCAAATAG |
| GGCCCCCGAAATCGATAATAATG | |
| HML promoter | CACTTCTAAGCTGATTTCAATCTCTC |
| GGATGCTTTGTTCTTAATTTTGAAAGCAG | |
| HML-I | CGATGCTTATTGTGCTTTGTTGGG |
| GTTTGCCATTTCCAGCACCTC | |
| K. lactis | |
| RRP7 | GCAACAACAGATACTGT GG |
| CCTACTACTAATGTGAAACCATC | |
| CDA2 | CGATGTATCGGCTAGTAATATTTCG |
| GGATCAATGGGGAGGCTGTAG | |
| SPS4 | CGGCCTACAGAAATGACTACTG |
| CTCGCTTAATATCGGTTGACAC | |
| Telomere BR-A | TCGAGACCCCAGAGTTTAAGAC |
| ATATACGGTACCGGTCCAAGGA | |
| Telomere BR-B | CAAACACCAGAAATTGAAACTGCC |
| GAGTAAACACCGTTGTGGTAGGA | |
| Telomere BR-C | TGGAGAGTTCTATTACTTCCGCC |
| GTGAACGAATCCGATGTCTGTG | |
| Telomere BR-D | AGCTCTAGTGTTGTTGTTGGCTC |
| CTTCTGGGGTATTAATGCTGCTG | |
| Telomere BR-E | ATCACGTGACTGGAAGTCGAGT |
| TTGCAACGATTCGAACATGCTGT | |
| Telomere BR-F | ACAGGAAAGAAAGGAGTAGAGGTG |
| CATCCCCCAGCATAAATTCATCA | |
| Telomere BR-G | AACAAAGGAGAATGCAGGGAGAGT |
| CCCGCTATATTTGGTCCATCATC | |
Mating and reporter assays.
One OD equivalent of cells was collected from logarithmically growing cultures by centrifugation and resuspended in 100 μl minimal medium. For each transformed strain, 10-fold serial dilutions were prepared, and 2 to 3 μl of each sample in the dilution series was spotted onto a YPD plate to confirm equivalent dilutions. To assay expression of the pPES4–HIS3 reporter, 3 μl of each sample in the dilution series was spotted onto medium lacking histidine and uracil. Uracil was omitted to maintain the plasmids. To assay mating, an equal volume of the tester strain LRY1022 at 10 OD equivalents/ml in YPD was mixed with each sample in the dilution series, and 3 μl of this mixture was spotted onto minimal plates to select for the growth of prototrophic diploids. Yeast cells were grown at 30°C for 2 days and imaged.
RESULTS
ScSir2 and ScHst1 have distinct domains for interacting with their respective complexes.
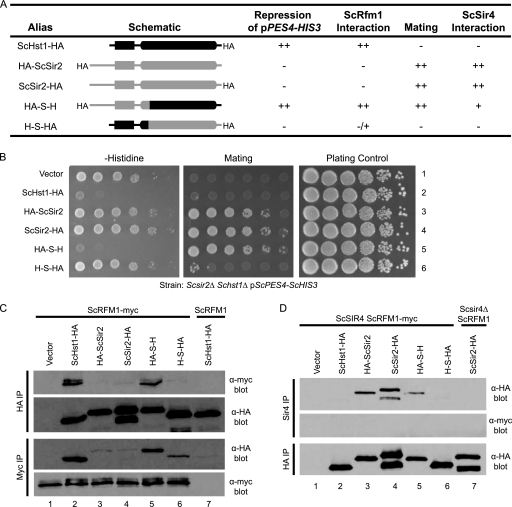
We postulated that Sir2 and Hst1 subfunctionalized by acquiring complementary mutations in interaction domains that are limited to either the N-terminal portion (Sir2-Sir4) or the C-terminal portion (Hst1-Rfm1) (Fig. 1B) of the protein. However, an alternative possibility is that the N- and C-terminal portions both contribute to both interactions by folding together to form an interaction surface. The first possibility is consistent with the previous finding that a chimeric protein containing the N terminus of ScSir2 and the C terminus of ScHst1 (S-H) interacts with both the Sir and Sum1 complexes (17, 30). Yet, these results do not exclude the second possibility, in which either the N or the C terminus might be sufficient to allow the protein to interact with the Sir or Sum1 complex, as long as the remainder of the domain is provided by the paralog. To distinguish between these possibilities, we examined the function of a previously untested reverse chimera (H-S) containing the N terminus of ScHst1 and the C terminus of ScSir2 (Fig. 2 A). If interaction domains are limited to the N-terminal portion of ScSir2 and the C-terminal half of ScHst1, the H-S chimera should not interact with either complex. On the other hand, if ScSir2 and ScHst1 have interaction domains incorporating regions from both the N and C termini, the H-S chimera should interact with both complexes.
Fig. 2.
Distinct portions of ScSir2 and ScHst1 are required for specificity. (A) Summary of chimeric proteins and their properties. (B) Hst1-mediated repression and Sir2-mediated silencing were assessed using a pPES4–HIS3 reporter and a mating assay. A Scsir2Δ Schst1Δ mutant strain (LRY2083) was transformed with an empty vector (pRS416) or plasmids expressing the constructs shown in panel A (Table 2). To assess ScHst1-mediated repression (left side), yeast cells were spotted onto medium lacking histidine and uracil (for plasmid maintenance) in 10-fold serial dilutions. To assess ScSir2-mediated silencing (middle), the same dilutions were mixed with a tester strain of the opposite mating type (LRY1022) and spotted onto minimal medium to select for prototrophic diploid cells. As a plating control (right side), yeast cells were spotted onto rich medium. (C) The association of chimeric proteins with ScRfm1-myc was examined by co-IP. HA-tagged protein or ScRfm1-myc was immunoprecipitated from an Scsir2Δ Schst1Δ ScRFM1–myc mutant strain (LRY2507) transformed with plasmids expressing the constructs in panel A or an Scsir2Δ Schst1Δ ScRFM1 mutant strain (LRY2083) transformed with ScHST1-HA (lane 7). The precipitated material was examined by immunoblotting with mouse anti-HA or anti-myc antibody. Two major bands were observed for C-terminally HA-tagged ScSir2 but not the N-terminally tagged protein. The larger band (∼70 kDa) is close to the predicted size of full-length ScSir2-HA. Others have also observed multiple bands for C-terminally tagged ScSir2 constructs (14a, 34) and have postulated that the smaller bands are degradation products. However, as multiple bands are not seen for N-terminally tagged ScSir2, the smaller band could result from internal translation initiation. (D) The association of chimeric proteins with ScSir4 was examined by co-IP. ScSir4 was immunoprecipitated from the same strains used in panel C or an Scsir2Δ Schst1Δ Scsir4Δ mutant strain (LRY2590) transformed with ScSIR2-HA (lane 7).
