Abstract
Disruption of the BRCA1 tumor suppressor can be caused not only by inherited mutations in familial cancers but also by BRCA1 gene silencing in sporadic cancers. Hypoxia, a key feature of the tumor microenvironment, has been shown to downregulate BRCA1 at the transcriptional level via repressive E2F4/p130 complexes. Here we showed that hypoxia also drives epigenetic modification of the BRCA1 promoter, with decreased H3K4 methylation as a key repressive modification produced by the lysine-specific histone demethylase LSD1. We also observed increased H3K9 methylation coupled with decreased H3K9 acetylation. Similar modifications were seen in the RAD51 promoter, which is also downregulated by hypoxia, whereas exactly opposite changes were seen in the promoter of the hypoxia-inducible gene VEGF. In cells containing the BRCA1 promoter driving a selectable HPRT gene, long-term silencing of the promoter was observed following exposure to hypoxic stress. Clones with silenced BRCA1 promoters were detected at frequencies of 2% or more following hypoxia, but at less than 6 × 10−5 without hypoxia. The silenced clones showed decreased H3K4 methylation and decreased H3K9 acetylation in the BRCA1 promoters, consistent with the acute effects of hypoxic stress. Hypoxia-induced BRCA1 promoter silencing persisted in subsequent normoxic conditions but could be reversed by treatment with a histone deacetylase (HDAC) inhibitor but not with a DNA methylation inhibitor. Interestingly, treatment of cells with inhibitors of poly(ADP-ribose) polymerase (PARP) can cause short-term repression of BRCA1 expression, but such treatment does not produce H3K4 or H3K9 histone modification or BRCA1 promoter silencing. These results suggest that hypoxia is a driving force for long-term silencing of BRCA1, thereby promoting genome instability and tumor progression.
INTRODUCTION
Solid tumors constitute a unique tissue type, characterized by hypoxia, low pH, and nutrient deprivation. Previous work has shown that hypoxic stress is a source of genetic instability in tumors (3, 5, 7, 40, 58), causing increased point mutations (40), gene amplification (11, 58), and fragile-site induction (12). Our previous work revealed that BRCA1 and RAD51, key genes in the homology-dependent repair (HDR) pathway, and MLH1, a key DNA mismatch (MMR) repair gene, are downregulated at the mRNA and protein levels in response to hypoxia via specific pathways of transcriptional regulation (3–5, 7). Moreover, BRCA1 and MLH1 have been found to be silenced in many sporadic cancers of multiple sites (8, 14, 16). The silencing of BRCA1 and MLH1 has been attributed primarily to promoter DNA hypermethylation at CpG sites (14). However, recent studies suggest that silenced promoters in cancer cells are also marked by characteristic histone modifications (9, 33, 48), and evidence is emerging that histone methylation may be a mediator of silencing that is independent of DNA methylation (26, 29, 30).
Posttranslational modification of histones is widely recognized as an important epigenetic mechanism in the organization of chromosomal domains and in gene regulation (31, 32, 36, 39). Methylation of lysine 4 and acetylation of lysine 9 of H3 have been associated with regions of active transcription, whereas methylation of H3K9 and methylation of H3K27 are generally associated with gene repression (31, 32, 36, 39, 48). Hypoxia-induced histone modifications have recently been reported, and these can be found in both hypoxia-activated and hypoxia-repressed genes (20). The regulation of gene expression by hypoxia through covalent modification of histones is also supported by evidence that histone deacetylase (HDAC) activity plays a role in the activation of many hypoxia-inducible factor 1 (HIF-1)-responsive genes (22). In addition, certain histone demethylases and histone methyltransferases, including JMJD1A, JMJD2B, JARID1B, and G9a histone methyltransferase, have been identified as hypoxia- or HIF-1-regulated genes (2, 9, 27, 29, 53, 56).
In previous work, we found that hypoxia causes decreased expression of BRCA1 and RAD51 at the mRNA and protein levels, and we demonstrated that this repression is mediated, in part, by hypoxia-induced dephosphorylation and nuclear accumulation of p130, one of the retinoblastoma (Rb)-related pocket proteins, leading to the formation of repressive E2F4/p130 complexes and increased binding of these complexes to the BRCA1 and RAD51 promoters (3, 6). We also demonstrated that downregulation of these factors is linked to decreased DNA repair capacity, establishing a mechanism by which hypoxia can drive genetic instability in cancer cells (5, 7).
Since hypoxia occurs early in neoplastic growth and is a common feature of solid tumors, we asked in this work whether hypoxia might also be a driving force in the silencing of the BRCA1 promoter. Recent evidence has shown that silenced BRCA1 alleles found in sporadic cancers are associated not only with promoter DNA hypermethylation (14, 35, 50) but also with histone modifications in the promoter region (19). Hence, we hypothesized that hypoxia-induced downregulation of BRCA1 might cause epigenetic histone modifications that mark the locus for potential silencing. We report here a series of chromatin immunoprecipitation (ChIP) studies revealing that hypoxia induces a set of repressive histone modifications at both the BRCA1 and RAD51 promoters, including H3K4 demethylation, H3K9 methylation, and H3K9 deacetylation. In contrast, exactly opposite changes were seen in the histone code at the promoter of the hypoxia-inducible VEGF gene. We showed that a key histone modification at the BRCA1 and RAD51 promoters in response to hypoxia, H3K4 demethylation, is mediated by the histone demethylase LSD1. We also showed that prolonged exposure of cells to hypoxia can promote the emergence of subclones in which the BRCA1 promoter has undergone long-term silencing that persists even when cells are no longer in hypoxic conditions. In these silenced clones, the BRCA1 promoter is characterized by H3K4 demethylation and H3K9 deacetylation. Such histone changes are mechanistically linked to the silencing because exposure of cells to an HDAC inhibitor after hypoxia reverses the BRCA1 silencing. This work suggests that hypoxia drives not only genetic instability but also epigenetic alteration and tumor suppressor gene silencing in malignant cells.
MATERIALS AND METHODS
Cells.
MCF-7, A549, RKO, and HCC 38 cells were obtained from the American Type Culture Collection (Manassas, VA) and grown according to supplier instructions. MCF-7 PLU-1 knockdown cell lines, MCF-7 985, MCF-7 1000, and MCF-7 1100, with control MCF-7 SLR cells, were obtained from Qin Yan (Department of Pathology, Yale University) and cultured in RPMI 1640 supplemented with 10% fetal bovine serum (FBS) and 1.5 μg/ml of puromycin. RKO Neo and RKO E7 cells were obtained from Kathleen Cho (Department of Pathology, University of Michigan, Ann Arbor, MI) and cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS and 400 μg/ml of G418. Dif-6-derived cells (described below) were cultured in DMEM supplemented with 5% FBS and 5% Serum Plus (SABC Bioscience). BSH8 cells were grown in 10 μg/ml of azaserine (Sigma), 10 μg/ml of hypoxanthine (Sigma), and 1.5 μg/ml of puromycin (Invitrogen). BSH8 TGH22 and TGH31 cells were grown in 2.5 μg/ml of 6-thioguanine (6-TG; Sigma) and 1.5 μg/ml of puromycin. BSH8 2XTGH22 and 2XTGH31 cells were grown in 20 μg/ml of 6-TG and 1.5 μg/ml of puromycin.