To test for ScHst1 function, a previously described (17) reporter was used in which the ScHst1-regulated PES4 promoter drives the expression of HIS3, which is required for growth in the absence of histidine. Wild-type Hst1 function is indicated by lack of growth in the absence of histidine. To test for ScSir2 function, a mating assay was used. ScSir2 is required for silencing of the cryptic mating-type loci, HML and HMR. If these loci are not silenced, cells express both a and α mating information and cannot mate. Wild-type Sir2 function is indicated by growth on medium selecting for diploids. For these experiments, ScSIR2 and ScHST1 were deleted from the genome and ScHst1, ScSir2, and chimeric proteins were expressed from plasmids.
As previously reported, a chimera containing the N terminus of ScSir2 and the C terminus of ScHst1 (S-H) was able to function as both ScSir2 and ScHst1 (Fig. 2B). In contrast, the new reverse chimera, H-S, was not able to function as either ScSir2 or ScHst1 although it was stably expressed (Fig. 2C). Therefore, different portions of the deacetylases confer specificity on ScSir2 and ScHst1.
To assess the interactions of these chimeric proteins with ScSir4 and ScRfm1, a co-IP assay was used. The S-H chimera was able to interact with ScRfm1 robustly (Fig. 2C) and with ScSir4 weakly (Fig. 2D). In contrast, the H-S chimera was unable to interact with ScSir4 and had a weaker interaction with ScRfm1 (Fig. 2C and D), indicating that the N-terminal domain of ScSir2 is necessary for association with ScSir4 and the C-terminal domain of ScHst1 is required for optimal association with ScRfm1. The modest interaction between the H-S chimera and ScRfm1 is consistent with previous work indicating that ScSir2 itself interacts weakly with the ScSum1 complex (17). This interaction may be increased in the chimeric protein, which lacks the N-terminal Sir4 interaction domain and hence is available to interact with Rfm1. In summary, although both portions of these deacetylases are required for maximum interaction with ScSir4 and ScRfm1, the N terminus is more critical for ScSir2 and the C terminus is more critical for ScHst1.
The ability of S-H to interact with both ScRfm1 and ScSir4 raises the possibility that both interactions occur simultaneously. However, ScSir4 did not coimmunoprecipitate with ScRfm1 in the presence of S-H (Fig. 2D), suggesting that a single molecule of S-H cannot interact with both complexes simultaneously.
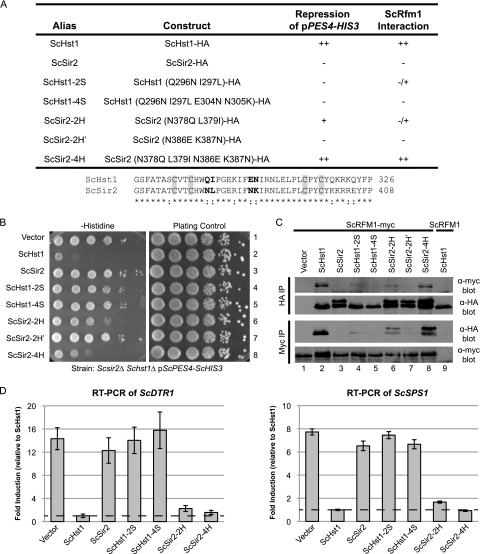
Four amino acid changes enable ScSir2 to function as ScHst1.
The proposed model for the subfunctionalization of Sir2 and Hst1 (Fig. 1B) postulates that the domains that interact with Sir4 and Rfm1 were present in the ancestral, nonduplicated Sir2 protein. To test this idea, we identified mutations in ScHst1 that would disrupt its interaction with ScRfm1 and then determined whether mutations in the homologous residues of KlSir2 disrupt its interaction with KlRfm1. To facilitate the identification of such mutations, we first determined the minimal portion of ScHst1 that, when placed in ScSir2, would confer the specificity of ScHst1. The C-terminal portion of ScHst1, which enables it to interact with ScRfm1 (Fig. 2), has high homology with ScSir2, suggesting that small differences between these deacetylases determine specificity. Indeed, replacing two amino acids in the zinc-binding module of ScSir2 with the amino acids from ScHst1, N378Q and L379I, enables ScSir2 to interact with ScRfm1 and repress a promoter containing a heterologous ScSum1 binding site (30).
To determine whether one of these two amino acids is sufficient for ScSir2 to interact with ScRfm1, we generated separate N378Q and L379I mutations and tested them using the pPES4–HIS3 reporter (Fig. 3 A). Neither single mutation conferred the ability to repress this reporter (data not shown). Moreover, the two-amino-acid mutation (ScSir2-2H) was less effective than ScHst1 (Fig. 3B) and had a lower affinity for ScRfm1-myc (Fig. 3C). Therefore, although the previously described two-amino-acid substitution increases the affinity of ScSir2 for ScRfm1, it does not match the affinity of wild-type ScHst1.
Fig. 3.
Four amino acid substitutions confer ScHst1 function on ScSir2. (A) Summary of mutations and their properties. An alignment of the zinc-binding domains of ScHst1 and ScSir2 is shown, with mutated residues in bold and the zinc-binding cysteines in gray boxes. (B) ScHst1-mediated repression was assessed using the pPES4–HIS3 reporter. A Scsir2Δ Schst1Δ ScRFM1–myc mutant strain (LRY2507) was transformed with an empty vector or plasmids expressing the constructs shown in panel A and analyzed as described in the legend to Fig. 2B. (C) The association of mutated proteins with ScRfm1-myc was examined by co-IP. HA epitope-tagged protein or ScRfm1-myc was immunoprecipitated from the same strains as used in panel B or an Scsir2Δ Schst1Δ ScRFM1 mutant strain (LRY2083) transformed with ScHST1-HA (lane 9). (D) Expression of ScDTR1 and ScSPS1 was assessed by RT-PCR in the same strains as in panel B. Levels of ScDTR1 and ScSPS1 mRNAs were first normalized to ScACT1 and then expressed relative to those in the strain containing wild-type ScHst1-HA.
To determine whether additional mutations in ScSir2 could enhance its interaction with ScRfm1, two other amino acids not conserved between ScSir2 and ScHst1 were mutated. These two mutations, N386E and K387N, were made by themselves (ScSir2-2H′) and with the original pair of mutations (ScSir2-4H) (Fig. 3A). ScSir2-2H′ had no ScHst1-like function (Fig. 3B) and did not coimmunoprecipitate with ScRfm1 (Fig. 3C). However, in combination with the two previously identified mutations, N386E and K387N increased the ability of ScSir2 to repress the reporter (Fig. 3B) and increased the co-IP with ScRfm1 (Fig. 3C).
The pPES4–HIS3 reporter provides only a qualitative assessment of repression. Therefore, quantitative reverse transcription (RT)-PCR was used to assess the levels of mRNA of two ScHst1-regulated genes, DTR1 and SPS1 (Fig. 3D). In the presence of ScSir2-2H and ScSir2-4H, DTR1 and SPS1 were increased in expression only slightly compared to wild-type ScHst1, indicating that even the two-amino-acid substitution confers Hst1-like properties on ScSir2. Thus, modest amino acid changes in ScSir2 were sufficient to confer ScHst1-like function.
The identification of amino acid substitutions that enhance the affinity of ScSir2 for ScRfm1 pinpointed the interaction domain and enabled us to test whether a small number of mutations in this domain would disrupt the interaction, as might have occurred during the process of subfunctionalization. Therefore, mutations at the two originally identified residues (ScHst1-2S) and all four residues (ScHst1-4S) were made in ScHst1 and examined for their effects on the function of ScHst1. Indeed, neither ScHst1-2S nor ScHst1-4S repressed the HIS3 reporter (Fig. 3B) or the DTR1 and SPS1 genes (Fig. 3D). A weak interaction with ScRfm1 was detected for ScHst1-2S (Fig. 3D), but the four amino acid mutations in ScHst1-4S were sufficient to eliminate the interaction with ScRfm1 (Fig. 3C).
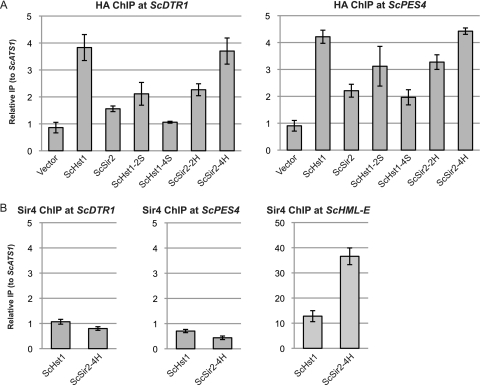
ScSir4 was not recruited to promoters repressed by ScSir2-4H.
The ability of ScSir2-4H to interact with both ScSir4 and ScRfm1 raises the possibility that ScSir2-4H can bring the Sir complex to ScHst1-regulated genes. If it can, the Sir complex could be responsible for repressing those genes. To examine this possibility, the presence of ScSir4 at the DTR1 and PES4 promoters was examined using chromatin IP. Consistent with the functional and co-IP results discussed above, ScSir2-2H was more enriched than ScSir2 at the DTR1 and PES4 promoters, and ScSir2-4H had an enrichment comparable to that of ScHst1 (Fig. 4 A). However, even in the presence of ScSir2-4H, there was no enrichment of ScSir4 at either promoter (Fig. 4B). ScSir4 was detected at HML (Fig. 4B), indicating that the IP was successful. Therefore, ScSir4 was not recruited to these promoters by ScSir2-4H and was not responsible for the repression mediated by ScSir2-4H.
Fig. 4.
ScSir4 is not associated with promoters of sporulation genes. (A) The association of ScHst1, ScSir2, and derivatives with the ScDTR1 and ScPES4 promoters was examined by chromatin IP (ChIP). HA-tagged proteins were precipitated from an Scsir2Δ Schst1Δ ScRFM1–myc mutant yeast strain (LRY2507) transformed with the empty vector or plasmids containing the constructs shown in Fig. 3A. Associated DNA was analyzed by quantitative PCR, and enrichment is expressed relative to the ScATS1 locus, which is not associated with ScSir2 or ScHst1. (B) The association of ScSir4 with the ScDTR1 and ScPES4 promoters and the HML–E silencer was examined by ChIP as in panel A.
As a control, the presence of ScRfm1-myc at these promoters was also examined. ScRfm1 was enriched regardless of whether a deacetylase was present (data not shown), consistent with the view that ScRfm1 is recruited through an interaction with ScSum1 independently of its interaction with ScHst1.
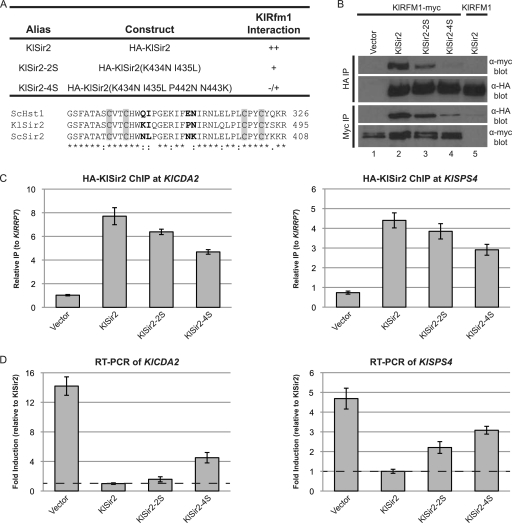
The interaction domain for Rfm1 is conserved between ScHst1 and KlSir2.
To determine whether the interaction domain between Hst1 and Rfm1 predates the duplication, we used nonduplicated Sir2 from K. lactis as a proxy for the ancestral deacetylase. If the interaction domain is ancestral, mutating amino acids in KlSir2 homologous to those that disrupt the interaction in S. cerevisiae should also disrupt the interaction in K. lactis. Therefore, we generated mutations equivalent to ScHst1-2S (KlSir2-2S) and ScHst1-4S (KlSir2-4S) and examined their abilities to interact with KlRfm1-myc by co-IP. The affinity of KlSir2-2S for KlRfm1-myc was slightly reduced compared to that of KlSir2 (Fig. 5 B), and the four-amino-acid mutation, KlSir2-4S, had a much weaker interaction with KlRfm1-myc. To confirm that the loss of interaction was specific for these amino acids, three other pairs of mutations in KlSir2 were generated targeting residues that differ between KlSir2 and ScSir2. The T358M/E359H and S457K/L460R mutations fall on the surface of the zinc-binding domain some distance from the 4S mutations, and the E412S/P413T mutation falls on the surface of the main catalytic domain on the same face of the protein as the 4S mutations. These mutations had either no effect or very slight effects on the interaction between KlSir2 and KlRfm1 (data not shown). Thus, the KlSir2-2S mutation has a considerably greater impact on the interaction with KlRfm1 and likely identifies an interaction surface that is homologous to the interaction surface in ScHst1. Therefore, the interaction domain for Rfm1 is ancestral rather than derived after the duplication. Moreover, the capacity of this interaction to be reduced by simple substitution mutations is consistent with the DDC model of subfunctionalization. The duplicate that became SIR2 needed to acquire only a few mutations in this region to be outcompeted by Hst1 and largely excluded from the Sum1 complex.
Fig. 5.