Constructs.
Lentivirus vectors for delivery of short hairpin RNAs (shRNAs) directed against JARID1A/RBP2-sh-1 and RBP2-sh-3 and control vector LLP were obtained from Marie Classon (Massachusetts General Hospital Cancer Center). Lentivirus shRNA vectors for LSD1 knockdown were obtained from Sigma-Aldrich (LSD1-1, catalogue number TRCN0000046068; LSD1-2, catalogue number TRCN0000046071; LSD1-3, catalogue number TRCN0000046072). The lentivirus shRNA vector for HIF-1α knockdown was obtained from Zhong Yun (Department of Therapeutic Radiology, Yale University). The BRCA1-HPRT construct was made by cloning 1.3-kb full-length human hypoxanthine phosphoribosyltransferase (HPRT) cDNA (OriGene) downstream of the 218-bp human BRCA1 promoter, which has been described previously (3).
Hypoxia.
For hypoxia exposure, cells received a continuous flow of a humidified mixture of 95% N2 and 5% CO2 gas certified to <10 ppm O2 for 48 h at 37°C, as previously described (40).
ChIP assays.
ChIP assays were done essentially as described previously (3, 6). The primer sequences for the BRCA1 and RAD51 promoter regions have also been reported (3, 6). All the antibodies for the ChIP assays are available on request.
Assays for cells with BRCA1 promoter silencing.
Dif-6 cells were transfected by electroporation as described previously (47) with 10 μg of BRCA1-HPRT plasmid and 2 μg of a plasmid carrying puromycin resistance (Clontech). BSH8, a stable transfectant expressing functional HPRT, was established by selection first with medium containing 1.5 μg/ml of puromycin and 1 week later with medium containing 10 μg/ml of azaserine and 10 μg/ml of hypoxanthine. The BSH8 clone was screened for physical linkage between puromycin resistance and BRCA1-HPRT by demonstrating low-frequency loss of HPRT expression in the presence of puromycin and 2.5 μg/ml of 6-TG but high-frequency loss when puromycin selection was omitted.
To test the impact of hypoxia on silencing of the BRCA1 promoter in the BSH8 cells, the cells were plated in 100-mm dishes at 100,000 cells per dish and grown under 0.5% hypoxic conditions (or under normoxic conditions for the controls) for 2 weeks with passage twice per week. The cell population was then subject to selection for two additional weeks in the presence of 2.5 μg/ml of 6-TG. This selection induced substantial cell death, and clones formed by surviving cells were counted for each condition tested. The TGH22 and TGH31 cell lines were isolated from the BSH8 cells as part of this process.
To measure BRCA1-HPRT reactivation by trichostatin A (TSA) or 5-aza-2′-deoxycytidine (5-Aza-dC), we incubated selected clones with silenced BRCA1 promoters, BSH8 THG22, BSH 2XTGH22, and BSH8 TGH31, in medium containing 100 nM TSA (Wako, Richmond, VA) for 16 h to inhibit histone deacetylation or with medium containing 300 nM 5-aza-dC (Sigma) for 16 h to inhibit DNA methylation. After an additional 24 h, the cells were plated in 100-mm dishes (100,000 cells per dish) and the next day were exposed to medium containing 10 μg/ml of azaserine and 10 μg/ml of hypoxanthine to select for cells that had regained HPRT expression. Silencing or reactivation frequencies were calculated by dividing the number of clones growing under selection by the effective number of cells plated (as determined with the cloning efficiency plates).
To test the impact of TSA on initial BRCA1 promoter silencing by hypoxia, we plated BSH8 cells in 100-mm dishes at 100,000 cells per dish and grew them under 0.5% oxygen hypoxic conditions for 2 weeks (as described above). The cells were treated with 100 nM TSA for 24 h when first removed from hypoxia. Then the cell population was selected in the presence of 2.5 μg/ml of 6-TG for an additional 2 weeks under normoxic conditions.
To test for silencing of the BRCA1 promoter by the poly(ADP-ribose) polymerase (PARP) inhibitor, 6(5H)-phenanthridinone (PHEN), we plated BSH8 cells in 100-mm dishes at 100,000 cells per dish and treated them with selected concentrations of PHEN for 2 weeks. Then the cells were selected in the presence of 2.5 μg/ml of 6-TG for two additional weeks. Cells without selection were assayed as controls for cloning efficiency.
Western blotting.
After the designated treatments, cells were lysed in RIPA buffer (25 mM Tris-HCl [pH 7.6], 150 mM NaCl, 1% Igepal CA-630, 1% sodium deoxycholate, 0.1% SDS) with protease inhibitor cocktail (Clontech). The primary antibodies used for Western blotting were as follows: monoclonal anti-BRCA1 (D-9; Santa Cruz Biotechnology), β-actin (C4; Santa Cruz Biotechnology), anti-JARID1B/PLU-1 (ab50958; Abcam), anti-Rad51 (3C10; Upstate), anti-JARID1A/RBP2 (catalogue number 3867; Cell Signaling), and anti-LSD1 (catalogue number 4218s; Cell Signaling).
Quantitative real-time PCR analysis.
For quantitative PCR analysis of BRCA1 and RAD51 mRNA expression, total RNA was prepared using an Absolutely RNA miniprep kit (Agilent Technologies). Two micrograms of RNA was used to synthesize cDNA using a High Capacity cDNA reverse transcription kit (Applied Biosystems). The resulting cDNA was used in PCRs containing TaqMan universal PCR master mix (Applied Biosystems), premixed TaqMan probes and primers for BRCA1 and RAD51, and 18S (Applied Biosystems). The Mx3000P real-time PCR system (Stratagene) was used to monitor fluorescence intensity in real time to allow quantitative comparisons, as described previously (3, 6).
For quantitative PCR analysis of BRCA1 promoter-driven HPRT mRNA expression, total RNA was isolated from cell culture with an RNeasy minikit (Qiagen) according to the manufacturer's instructions. Total RNA samples were converted to cDNA using a QuantiTect reverse transcription kit (Qiagen) with removal of genomic DNA contamination. One hundred nanograms of cDNA was used as input in a subsequent quantitative-PCR analysis for either HPRT (Applied Biosystems) or GAPDH (Applied Biosystems) with iQ supermix (Bio-Rad) and a Bio-Rad iCycler instrument. HPRT results were normalized in relation to GAPDH mRNA levels.
RESULTS
Hypoxia decreases H3K4 methylation at the BRCA1 and RAD51 promoters.