The interaction domain for Rfm1 is conserved between KlSir2 and ScHst1. (A) Summary of mutations and their properties. An alignment of the zinc-binding domains of ScHst1, KlSir2, and ScSir2 is shown, with mutated residues in bold and the zinc-binding cysteines in gray boxes. (B) The association of KlSir2 with KlRfm1 was examined by co-IP. HA-KlSir2 or KlRfm1-myc was immunoprecipitated from a Klsir2Δ KlRFM1–myc mutant strain (LRY2654) transformed with an empty vector (pLR849) or plasmids expressing the constructs shown in panel A or a Klsir2Δ mutant strain with untagged KlRFM1 (LRY2128) transformed with HA–KlSIR2. (C) The association of HA-KlSir2 with the promoters of KlCDA2 and KlSPS4 was assessed by ChIP using the same strains as in panel B. Enrichment at the promoters is expressed relative to the KlRRP7 locus, which is not associated with KlSir2. (D) Expression of KlCDA2 (KLLA0C17226g) and KlSPS4 (KLLA0F08679g) was assessed by RT-PCR in the same strains as in panel B. Levels of KlCDA2 and KlSPS4 mRNA were first normalized to KlACT1 and then expressed relative to those of the strain containing wild-type HA-KlSir2.
To determine whether the decreased interaction with KlRfm1 was sufficient to reduce the recruitment of KlSir2-2S and KlSir2-4S to repressed loci, chromatin IP was conducted at the KlCDA2 and KlSPS4 promoters. KlSir2-4S was still present at both promoters but to a lesser extent than KlSir2 or KlSir2-2S, consistent with its reduced interaction with KlRfm1 (Fig. 5C). A potential explanation for the residual association of KlSir2-4S with repressed promoters, despite the apparent lack of interaction with KlRfm1, is that the chromatin IP is stabilized by cross-linking. In addition, KlSir2-4S may be recruited to these promoters by other proteins, as suggested by previous observations that wild-type KlSir2 is recruited to KlCDA2 and KlSPS4 in the absence of KlRfm1 (18). As expected, the association of KlRfm1 with KlCDA2 and KlSPS4 was not significantly affected in the presence of KlSir2-2S and KlSir2-4S (data not shown).
To assess the functional consequence of the reduced association of KlSir2-2S and KlSir2-4S with target promoters, the expression of KlCDA2 and KlSPS4 was examined by quantitative RT-PCR (Fig. 5D). Both KlCDA2 and KlSPS4 were induced in the presence of KlSir2-4S, although not to the levels of a strain lacking KlSir2. These results are consistent with the lower enrichment of KlSir2 at the promoters of these genes and the loss of affinity for KlRfm1.
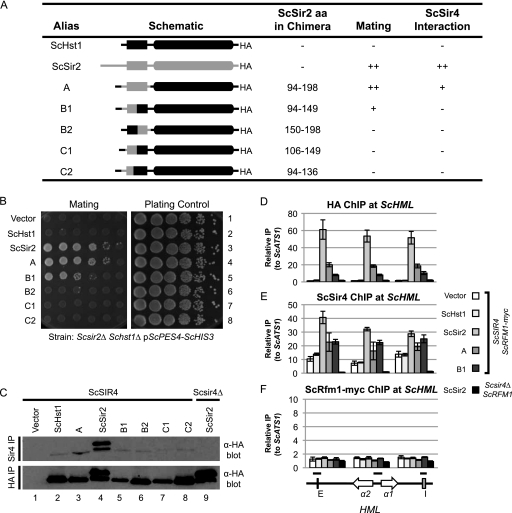
Amino acids 94 to 149 of ScSir2 enable ScHst1 to function as ScSir2.
The results above indicate that, after duplication, the paralog that became Sir2 lost affinity for the Sum1 complex by acquiring mutations in the zinc-binding module. To determine whether a similar process reduced the affinity of the paralog that became Hst1 for the Sir complex, the region important for the interaction of ScSir2 with ScSir4 was identified and this information was used to determine whether this interaction domain is conserved between ScSir2 and KlSir2. The sequences of the N-terminal domains of ScSir2, ScHst1, and KlSir2 are less well conserved than the zinc-binding module, and consequently, it is unlikely that a few amino acid substitutions in ScHst1 could confer the specificity of ScSir2. Therefore, we identified the minimal region of ScSir2 necessary for its function by generating a series of chimeric proteins in which a region of ScSir2 replaced the homologous region of ScHst1 (Fig. 6 A).
Fig. 6.
ScSir2 aa 94 to 149 confer ScSir2 function on ScHst1. (A) Summary of chimeric proteins and their properties. (B) ScSir2-mediated silencing was assessed using a mating assay. A Scsir2Δ Schst1Δ mutant strain (LRY2083) was transformed with an empty vector or plasmids expressing the constructs shown in panel A. Mating was assessed as described for Fig. 2B. (C) The association of the chimeric proteins with ScSir4 was examined by co-IP. ScSir4 was immunoprecipitated from the same strains used in panel B or an Scsir2Δ Schst1Δ Scsir4Δ mutant strain (LRY2590) transformed with ScSIR2-HA. (D) The association of ScHst1, ScSir2, and chimeric proteins with HML was examined by ChIP. HA epitope-tagged proteins were immunoprecipitated from an Scsir2Δ Schst1Δ ScRFM1–myc mutant yeast strain (LRY2507) transformed with the empty vector or plasmids expressing the constructs shown in panel A or an Scsir2Δ Schst1Δ Scsir4Δ ScRFM1 mutant strain (LRY2590) transformed with ScSIR2-HA. (E) The association of ScSir4 with HML in the same strains as in panel D was assessed by ChIP. (F) The association of ScRfm1-myc with HML in the same strains as in panel D was assessed by ChIP. Black bars above HML indicate the locations of PCR amplicons.
The design of chimeric proteins was guided by an observation that deleting the first 93 aa of ScSir2 has no effect on its interaction with ScSir4, whereas deleting the first 198 aa abolishes the interaction (7). Chimera A (containing ScSir2 aa 94 to 198) functioned as well as ScSir2 in a mating assay (Fig. 6B), suggesting that the interaction domain for ScSir4 is contained within aa 94 to 198. Indeed, chimera A coimmunoprecipitated with ScSir4 (Fig. 6C) and associated with the silenced HML locus (Fig. 6D). However, the affinity of chimera A for ScSir4 was reduced compared to that of ScSir2, consistent with the reduced affinity of the S-H construct (Fig. 2D). Chimera B1, containing a smaller portion of ScSir2 (aa 94 to 149), restored mating to about 1/10 of the level of ScSir2 and chimera A and associated with HML (Fig. 6B and D). However, an interaction between chimera B1 and ScSir4 was not detected (Fig. 6C). The interaction of chimera B1 with ScSir4 must be sufficient for some silencing of HML but not stable enough to be maintained through co-IP. In contrast, chimera B2 (ScSir2 aa 150 to 198) was unable to restore mating, although it was expressed and retained the ability to repress ScHst1-regulated promoters (Fig. 6B and C and data not shown). Therefore, aa 94 to 149 of ScSir2 are critical for the interaction of ScSir2 with ScSir4 and aa 150 to 198 increase this interaction.
Chimeric proteins containing smaller fragments of ScSir2 had no ScSir2 function, as neither chimera C1 (ScSir2 aa 106 to 149) nor C2 (ScSir2 aa 94 to 136) conferred the ability to mate (Fig. 6B) or associate with ScSir4 (Fig. 6C). Therefore, the minimal region required for ScSir2 function is aa 94 to 149.
Another way to determine whether the chimeric deacetylases functioned at HML was to determine whether they enabled the spreading of ScSir4. ScSir4 is recruited to silencers independently of ScSir2 but cannot spread without ScSir2 (21, 27, 35). Therefore, the distribution of ScSir4 across HML was examined by chromatin IP. ScSir4 was more enriched at all three HML loci queried in the presence of chimeras A and B1 than in the presence of ScHst1 but was less enriched than in the presence of ScSir2 (Fig. 6E). Thus, chimeras A and B1 promoted the spreading of ScSir4.
Given that chimeras A and B1 are also recruited to sporulation genes (data not shown) and therefore must interact with the Sum1 complex, it was possible that these proteins were bringing the Sum1 complex to HML and that the Sum1 complex was contributing to the silencing of HML. However, ScRfm1-myc was not enriched at HML when either chimera A or B1 was associated with HML (Fig. 6F). This result, in conjunction with Fig. 2D and 4B, suggests that ScSir2 and ScHst1 do not interact simultaneously with the Sir and Sum1 complexes.
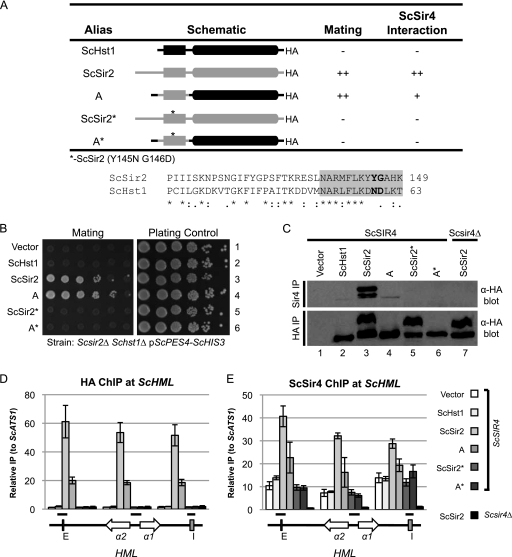
Two mutations in ScSir2 disrupt its interaction with ScSir4.
Having identified the region of ScSir2 that interacts with ScSir4, we investigated whether this interaction domain for Sir4 is the same in KlSir2. To do so, we sought simple substitution mutations that would disrupt the interactions of both ScSir2 and KlSir2 with Sir4. Given that chimeras C1 and C2 lack ScSir2 function, aa 94 to 105 and 137 to 149 must be required. Guided by an alignment of the Sir2 and Hst1 proteins from both nonduplicated and duplicated yeast species, aa Y145 and G146 of ScSir2 were identified as good candidates to confer specificity based on their position and conservation among duplicated Sir2 proteins but not all duplicated Hst1 proteins. This pair of amino acids was mutated to the homologous amino acids from ScHst1 in both ScSir2 (ScSir2*) and chimera A (A*).
The Y145N G146D mutations in both ScSir2 and chimera A completely abolished mating (Fig. 7 B). However, A*, like chimera A, was still able to repress the ScHst1-repressed reporter (data not shown), indicating that the Y145N G146D mutations did not affect the overall stability or catalytic function of the protein. In addition, ScSir2* and A* were unable to interact with ScSir4 (Fig. 7C) and did not associate with HML (Fig. 7D). Furthermore, enrichment of ScSir4 at HML was reduced in the presence of ScSir2* and A* to the same levels as when the vector alone or ScHst1 was present (Fig. 7E). Thus, the Y145N G146D mutation eliminated the interaction between ScSir2 and ScSir4.
Fig. 7.
Mutations that disrupt the ScSir2-ScSir4 interaction are identified. (A) Summary of mutations and their properties. An alignment of the relevant portion of ScSir2 and ScHst1 is shown. Positions that were mutated in ScSir2 are in bold. The gray box indicates aa 137 to 149, which are essential for ScSir2 function. (B) ScSir2-mediated silencing was assessed using a mating assay. A Scsir2Δ Schst1Δ mutant strain (LRY2083) was transformed with the empty vector or plasmids expressing the constructs in panel A. Mating was assessed as described in the legend to Fig. 2B. (C) The association of the mutant proteins with ScSir4 was examined by co-IP. ScSir4 was immunoprecipitated from the same strains used in panel B or an Scsir2Δ Schst1Δ Scsir4Δ mutant strain (LRY2590) transformed with ScSIR2-HA. (D) The association of ScHst1, ScSir2, and chimeric proteins with HML was examined by chromatin IP (ChIP). HA epitope-tagged proteins were immunoprecipitated from an Scsir2Δ Schst1Δ mutant yeast strain (LRY2507) transformed with the empty vector or plasmids expressing the constructs in panel A or an Scsir2Δ Schst1Δ Scsir4Δ mutant strain (LRY2590) transformed with ScSIR2-HA. (E) The association of ScSir4 with HML in the same strains as in panel D was assessed by ChIP.
Interaction domains for KlSir4 were conserved in KlSir2 and ScSir2.
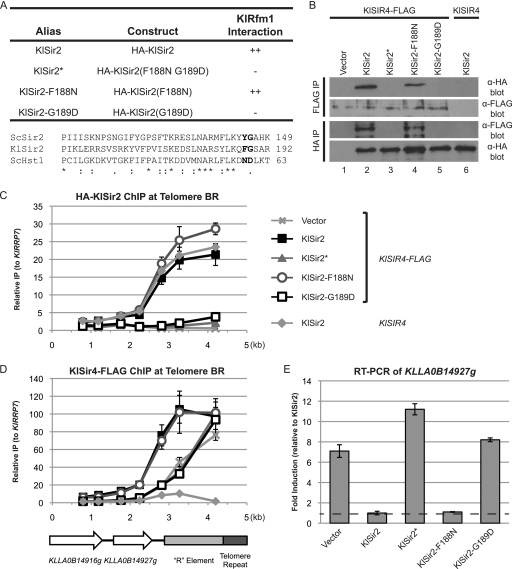
To determine whether the interaction domain between Sir2 and Sir4 was ancestral, we investigated whether mutations homologous to the ScSir2* mutations would disrupt the interaction of the nonduplicated KlSir2 with KlSir4. Residues F188 and G189 of KlSir2 were mutated to the same amino acids that were disruptive in ScSir2 (Fig. 8 A), and this protein (KlSir2*) was tested for its interaction with KlSir4-FLAG. Whereas wild-type KlSir2 had a robust interaction with KlSir4, KlSir2* did not interact (Fig. 8B). Thus, homologous amino acids are necessary for KlSir2 and ScSir2 to interact with Sir4, consistent with the interaction domain being ancestral.
Fig. 8.