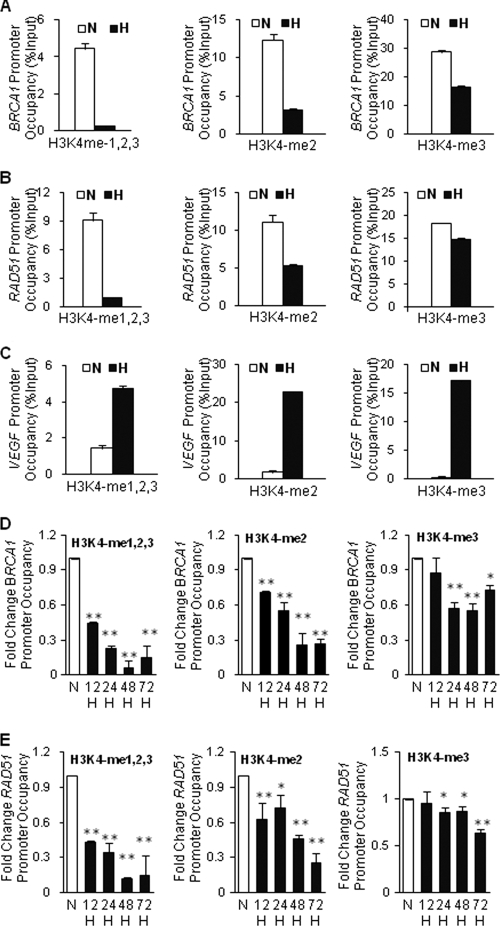
To test the hypothesis that downregulation of BRCA1 and RAD51 during hypoxia is linked to histone modifications, we used the technique of quantitative chromatin immunoprecipitation (qChIP) to probe histone changes at BRCA1 and RAD51 promoters during hypoxia. We first examined histone H3K4 methylation levels at the promoters because the methylation status of this residue is a key histone marker that is associated with increased transcriptional activity. MCF-7 breast cancer cells were exposed or not exposed to hypoxia at 0.01% oxygen for 48 h, and chromatin was prepared for analysis. ChIP analysis was performed using either an antibody that simultaneously recognizes the mono-, di-, and trimethylated forms of H3K4 (H3K4me1,2,3) or antibodies that individually recognize either H3K4me1, H3K4me2, or H3K4me3. Hypoxia was found to cause substantial decreases in the levels of H3K4 me1,2,3 at the BRCA1 (Fig. 1A) and RAD51 (Fig. 1B) promoters. Similarly, the levels of H3K4me2 and H3K4me3 were also decreased at these promoters under hypoxic conditions (Fig. 1A and B). Representative agarose gel images corresponding to panels A and B are available on request. H3K4me1 levels were low preexposure and remained low under hypoxic conditions (data not shown). Decreased H3K4 methylation levels were also induced by hypoxia in other cell lines tested, including RKO colon cancer cells (data available on request). In contrast, at the promoter of the hypoxia-induced VEGF gene, levels of all forms of H3K4 methylation, H3K4me1,2,3, H3K4me2, and H3K4me3, were increased by hypoxia in both MCF-7 (Fig. 1C; also data available on request) and RKO (data available on request) cells. We also conducted a time course experiment in which we placed MCF-7 cells under hypoxia and then collected the cells 12, 24, 48, and 72 h later for analysis by ChIP of H3K4 methylation at the BRCA1 and RAD51 promoters (Fig. 1D and E). In both cases, promoter H3K4 demethylation occurred by 12 h and persisted through 72 h, although there was a slight uptick in H3K4 methylation at the 72-h time point. Collectively, these results demonstrate that hypoxic stress causes decreased H3K4 methylation levels at the BRCA1 and RAD51 promoters.
Fig. 1.
Decreased H3K4 methylation levels at the BRCA1 and RAD51 promoters in response to hypoxia in MCF-7 cells. ChIP assays were performed on MCF-7 cells following exposure to normoxia (N; white bars) or to hypoxia at 0.01% oxygen (H; black bars) for various times, as indicated. Cells were then collected for ChIP analyses using antibodies to the indicated H3K4 methylation forms to determine H3K4 methylation levels at the BRCA1, RAD51, and VEGF promoters. (A) ChIP analysis of H3K4 methylation levels at the BRCA1 promoter after 48 h of normoxia or hypoxia as quantified by real-time PCR. Specific antibodies that either simultaneously recognize the mono-, di-, and trimethylation forms of H3K4 (H3K4me1,2,3) or that individually recognize either the dimethylated form of H3K4 (H3K4me2) or the trimethylated form of H3K4 (H3K4me3) were used. Relative promoter occupancies (% input) are shown with error bars based on standard errors (SEs) calculated from at least three replicates. The input signal is set as 100% (not depicted in graphs) for each set of assays. (B) ChIP analysis of H3K4 methylation levels at the RAD51 promoter after 48 h of N or H as quantified by real-time PCR. Relative promoter occupancies (% input) by the indicated H3K4 methylated forms are shown with error bars based on SEs calculated from at least three replicates. (C) ChIP analysis of H3K4 methylation levels at the VEGF promoter after 48 h of N or H as quantified by real-time PCR. Relative promoter occupancies (% input) by the indi- cated H3K4 methylation forms are shown with error bars based on SEs calculated from at least three replicates. Representative agarose gel images corresponding to panels A, B, and C are shown in data available on request. (D) Time course assay of H3K4 methylation at the BRCA1 promoter in MCF-7 cells placed under hypoxia and collected at the indicated times for analysis by ChIP and quantification by real-time PCR. Promoter occupancy levels are expressed as the fold change relative to those for normoxia, based on three independent ChIP assays, with error bars based on SEs. Significant differences were identified as P < 0.05 (indicated by *) or P < 0.01 (indicated by **) compared to normoxic levels. (E) Time course assay of H3K4 methylation at the RAD51 promoter in MCF-7 cells placed under hypoxia and collected at the indicated times for analysis by ChIP and quantification by real-time PCR, as for panel D.
The lysine-specific demethylase LSD1, but not JARID1B/PLU-1 or JARID1A/RBP2, is required for hypoxia-induced H3K4 demethylation at the BRCA1 and RAD51 promoters.
The identification of specific histone demethylases has changed the understanding of histone methylation, which had been considered a static modification but is now understood to be dynamically regulated (10, 25). Several H3K4-specific demethylases have been identified, including LSD1 (42), JARID1A/RBP2 (24), and JARID1B/PLU-1 (55). We sought to determine whether one or more of the above H3K4 demethylases might be involved in hypoxia-decreased H3K4 methylation levels at the BRCA1 and RAD51 promoters.
We first examined the changes in protein levels of LSD1, RBP2, and PLU-1 in response to hypoxia treatment. We found that there was no change in the levels of LSD1, whereas RBP levels were decreased and PLU-1 levels were increased in MCF-7, RKO, and A549 cells (data available on request). Hypoxia-induced upregulation of PLU-1 had also been seen in another study (27).
Based on its upregulation in response to hypoxia, PLU-1 therefore seemed to be a good candidate for regulation of BRCA1 and RAD51 in hypoxia. To test the role of PLU-1 in hypoxia-decreased BRCA1 and RAD51 expression, we used stable shRNA-mediated knockdown of PLU-1 in MCF-7 cell lines. Three cell line subclones with shRNA knockdown of PLU-1 were established (MCF-7 985, MCF-7 1000, and MCF-7 1100) along with vector control cells (MCF-7 SLR). Reduction in the baseline expression of PLU-1 in normoxia and attenuation of PLU-1 induction in hypoxia were seen in all three shRNA-expressing lines compared to those of the vector control cells (data available on request). Using these engineered MCF-7-derived cell lines, we found that knockdown of PLU-1 had no effect on hypoxia-mediated suppression of BRCA1 and RAD51 protein levels (data available on request). Nonetheless, we also examined BRCA1 and RAD51 mRNA levels in response to hypoxia in MCF-7 SLR and MCF-7 1000 cells, the latter chosen because they showed the most robust knockdown of PLU-1. We found that knockdown of PLU-1 in the MCF-7 1000 cell line yielded increased expression of BRCA1 mRNA under normoxic conditions relative to that of the vector control cells (data available on request), consistent with a prior report (55). However, hypoxia-induced downregulation of BRCA1 mRNA was still seen in the MCF-7 1000 cell line in spite of the PLU-1 knockdown (data available on request). In like fashion, RAD51 mRNA expression levels were downregulated in hypoxia in both the MCF-7 SLR and MCF-7 1000 cells (data available on request). Turning to chromatin analysis, we found that there was no significant difference in hypoxia-induced H3K4 demethylation at the BRCA1 and RAD51 promoters between the vector control cells and the MCF-7 1000 cells with PLU-1 knockdown (data available on request), further ruling out a role for PLU-1 in this process. Similar negative results were also seen in cells with shRNA-mediated knockdown of JARID1A/RBP2 (data available on request).
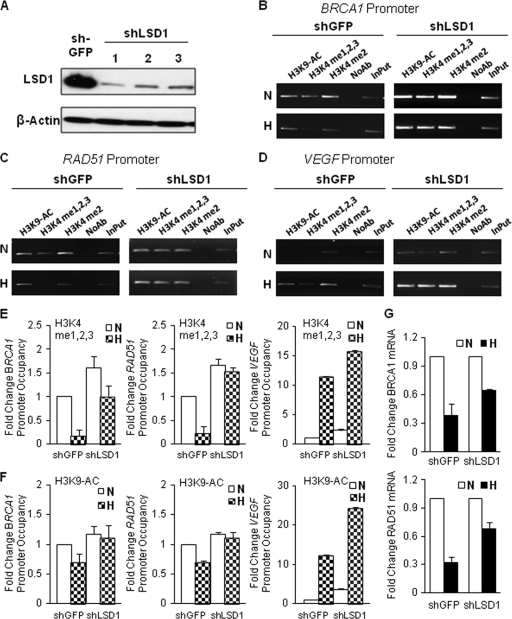
Turning to LSD1, we established three MCF-7-derived cell lines with stable shRNA-mediated knockdown of LSD1 (Fig. 2A). As a control, a cell line was established with shRNA-targeting green fluorescent protein (GFP), which had no effect on LSD1 levels (Fig. 2A). We chose one LSD1 knockdown line, MCF-7 shLSD1-1, for further analysis. These cells were exposed or not exposed to hypoxia for 48 h and analyzed for H3K4 methylation status in comparison to that of the shGFP-expressing control cells. We found that knockdown of LSD1 (Fig. 2A) clearly prevented the hypoxia-induced demethylation of H3K4 at both the BRCA1 and RAD51 promoters (Fig. 2B and C; quantified in Fig. 2E). However, at the VEGF promoter, H3K4 methylation was still seen to increase in response to hypoxia, consistent with hypoxia-induced VEGF gene activation, which is not affected by LSD1 knockdown (Fig. 2D; quantified in Fig. 2E).
Fig. 2.
The histone demethylase LSD1 mediates hypoxia-induced H3K4 demethylation at the BRCA1 and RAD51 promoters. MCF-7 cells were transduced with lentiviral expression constructs for a GFP shRNA (control) or for one of three different shRNAs targeting LSD1, and stable shRNA-expressing cell lines were established: MCF-7 shGFP and MCF-7 shLSD1-1, MCF-7 shLSD1-2, and MCF-7 shLSD1-3. (A) Western blot analyses to determine LSD1 expression levels in MCF-7 shGFP, MCF-7 shLSD1-1, shLSD1-2, and shLSD1-3 cells. Substantially reduced expression of LSD1 was detected in all three stable LSD1 shRNA-expressing MCF-7 cell lines. (B, C, and D) ChIP analyses of H3K4 methylation and H3K9 acetylation levels at the BRCA1 (B), RAD51 (C), and VEGF (D) promoters following 48-h exposure to normoxia or hypoxia in MCF-7 shGFP cells compared to those for MCF-7 shLSD1-1 cells. Representative agarose gels showing PCR amplification products corresponding to the BRCA1 (B), RAD51 (C), or VEGF (D) promoter regions are shown. (E) Quantification of H3K4 methylation levels by real-time PCR at the BRCA1 (left), RAD51 (middle), and VEGF (right) promoters following 48-h exposure to normoxia or hypoxia in MCF-7 shGFP and MCF-7 shLSD1-1 cells. Promoter occupancy levels are expressed as the fold change relative to the promoter occupancy levels of the normoxic MCF-7 shGFP cells, based on three independent ChIP assays, with error bars based on SEs. (F) Quantification of H3K9 acetylation levels by real-time PCR at the BRCA1 (left), RAD51 (middle), and VEGF (right) promoters in MCF-7 shGFP and MCF-7 shLSD1-1 cells under the same normoxic or hypoxic treatment. Promoter occupancy levels are expressed as the fold change relative to the acetylation levels of the normoxic MCF-7 shGFP cells, based on three independent ChIP assays, with error bars based on SEs. (G) Quantitative real-time PCR analysis of endogenous BRCA1 (top) and RAD51 (bottom) mRNA expression levels in normoxic and hypoxic MCF-7 shGFP cells and in LSD1 knockdown MCF-7 shLSD1-1 cells, normalized to 18S rRNA expression. mRNA levels are expressed as the fold change relative to those of the corresponding normoxic control cells.
Interestingly, LSD1 knockdown also blocked the reduction in H3K9 acetylation that otherwise occurs at the BRCA1 and RAD51 promoters in response to hypoxia (Fig. 2B and C; quantified in Fig. 2F), consistent with cross talk between the H3K4 and H3K9 modifications. LSD1 knockdown again had no effect at the VEGF promoter with respect to the dynamics of H3K9 acetylation.
We next examined the changes in BRCA1 and RAD51 mRNA levels in response to hypoxia with or without LSD1 knockdown, using the same set of cell lines as above (MCF-7 shGFP and MCF-7 shLSD1). After the cells were exposed or not exposed to hypoxia for 48 h, mRNA levels for BRCA1 and RAD51 were evaluated by quantitative real-time reverse transcription (RT)-PCR (Fig. 2G). LSD1 knockdown blocked about half of the reduction in mRNA levels for these factors that otherwise occurs in response to hypoxia, again consistent with a key role for LSD1 and H3K4 promoter methylation status in the regulation of BRCA1 and RAD51 in hypoxia. Together, these results identify LSD1 as the key histone demethylase that mediates epigenetic regulation of BRCA1 and RAD51 in response to hypoxia.