The interaction domain for Sir4 is conserved between KlSir2 and ScSir2. (A) Summary of mutations and their properties. An alignment of the relevant portions of ScSir2, KlSir2, and ScHst1 is shown. Positions mutated in KlSir2 are in bold. (B) The association of HA-KlSir2 with KlSir4-FLAG was examined by co-IP. HA-KlSir2 and KlSir4-FLAG were immunoprecipitated from a Klsir2Δ KlSIR4–FLAG mutant strain (LRY2388) transformed with the empty vector or plasmids expressing the constructs in panel A or from a Klsir2Δ KlSIR4 mutant strain (LRY2333) transformed with HA-KlSIR2. (C) The distribution of HA-KlSir2 at Telomere BR was assessed in the same strains as in panel B by ChIP. (D) The distribution of KlSir4-FLAG at Telomere BR was assessed as described for panel C. (E) Expression of KLLA0B14927g was assessed by quantitative RT-PCR in the same strains as in panel B. Levels of KLLA0B14927g mRNA were first normalized to KlACT1 and then expressed relative to those of the strain containing wild-type HA-KlSir2.
To examine the functional consequences of eliminating the interaction between KlSir2 and KlSir4, the association of KlSir2* with Telomere BR was examined by chromatin IP. We previously found that KlSir2 and KlSir4 are associated with Telomere BR but KlSum1 is not (19), suggesting that recruitment of KlSir2 is dependent on KlSir4 but not KlSum1. KlSir2 was recruited to the end of the chromosome, and its enrichment tapered off with increased distance from the telomere (Fig. 8C). In contrast, KlSir2*, which does not interact with KlSir4, was not enriched at Telomere BR above background levels.
In S. cerevisiae, Sir4 can be recruited to the telomeres in the absence of Sir2 but requires Sir2 for spreading (21, 27, 35). If a similar dependency exists in K. lactis, the association of KlSir4 with Telomere BR should be reduced in the presence of KlSir2*. Indeed, the distribution of KlSir4 was restricted at Telomere BR in the presence of KlSir2* compared to KlSir2 and had the same sharp drop-off as in the absence of KlSir2 (Fig. 8D). Therefore, KlSir4 cannot spread at the telomere without an interaction with KlSir2.
The impact of the KlSir2* mutation on transcription was assessed by examining the expression of the telomere-proximal gene KLLA0B14927g by quantitative RT-PCR (Fig. 8E) (12). KLLA0B14927g was induced in the absence of KlSir2 about 7-fold compared to its level in the presence of KlSir2. In the presence of KlSir2*, KLLA0B14927g was induced approximately 11-fold, indicating that KlSir2 must interact with KlSir4 to silence at least this subtelomeric gene.
A single nucleotide change disrupted the interaction between KlSir2 and KlSir4.
The results outlined above support a DDC mechanism for the subfunctionalization of Sir2. In particular, the retention of the interaction domains for the Sir and Sum1 complexes between S. cerevisiae and K. lactis implies that the ancestral deacetylase that became duplicated also employed these domains. Thus, all that would be required for subfunctionalization would be for the duplicates to acquire complementary mutations in these two interaction domains.
To determine whether the simplest type of mutation, a single nucleotide change, could be sufficient to differentiate the functions of the duplicate deacetylases and initiate subfunctionalization, the F188N and G189D mutations were made singly (Fig. 8A). KlSir2-F188N still interacted with KlSir4 similarly to KlSir2 (Fig. 8B). However, the G189D mutation abolished this interaction. Moreover, KlSir2-G189D was absent from Telomere BR (Fig. 8C), resulting in the inability of KlSir4 to spread (Fig. 8D) and the induction of KLLA0B14927g (Fig. 8E). In contrast, KlSir2-F188N was recruited to and spread across Telomere BR in a fashion similar to that of KlSir2, thereby enabling the spreading of KlSir4 and the silencing of KLLA0B14927g. Thus, the single amino acid change G189D, which results from a single nucleotide change, was sufficient to disrupt the interaction of KlSir2 with KlSir4. This glycine is preserved in some duplicated Hst1 orthologs and therefore is unlikely to have been a key mutation that distinguished Hst1 from Sir2. However, a mutation with a similar impact could have occurred in one paralog after duplication, forcing the other paralog to maintain its interaction with Sir4 and become Sir2.
DISCUSSION
Subfunctionalization through the acquisition of complementary inactivating mutations.
This work strongly supports the model in which Sir2 and Hst1 subfunctionalized through the acquisition of complementary inactivating mutations in specificity-determining domains (Fig. 1B). It was previously concluded that the ancestral deacetylase that underwent duplication had both Sir2-like and Hst1-like functions (17, 18), indicating that subfunctionalization occurred. In addition, distinct regions of ScSir2 and ScHst1 were found to confer specificities for their respective complexes (17, 30). However, it was not known whether these specificity-determining regions were ancestral, in which case inactivating mutations would lead to subfunctionalization, or whether these specificity-determining regions were derived after duplication. We now demonstrate that the interactions of Sir2/Hst1 with Sir4 or Rfm1 are disrupted by homologous mutations in the duplicated species S. cerevisiae and the nonduplicated species K. lactis, strongly implying that the common ancestor of ScSir2, ScHst1, and KlSir2 utilized these same interaction domains. Thus, the interaction domains were ancestral, making it probable that inactivating mutations in these domains led to subfunctionalization. Moreover, the interaction between KlSir2 and KlSir4 can be disrupted by a single amino acid change, resulting from a single nucleotide change, indicating that relatively minor sequence changes would be sufficient to generate functionally distinct paralogs and initiate subfunctionalization.
The specificity of Sir2/Hst1 for the Sum1 complex is determined by amino acids in the zinc-binding module, a structural element found in all Sir2 family members but not directly involved in catalysis. An alignment of this module from nonduplicated and duplicated yeast species reveals few sequence differences that distinguish Hst1 and Sir2 paralogs. Indeed, the previously identified specificity-determining amino acids (ScHst1 Q296 and I297) (30) are the only two residues that consistently differ between duplicated Hst1 and Sir2 paralogs. Moreover, the Q296 position is conserved as a lysine in nonduplicated orthologs but mutated in almost all duplicated orthologs. Thus, mutations at this position are likely to have contributed to subfunctionalization and may have been coupled with compensatory mutations in the interacting partner Rfm1.
The specificity of Sir2 for the Sir complex is determined by a domain N terminal to the catalytic core. An alignment of this region reveals much greater sequence variation than in the zinc-binding module. Strikingly, changes in the duplicated Sir2 sequences compared to the nonduplicated orthologs tend to occur in fixed positions and to be found in all orthologs. In contrast, changes in the Hst1 orthologs are more sporadic and frequent. These patterns are consistent with the greater importance of this domain for the interaction with Sir4 than Rfm1. Thus, there are multiple positions at which mutations might initially have occurred to reduce the affinity for the Sir complex but not the Sum1 complex, thereby placing a duplicate on the path to becoming Hst1. The duplicate that became Sir2 also acquired mutations, but these were fixed early in the lineage.