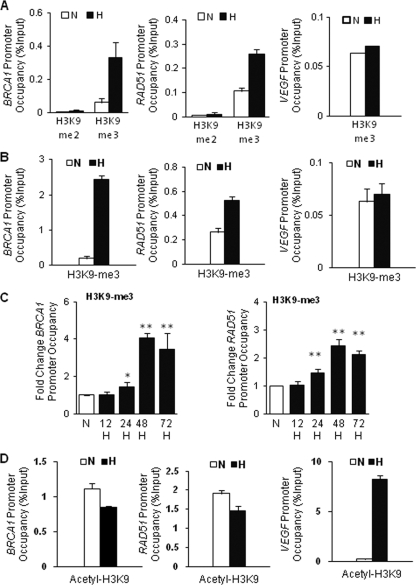
Hypoxia increases H3K9 methylation and decreases H3K9 acetylation at the BRCA1 and RAD51 promoters.
H3K9 methylation is a critical epigenetic mark for gene repression and silencing (13, 17). It has been reported that hypoxia induces H3K9 methylation at different gene promoters, which is correlated with hypoxic repression and silencing of those genes (9, 20). For this reason, we examined the effect of hypoxia on H3K9 methylation levels at the BRCA1 and RAD51 promoters. There were significant increases in H3K9 me3 levels at both the BRCA1 and RAD51 promoters in MCF-7 cells in response to hypoxia (Fig. 3A). Similar results were found in RKO cells (Fig. 3B). Conversely, there was no increase in H3K9me3 levels at the VEGF promoter in either cell line (Fig. 3A and B). We also examined the kinetics of H3K9 methylation at the BRCA1 and RAD51 promoters over time in response to hypoxic stress (Fig. 3C). We observed increased methylation beginning at about 24 h and peaking around 48 h, a slightly slower response than we observed for demethylation of H3K4 (Fig. 1D and E), suggesting that H3K4 demethylation is an earlier modification that may be upstream of H3K9 methylation at these promoters under hypoxic stress.
Fig. 3.
Hypoxia increases H3K9 methylation and decreases H3K9 acetylation at the BRCA1 and RAD51 promoters. ChIP assays were performed on MCF-7 or RKO cells following exposure to normoxia (N) or hypoxia (H; 0.01% O2) for 48 h using antibodies to the indicated H3K9 modifications at the BRCA1, RAD51, and VEGF promoters. (A) ChIP analysis of H3K9 methylation levels at the BRCA1 (left), RAD51 (middle), and VEGF (right) promoters as quantified by real-time PCR in MCF-7 cells. Specific antibodies that recognize either the dimethylated (H3K9me2) or trimethylated (H3K9me3) form of H3K9 were used. Relative promoter occupancies (% input) are shown with error bars based on standard errors calculated from at least three replicates. (B) ChIP analysis of H3K9 methylation levels at the BRCA1 (left), RAD51 (middle), and VEGF (right) promoters as quantified by real-time PCR in RKO cells. Specific antibodies that recognize H3K9me3 were used. Relative promoter occupancies (% input) by trimethylated H3K9 are shown with error bars based on standard errors calculated from at least three replicates. (C) Time course of hypoxia-induced H3K9 methylation at the BRCA1 (left) or RAD51 (right) promoter regions by ChIP analysis. MCF-7 cells were exposed to normoxia (N) or hypoxia (H) for the indicated times, and H3K9 methylation levels at the promoters were analyzed by quantification of real-time PCR of ChIP samples corresponding to the BRCA1 or RAD51 promoters. Promoter occupancy levels were expressed as the fold change relative to promoter occupancy levels for normoxia, based on three independent ChIP assays with error bars based on SEs. Significant differences were identified as P < 0.05 (indicated by *) or P < 0.01 (indicated by **) compared to normoxia control levels. (D) ChIP analysis of H3K9 acetylation levels at the BRCA1 (left), RAD51 (middle), and VEGF (right) promoters as quantified by real-time PCR in MCF-7 cells. Relative promoter occupancies (% input) by acetylated H3K9 are shown with error bars based on SEs calculated from at least three replicates.
We next tested for the putative role of specific H3K9 methyl-transferases, including G9a, SUV39-h, and SEDTB1 (28), by looking for interactions with either the BRCA1 or RAD51 promoters in response to hypoxia by ChIP. We could not detect any of these factors in association with either promoter (data not shown), and so the factor mediating the increased H3K9 methylation remains to be determined.
H3K9 acetylation (H3K9ac) is a marker of transcriptional activation. Following up the initial observations presented in Fig. 2F, we went on to confirm in multiple experiments that hypoxia produces slight decreases in H3K9 acetylation levels at both the BRCA1 and RAD51 promoters (Fig. 3D), whereas it substantially increases H3K9 acetylation at the VEGF promoter in MCF-7 (Fig. 3D). Similar results were obtained in RKO cells (data not shown).
H3K27 methylation is another mark of transcriptional repression, and so we analyzed the BRCA1 and RAD51 promoters in MCF-7 cells for this modification under normoxic and hypoxic conditions. However, there were no changes in either H3K27me1, H3K27me2, or H3K27me3 levels at the BRCA1 or RAD51 promoters under hypoxic conditions compared to those under normoxic conditions (data not shown). The Polycomb protein and histone methylase EZH2 is associated with H3K27 methylation. Consistent with the above results, we could not detect by ChIP any evidence for the presence of EZH2 at either the BRCA1 or RAD51 promoter in either normoxia or hypoxia (data available on request).
HIF-1 is not required for the H3K4 demethylation at the BRCA1 and RAD51 promoters.
In prior work, we have shown that the downregulation of RAD51 and BRCA1 at the transcriptional level in response to hypoxia is not dependent on the hypoxia-inducible factor HIF-1 (3, 6). Nonetheless, we still asked whether HIF-1 might be required for the histone modifications that we observed at the promoters in response to hypoxia. Using an shRNA expression vector to knock down expression of HIF-1α (data available on request), we found that H3K4 demethylation occurs at the BRCA1 and RAD51 promoters in response to hypoxia regardless of HIF-1 knockdown (data available on request). This result is consistent with the lack of a role for HIF-1 in regulating these factors at the transcriptional level. In contrast and in keeping with the known role of HIF-1 in stimulating VEGF expression, we observed that the hypoxia-induced increase in H3K4 methylation at the VEGF promoter was attenuated by HIF-1 knockdown (data available on request).
Hypoxia induces BRCA1 promoter silencing.
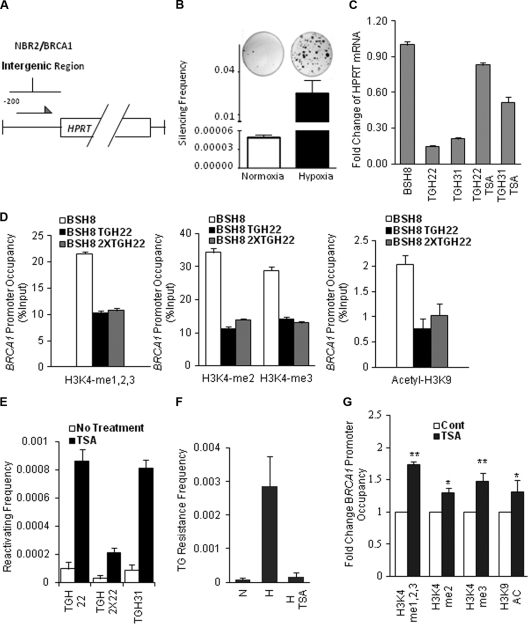
BRCA1 is frequently silenced in sporadic tumors of multiple sites (8, 14, 51, 52). To test whether hypoxia might play a role in the silencing of the BRCA1 promoter, we established a system to select for cells in which the BRCA1 promoter had been silenced. We engineered a construct with the 218-bp BRCA1 proximal promoter (essentially the entire NBR2-BRCA1 intergenic region) driving expression of the human HPRT cDNA. This construct (Fig. 4A) was transfected into the mouse Dif-6 cell line, chosen because it lacks expression of endogenous hprt, thereby allowing both positive and negative selection for HPRT function. One stable transfectant cell line, designated BSH8, was found to express high baseline levels of HPRT. To test whether hypoxia could silence the BRCA1 promoter in this construct, the BSH8 cells were exposed to hypoxia for 2 weeks and then subjected to selection for HPRT-deficient cells in medium supplemented with the purine analog 6-TG. 6-TG is toxic to cells with functional HPRT. The size of the fraction of surviving 6-TG-resistant clones reflects the frequency at which the BRCA1 promoter-HPRT construct is inactivated. We found that hypoxia exposure led to the emergence of HPRT-negative clones at a frequency of more than 2%, 500-fold more than the background frequency arising in cells grown under standard normoxic conditions (Fig. 4B) and more than 1,000-fold greater than the typical spontaneous mutation frequencies in the HPRT coding region (49).
Fig. 4.
Hypoxia induces silencing of the BRCA1 promoter that is correlated with specific histone modifications. (A) Schematic of the BRCA1-HPRT construct that was used to express HPRT in Dif-6 cells, which lack endogenous HPRT expression. A subclone containing a stable, single-copy integrant of the BRCA1-HPRT expression construct was isolated and designated BSH8. (B) Frequency of 6-TG-resistant clones (indicative of silenced BRCA1-HPRT expression) following exposure of BSH8 cells to normoxia or hypoxia for 2 weeks and then selection in 6-TG for two additional weeks. (Top) Visualization of differential 6-TG-resistant colony formation following growth in normoxia or hypoxia and subsequent 6-TG selection. (Bottom) Quantification of the frequency of 6-TG-resistant clones arising after growth in normoxia or hypoxia. Error bars represent standard errors from three replicates. (C) Quantitative real-time PCR analysis of HPRT mRNA expression in two randomly selected 6-TG-resistant clones, BSH8 TGH22 and BSH8 TGH31, compared with HPRT mRNA expression in the parental cell line BSH8. The TGH22 and TGH31 cells were also treated with the HDAC inhibitor TSA and then analyzed for HPRT expression. HPRT mRNA levels were normalized to those for GAPDH and presented as the fold change relative to HPRT mRNA levels of the parental BSH8 cells. Error bars represent standard errors from three replicates. (D) ChIP analysis of histone modifications at the BRCA1-HPRT promoters in the 6-TG-resistant clones, BSH8 TGH22 and BSH8 2XTGH22. (Left) Relative promoter occupancies (% input) by all three forms of H3K4 methylation (H3K4me1,2,3) are shown with error bars based on standard errors calculated from three replicates. (Middle) Relative promoter occupancies (% input) by dimethylated H3K4 and trimethylated H3K4 are shown with error bars based on standard errors calculated from three replicates. (Right) Relative promoter occupancies (% input) by acetylated H3K9 are shown with error bars based on standard errors calculated from three replicates. (E) Reactivation of HPRT expression in BRCA1 promoter-silenced clones by TSA treatment. Three 6-TG-resistant clones, BSH8 TGH22, BSH8 2XTGH22, and BSH8 TGH31, were treated with TSA or not treated and were placed under selective conditions (in the presence of 10 μg/ml of azaserine and 10 μg/ml of hypoxanthine) to assay for reactivation of functional HPRT expression. The reactivation frequency represents the frequency of colonies surviving after 2 weeks of azaserine/hypoxanthine selection. (F) The HDAC inhibitor TSA blocks the emergence of silenced clones following hypoxia treatment of BSH8 cells. The cells were grown in hypoxia or normoxia for 2 weeks. After exposure to hypoxia, BSH8 cells were treated with 100 nM TSA or not treated for 24 h when first removed from hypoxia. The cell populations (normoxia for 2 weeks, hypoxia for 2 weeks, or hypoxia for 2 weeks plus TSA for 24 h) were then placed in normoxia under selective conditions in the presence of 2.5 μg/ml of thioguanine for an additional 2 weeks. The frequency of 6-TG-resistant clones was determined as for panel B. (G) TSA partially reverses certain histone modifications that were produced by hypoxia in the BRCA1 promoter-silenced clones. The 6-TG-resistant clone TGH22 was treated with TSA and placed under selection for functional HPRT expression and therefore for BRCA1 promoter reactivation in the presence of 10 μg/ml of azaserine and 10 μg/ml of hypoxanthine. ChIP analyses were performed to examine histone modifications at the BRCA1-HPRT promoters in the silenced TGH22 cells compared to those in the TGH22-derived cells with reactivation of the BRCA1 promoter following TSA treatment. Promoter occupancy is expressed as the fold change relative to histone modifications in the TGH22 cells, based on three independent ChIP experiments, with error bars based on standard errors. Significant differences compared to control levels were identified as P < 0.05 (indicated by *) or P < 0.01 (indicated by **).
Several independent 6-TG-resistant clones were isolated for further analysis. These showed greatly reduced levels of HPRT mRNA expression (Fig. 4C; compare TGH22 and TGH31 to BSH8), consistent with silencing of the BRCA1 promoter in the construct. The silencing was demonstrated to be durable, since silenced clones maintained in normoxic conditions for up to 8 weeks after hypoxic exposure retained the silenced state (data not shown). However, we found that the silenced promoters could be reactivated by treatment with the HDAC inhibitor TSA (Fig. 4C), pointing to epigenetic silencing via histone modifications.
Hypoxia-induced BRCA1 promoter silencing is correlated with specific histone modifications.