In summary, the ancestral nature of the interaction domains for the Sir and Sum1 complexes, combined with the ability of these interactions to be disrupted by simple mutations, implies that the functions of the ancestral deacetylase were partitioned through the acquisition of complementary degenerative interactions. Both Sir2 and Hst1 have been maintained in all of the postduplication yeast species examined, and gene synteny for SIR2 and HST1 has been largely preserved (5). Thus, the fates of Sir2 and Hst1 were likely fixed early after the duplication.
Chimeric deacetylases did not interact simultaneously with the Sir and Sum1 complexes.
It was theoretically possible that the chimeric deacetylases capable of interacting with both Sir4 and Rfm1 could interact with these proteins simultaneously. However, although ScSir2-4H gained the ability to coprecipitate with ScRfm1 and associate with sporulation genes, its presence did not result in the recruitment of ScSir4 to these genes (Fig. 3 and 4). Thus, ScSir2-4H did not stably associate with ScSir4 when it was part of the Sum1 complex. Likewise, chimera A did not stably associate with ScRfm1 when it was part of the Sir complex (Fig. 6). These results suggest that the binding of one complex to a deacetylase prevents the other complex from gaining access. Presumably, the two complexes interfere sterically with one another, although how the N-terminal and zinc-binding modules are positioned relative to one another is unclear, as no structure for full-length Sir2 or Hst1 has been determined.
The inability of a single deacetylase to interact simultaneously with the Sir and Sum1 complexes probably reduces the chance of silenced chromatin forming in inappropriate locations. For example, the low-level interaction of ScSir2 with the Sum1 complex (17) would have the undesirable consequence of initiating the formation of Sir-silenced chromatin at Sum1-repressed genes if Sir2 could interact simultaneously with both complexes. This barrier against the formation of Sir-silenced chromatin at Sum1-repressed genes is even more critical in K. lactis, as the same deacetylase is normally part of both Sum1 and Sir complexes (18). Indeed, KlSir4 does not associate with Sum1-repressed sporulation genes, despite the presence of KlSir2 at these genes (18). Therefore, like its orthologs in S. cerevisiae, KlSir2 does not interact simultaneously with the Sir and Sum1 complexes.
The subfunctionalization of Sir2 and Hst1 is consistent with the DDC model.
The subfunctionalization of Sir2 and Hst1 through the acquisition of complementary inactivating mutations is a good example of the DDC model of subfunctionalization (13, 41). This model was originally discussed in the context of complementary expression patterns, and several examples of this type have been described (10, 13, 24, 32). However, to our knowledge, there are no other well-documented examples of complementary mutations partitioning protein functions.
An important aspect of the DDC model is that the initial fixation of the duplicate genes in a population does not involve adaptive mutations conferring new, beneficial properties. Rather, complementary loss-of-function mutations in the duplicates necessitate the maintenance of both genes to retain the full complement of ancestral functions. Indeed, there is no existing evidence that the combined activities of ScSir2 and ScHst1 provide any function not also present in nonduplicated KlSir2. However, it remains possible that dividing the ancestral functions between Sir2 and Hst1 was ultimately beneficial to the yeast. For example, ScHst1 has a lower affinity than ScSir2 for the cofactor NAD+ (2), and consequently, at certain cellular concentrations of NAD+, Hst1-repressed genes are induced but Sir2-repressed genes are not. Consequently, as NAD+ levels start to fall, the ScHst1-repressed NAD+ biosynthetic genes are induced to restore NAD+ pools without compromising ScSir2 function.
Quantitative subfunctionalization.
Subfunctionalization has been categorized as qualitative or quantitative (13, 38, 41). In the qualitative case, each duplicate experiences an irreversible loss of one function through an event such as the loss of a protein domain. In contrast, quantitative subfunctionalization involves the acquisition of reversible mutations that reduce but do not eliminate function. These reversible mutations have been described as “activity-reducing mutations” (38) that occur in cis regulatory elements and affect expression or in coding regions and impair catalysis or alter substrate specificity. Activity-reducing mutations are thought to promote the retention of duplicate genes until the occurrence of subfunctionalization, neofunctionalization, or loss of one copy of the gene. Although the types of mutations we have described in ScSir2 and ScHst1 should be reversible and meet many of the qualifications of quantitative subfunctionalization, the mutations that likely led to subfunctionalization differ conceptually from the previously described activity-reducing mutations, which largely affect the ability of the duplicates to perform their catalytic functions. Instead, disruptive mutations in Sir2 and Hst1 have occurred in the interaction domains, resulting in one deacetylase outcompeting the other for binding to a given repressive complex and thereby affecting the biological functions of Sir2 and Hst1 without impairing catalysis.
Conclusion.
This study demonstrates the value of yeast species for investigating the fates of duplicated genes. Because both duplicated and nonduplicated species are experimentally tractable, it is possible to infer the properties of the ancestral protein and the paths by which the duplicates have diverged. Given that roughly 10% of S. cerevisiae genes are duplicates retained from the whole-genome duplication, large numbers of duplicated genes could be studied experimentally to uncover general patterns of duplicate gene divergence.
ACKNOWLEDGMENTS
We thank M. Hickman and E. Sevastopoulos for plasmid and strain constructions; R. Kamakaka, J. Rine, and K. Nasmyth for strains, plasmids, and antibodies; and M. Hickman for comments on the manuscript.
This research was supported by a grant from the National Institutes of Health (GM073991) to L.N.R.
Footnotes
Published ahead of print on 20 June 2011.