Since we showed above that short-term hypoxia causes decreased H3K4 methylation and H3K9 acetylation as well as increased H3K9 methylation at the BRCA1 and RAD51 promoters in MCF-7 and RKO cells, we hypothesized that the BRCA1 promoter silencing in the BRCA1-HPRT system was likely the result of similar or related histone modifications. To test this, we compared histone marks at the BRCA1 promoter within the BRCA1-HPRT construct in the parental BSH8 cells with those of two 6-TG-resistant subclones, BSH8 TGH22 and BSH8 2XTGH22. qChIP analysis revealed markedly reduced H3K4 methylation at the BRCA1-HPRT promoter in both the BSH8 TGH22 and BSH8 2XTGH22 cell lines compared to that of the parental BSH8 cell line that highly expresses HPRT (Fig. 4D). The level of H3K9 acetylation in the promoter was also decreased in both of the silenced subclones (Fig. 4D). Similar results were found in two other silenced subclones (BSH8 TGH31 and BSH8 2XTGH31; data not shown). The H3K4 demethylation and the H3K9 deacetylation seen in the silenced clones are in keeping with the effects of short-term hypoxia on the endogenous BRCA1 promoter in the MCF-7 cells, suggesting that these modifications play key roles in both BRCA1 suppression and BRCA1 silencing.
Consistent with histone modifications at the BRCA1 promoter in silenced clones, we found that treatment of the silenced clones with the HDAC inhibitor TSA could produce reactivation of functional HPRT expression following treatment, manifested both by elevated levels of HPRT mRNA (Fig. 4C) and by the generation of azaserine- and hypoxanthine-resistant subclones, indicative of reactivated BRCA1 promoter-HPRT expression constructs (Fig. 4E). These results further indicated that inactivation and silencing of the BRCA1 promoter-HPRT cDNA constructs in response to hypoxia was due to durable, but still pharmacologically reversible, silencing of the promoter. We also found that treatment of the BSH8 cells with TSA immediately after the 2-week hypoxia exposure but before selection with 6-TG prevented the emergence of silenced clones (Fig. 4F). In addition, when the BSH TGH22 cells were treated with TSA, thereby reactivating the BRCA1 promoter (as in Fig. 4E), the repressive histone changes were partially reversed (Fig. 4G), including not only the deacetylation of H3K9 but also the demethylation of H3K4.
DNA promoter methylation is not involved in hypoxic BRCA1 promoter silencing.
Because DNA hypermethylation at CpG sites in promoter regions has been associated with silenced tumor suppressor genes, including BRCA1, in many human cancers (14, 35, 50), we entertained the possibility that methylation at CpG sites might also be playing a role in the hypoxia-induced silencing. We tested this by methyl-specific PCR analysis in hypoxia-treated MCF-7 cells. Cells were exposed to hypoxia (0.01% O2) for 48 h or 1% oxygen for 2 weeks or 5 cycles of 48 h with alternating normoxia and hypoxia (0.01% O2) and then analyzed for BRCA1 promoter methylation. As a positive control for BRCA1 promoter methylation, the HCC38 breast cancer cell line was included (52). We did not detect any evidence of cytosine methylation in any of the hypoxia-treated cell populations (data available on request), even though we used published primers and methodologies that have previously detected methylation in human tumor samples (51). Interestingly, prior work probing for hypoxia-induced DNA methylation at the downregulated MLH1 promoter was also negative (34).
It has been reported that the methyl DNA binding proteins, MDB2 and MeCP2, bind not just to densely methylated regions of DNA but also to isolated methyl CpG dinucleotides (45, 54). Hence, we also tested by ChIP for the presence of MDB2 and MeCP2 at the BRCA1 promoters in MCF-7 cells exposed to hypoxia. No evidence for the recruitment of these proteins to the BRCA1 promoter was detected (data available on request).
Finally, we tested whether inhibiting DNA methylation with 5-aza-dC treatment could reactivate the silenced BRCA1 promoter in the BSH8 TGH22 and BSH8 2XTGH22 cells (data available on request), as we had seen with TSA treatment in Fig. 4E. No evidence of reactivation by treatment with 5-aza-dC was seen (data available on request), nor did combining 5-aza-dC treatment with TSA treatment have any additional effect beyond the substantial effect of TSA alone (data available on request). Taken together, these results indicate that the downregulation and silencing of the BRCA1 promoter by hypoxia can be associated with specific histone modifications but are not dependent on DNA methylation.
Hypoxia-induced E2F4/p130 pathway is not required for hypoxia-induced histone modifications at the BRCA1 promoter and is not sufficient for silencing.
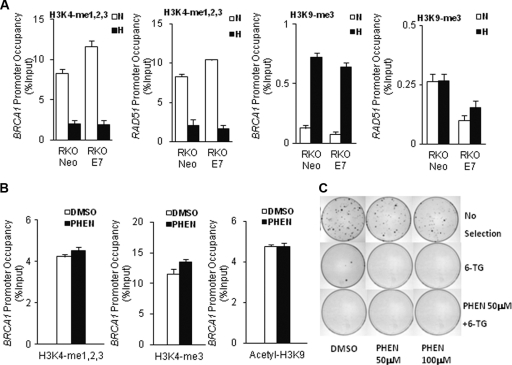
Histone modifications are regulated by many different enzymes, including histone methyltransferases, histone demethylases, and histone deacetylases (HDACs) (23, 41, 43, 57); however, the process by which histone-modifying enzymes are recruited to their target loci is not fully understood. Our previous work demonstrated that hypoxia-induced downregulation of both BRCA1 and RAD51 is mediated by binding of repressive E2F4/p130 complexes at the promoters (3, 6). We therefore hypothesized that these E2F4/p130 complexes might recruit histone-modifying enzymes to these promoters. To test this, we examined histone modifications at the BRCA1 and RAD51 promoters following exposure to normoxia or hypoxia in a matched pair of RKO-derived cell lines stably transfected with either a human papillomavirus (HPV) E7 cDNA construct (RKO E7) or an empty vector (RKO Neo). HPV E7 disrupts p130 function, and previous work established that E7 expression prevents downregulation of BRCA1 and RAD51 in hypoxia by disrupting the binding of p130/E2F4 complexes to the promoters (3, 6). However, as shown in Fig. 5A, the presence of E7 had no effect on the hypoxia-induced decreases in H3K4 methylation or increases in H3K9me3 at the BRCA1 and RAD51 promoters, even though E7 blocks the downregulation of these genes under the conditions tested (data not shown, but similar results were published in references 3 and 6).
Fig. 5.
The hypoxia-induced E2F4/p130 pathway is not required for hypoxia-induced histone modifications at the BRCA1 promoter and is not sufficient for silencing. (A) ChIP analyses for the presence of the indicated histone modifications at the BRCA1 or RAD51 promoters were performed in RKO cells expressing either E7 or a vector control (Neo) following a 48-h exposure to normoxia (N) or hypoxia (H). ChIP analyses of levels of H3K4 methylation (left two graphs) and levels of H3K9 methylation (right two graphs) are shown for the BRCA1 and RAD51 promoters, as quantified by real-time PCR. Relative promoter occupancies (% input) are shown, with error bars based on standard errors calculated from three replicates. (B) ChIP analyses of histone modifications at the BRCA1 promoter in cells treated or not treated with 200 μm of the PARP inhibitor PHEN. Quantification by real-time PCR of relative levels of BRCA1 promoter occupancy (% input) by the indicated factors is shown with error bars based on standard errors calculated from three replicates. (C) Visualization of 6-TG-resistant clones arising from BSH8 cells treated with PHEN or dimethyl sulfoxide (DMSO) (as a vehicle control). BSH8 cells were treated with PHEN at 50 μM or 100 μM or with DMSO (0.001%) for 2 weeks. Cells were then selected in 2.5 μg/m of 6-TG or 6-TG plus 50 μM PHEN for two additional weeks. The cells without selection were used to determine cloning efficiencies.