REFERENCES
- 1. Barsoum E., Sjostrand J. O., Astrom S. U. 2010. Ume6 is required for the MATa/MATalpha cellular identity and transcriptional silencing in Kluyveromyces lactis. Genetics 184:999–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bedalov A., Hirao M., Posakony J., Nelson M., Simon J. A. 2003. NAD+-dependent deacetylase Hst1p controls biosynthesis and cellular NAD+ levels in Saccharomyces cerevisiae. Mol. Cell. Biol. 23:7044–7054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brachmann C. B., et al. 1995. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev. 9:2888–2902 [DOI] [PubMed] [Google Scholar]
- 4. Braman J., Papworth C., Greener A. 1996. Site-directed mutagenesis using double-stranded plasmid DNA templates. Methods Mol. Biol. 57:31–44 [DOI] [PubMed] [Google Scholar]
- 5. Byrne K. P., Wolfe K. H. 2005. The Yeast Gene Order Browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res. 15:1456–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen X. J. 1996. Low- and high-copy-number shuttle vectors for replication in the budding yeast Kluyveromyces lactis. Gene 172:131–136 [DOI] [PubMed] [Google Scholar]
- 7. Cockell M. M., Perrod S., Gasser S. M. 2000. Analysis of Sir2p domains required for rDNA and telomeric silencing in Saccharomyces cerevisiae. Genetics 154:1069–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Conant G. C., Wolfe K. H. 2008. Turning a hobby into a job: how duplicated genes find new functions. Nat. Rev. Genet. 9:938–950 [DOI] [PubMed] [Google Scholar]
- 9. Cuperus G., Shafaatian R., Shore D. 2000. Locus specificity determinants in the multifunctional yeast silencing protein Sir2. EMBO J. 19:2641–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. de Martino S., et al. 2000. Expression of sox11 gene duplicates in zebrafish suggests the reciprocal loss of ancestral gene expression patterns in development. Dev. Dyn. 217:279–292 [DOI] [PubMed] [Google Scholar]
- 11. Des Marais D. L., Rausher M. D. 2008. Escape from adaptive conflict after duplication in an anthocyanin pathway gene. Nature 454:762–765 [DOI] [PubMed] [Google Scholar]
- 12. Fairhead C., Dujon B. 2006. Structure of Kluyveromyces lactis subtelomeres: duplications and gene content. FEMS Yeast Res. 6:428–441 [DOI] [PubMed] [Google Scholar]
- 13. Force A., et al. 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151:1531–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Goldstein A. L., McCusker J. H. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15:1541–1553 [DOI] [PubMed] [Google Scholar]
- 14a. Gotta M., et al. 1997. Localization of Sir2p: the nucleolus as a compartment for silent information regulators. EMBO J. 16:3243–3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Greiss S., Gartner A. 2009. Sirtuin/Sir2 phylogeny, evolutionary considerations and structural conservation. Mol. Cell 28:407–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hahn M. W. 2009. Distinguishing among evolutionary models for the maintenance of gene duplicates. J. Hered. 100:605–617 [DOI] [PubMed] [Google Scholar]
- 17. Hickman M. A., Rusche L. N. 2007. Substitution as a mechanism for genetic robustness: the duplicated deacetylases Hst1p and Sir2p in Saccharomyces cerevisiae. PLoS Genet. 3:e126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hickman M. A., Rusche L. N. 2009. The Sir2-Sum1 complex represses transcription using both promoter-specific and long-range mechanisms to regulate cell identity and sexual cycle in the yeast Kluyveromyces lactis. PLoS Genet. 5:e1000710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hickman M. A., Rusche L. N. 2010. Transcriptional silencing functions of the yeast protein Orc1/Sir3 subfunctionalized after gene duplication. Proc. Natl. Acad. Sci. U. S. A. 107:19384–19389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hittinger C. T., Carroll S. B. 2007. Gene duplication and the adaptive evolution of a classic genetic switch. Nature 449:677–681 [DOI] [PubMed] [Google Scholar]
- 21. Hoppe G. J., et al. 2002. Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation. Mol. Cell. Biol. 22:4167–4180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hughes A. L. 2005. Gene duplication and the origin of novel proteins. Proc. Natl. Acad. Sci. U. S. A. 102:8791–8792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Imai S., Guarente L. 2010. Ten years of NAD-dependent SIR2 family deacetylases: implications for metabolic diseases. Trends Pharmacol. Sci. 31:212–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jarinova O., et al. 2008. Functional resolution of duplicated hoxb5 genes in teleosts. Development 135:3543–3553 [DOI] [PubMed] [Google Scholar]
- 25. Kobayashi T., Horiuchi T., Tongaonkar P., Vu L., Nomura M. 2004. SIR2 regulates recombination between different rDNA repeats, but not recombination within individual rRNA genes in yeast. Cell 117:441–453 [DOI] [PubMed] [Google Scholar]
- 26. Lu S. P., Lin S. J. 2010. Regulation of yeast sirtuins by NAD(+) metabolism and calorie restriction. Biochim. Biophys. Acta 1804:1567–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Luo K., Vega-Palas M. A., Grunstein M. 2002. Rap1-Sir4 binding independent of other Sir, yKu, or histone interactions initiates the assembly of telomeric heterochromatin in yeast. Genes Dev. 16:1528–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lynch M., Katju V. 2004. The altered evolutionary trajectories of gene duplicates. Trends Genet. 20:544–549 [DOI] [PubMed] [Google Scholar]
- 29. McCord R., et al. 2003. Rfm1, a novel tethering factor required to recruit the Hst1 histone deacetylase for repression of middle sporulation genes. Mol. Cell. Biol. 23:2009–2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mead J., et al. 2007. Swapping the gene-specific and regional silencing specificities of the Hst1 and Sir2 histone deacetylases. Mol. Cell. Biol. 27:2466–2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Reference deleted.
- 32. Navratilova P., Fredman D., Lenhard B., Becker T. S. 2010. Regulatory divergence of the duplicated chromosomal loci sox11a/b by subpartitioning and sequence evolution of enhancers in zebrafish. Mol. Genet. Genomics 283:171–184 [DOI] [PubMed] [Google Scholar]
- 33. North B. J., Verdin E. 2004. Sirtuins: Sir2-related NAD-dependent protein deacetylases. Genome Biol. 5:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ozaydin B., Rine J. 2010. Expanded roles of the origin recognition complex in the architecture and function of silenced chromatin in Saccharomyces cerevisiae. Mol. Cell. Biol. 30:626–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rusché L. N., Kirchmaier A. L., Rine J. 2002. Ordered nucleation and spreading of silenced chromatin in Saccharomyces cerevisiae. Mol. Biol. Cell 13:2207–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rusché L. N., Kirchmaier A. L., Rine J. 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72:481–516 [DOI] [PubMed] [Google Scholar]
- 37. Rusché L. N., Rine J. 2001. Conversion of a gene-specific repressor to a regional silencer. Genes Dev. 15:955–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Scannell D. R., Wolfe K. H. 2008. A burst of protein sequence evolution and a prolonged period of asymmetric evolution follow gene duplication in yeast. Genome Res. 18:137–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schmitt M. E., Brown T. A., Trumpower B. L. 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 18:3091–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Reference deleted.
- 41. Stoltzfus A. 1999. On the possibility of constructive neutral evolution. J. Mol. Evol. 49:169–181 [DOI] [PubMed] [Google Scholar]
- 42. Straight A. F., et al. 1999. Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell 97:245–256 [DOI] [PubMed] [Google Scholar]
- 43. van Hoof A. 2005. Conserved functions of yeast genes support the duplication, degeneration and complementation model for gene duplication. Genetics 171:1455–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wolfe K. H., Shields D. C. 1997. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387:708–713 [DOI] [PubMed] [Google Scholar]
- 45. Xie J., et al. 1999. Sum1 and Hst1 repress middle sporulation-specific gene expression during mitosis in Saccharomyces cerevisiae. EMBO J. 18:6448–6454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang T., Kraus W. L. 2010. SIRT1-dependent regulation of chromatin and transcription: linking NAD(+) metabolism and signaling to the control of cellular functions. Biochim. Biophys. Acta 1804:1666–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zill O. A., Rine J. 2008. Interspecies variation reveals a conserved repressor of alpha-specific genes in Saccharomyces yeasts. Genes Dev. 22:1704–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]