The above results suggest that active E2F4/p130 complexes are not required to produce the repressive histone modifications at the BRCA1 promoter that are associated with silencing. We next sought to test the converse: whether such complexes are sufficient to promote these histone changes. We knew from previous work that inhibitors of the enzyme PARP could cause BRCA1 and RAD51 suppression and also do so via an E2F4/p130-dependent pathway (15), in partial similarity to the effect of hypoxia. Interestingly, we found that treatment of cells with the PARP inhibitor PHEN did not induce any changes in H3K4 methylation, H3K9 methylation, or H3K9 acetylation at the BRCA1 promoter (Fig. 5B; also data available on request), even though PHEN treatment suppresses expression of BRCA1 and induces E2F4/p130 binding to the promoter (data not shown here but published in reference 15). Taken together, the above results suggest that E2F4/p130 complexes are neither necessary nor sufficient for the hypoxia-induced histone modifications at the BRCA1 promoter.
Because inhibition of PARP causes downregulation of BRCA1 in a manner somewhat similar to the effect of hypoxia, we also tested whether prolonged treatment of the BSH8 cells (containing the BRCA1 promoter-HPRT construct) with PHEN could promote BRCA1 promoter silencing. The BSH8 cells were exposed to PHEN for 2 weeks and then selected for possible BRCA1-HPRT silencing by the addition of 6-TG to the medium, as above. Unlike the prolonged hypoxia exposure, the PHEN treatment did not produce any 6-TG-resistant clones above the background (Fig. 5C). Hence, our data suggest that while hypoxia-induced E2F4/p130 complexes can suppress BRCA1 expression, they do not, by themselves, mediate BRCA1 silencing.
DISCUSSION
The work presented here shows that hypoxia-induced downregulation of BRCA1 and RAD51 is associated with specific histone modifications, including H3K4 demethylation, H3K9 methylation, and H3K9 deacetylation, with the opposite changes seen in the promoter of the hypoxia-inducible gene VEGF. In testing a number of histone-modifying enzymes, we identified LSD1 as the histone demethylase responsible for H3K4 demethylation at the BRCA1 and RAD51 promoters in response to hypoxic stress. By using a cell line with the BRCA1 promoter driving a selectable marker, we found that hypoxia can cause epigenetic silencing of the promoter at frequencies of up to 2% in a cell population. Importantly, the hypoxia-induced silencing was associated with the same histone modifications at the BRCA1 promoter as seen after acute hypoxic exposure (H3K4 demethylation and H3K9 deacetylation), providing a mechanistic link between downregulation of the promoter and its silencing in response to hypoxia.
The hypoxia-induced silencing was found to be reversible by treatment with an HDAC inhibitor (TSA), consistent with the role of histone modifications in the silencing. In contrast, hypoxia-induced silencing of the BRCA1 promoter was not associated with promoter DNA methylation, as silenced clones showed neither methylation at CpG sites nor binding of methyl DNA binding proteins to the promoter, and 5-aza-dC treatment of the silenced clones was unable to reverse the silencing. Manipulation of the E2F4/p130 pathway via ectopic HPV E7 expression revealed that this complex is not necessary for the hypoxia-mediated histone modifications at H3K4 and H3K9. In addition, although inhibition of PARP downregulates BRCA1 and does so through E2F4/p130 (15), it does not induce the same histone modifications as hypoxia and does not cause promoter silencing. Taken together, the above results suggest that short-term downregulation of BRCA1, in the absence of coordinated histone modifications, is not sufficient, by itself, to produce long-term epigenetic silencing of the promoter. The difference between hypoxic stress and PARP inhibition points to a key role for specific hypoxia-induced histone modifications in the epigenetic repression and silencing of the BRCA1 promoter.
The most dramatic histone modifications induced by hypoxia at the BRCA1 and RAD51 promoters are H3K4 demethylation and H3K9 methylation. H3K4 demethylation has been previously seen in hypoxic repression of the tumor suppressor RUNX3 in gastric cancer cells (29), while H3K4 methylation has been linked to hypoxia-activated genes, such as EGR1 (20) and VEGF (Fig. 1C). Prompted by these observations, we tested for the potential roles of candidate histone demethylases, JARID1B/PLU-1, JARID1A/RBP2, and LSD1 (KDM1), in BRCA1 regulation in response to hypoxia. We found that shRNA-mediated knockdown of LSD1 but not of PLU-1 or RBP2 abrogated the hypoxia-induced demethylation of H3K4 at the promoters, establishing LSD1 as the key mediator of epigenetic regulation of BRCA1 and RAD51 in response to hypoxic stress.
In prior work, we had shown that exposure of cells to hypoxia can stimulate E2F4/p130 binding to the BRCA1 and RAD51 promoters, which leads to suppression of gene expression (3, 6). However, our present results suggest that this hypoxia-induced pathway is not coordinated with hypoxia-induced histone modification at those loci. We showed that E7-mediated disruption of p130 does not prevent hypoxia-induced H3K4 demethylation or H3K9 methylation at the BRCA1 and RAD51 promoters. With respect to the BRCA1 silencing produced by hypoxia in BRCA1 promoter-HPRT-transfected cells, we showed that histone modifications happen at silenced BRCA1 promoters that are similar to those at the hypoxia-downregulated endogenous BRCA1 promoter. Together, our results suggest that regulation of BRCA1 by hypoxia occurs through at least two mechanisms: activation of the E2F/p130 pathway to mediate acute downregulation (1) and induction of repressive histone modifications, especially H3K4 demethylation by LSD1, as a step toward epigenetic silencing (2).
Silencing of BRCA1 has previously been associated with promoter DNA hypermethylation at CpG islands, based on observations in human tumor specimens (8, 14, 21, 35, 50). However, we could not detect any cytosine methylation either at the endogenous BRCA1 promoter in MCF-7 cells after hypoxia exposure or at hypoxia-induced, silenced BRCA1 promoters in BSH8-derived cells. Also, treatment of cells in which the BRCA1 promoter was already silenced by prior hypoxic stress with the DNA methylation inhibitor 5-aza-dC had no effect on BRCA1 promoter reactivation frequencies, in contrast to the potent effect of the HDAC inhibitor TSA. These observations point to a pathway of gene silencing initially driven by histone modifications and are in keeping with those of other recent studies (1, 26, 29, 38).
Nonetheless, while our results are consistent with a model in which progressive changes in promoter-associated histones are the key drivers of silencing at the BRCA1 promoter, they do not rule out subsequent accumulation of DNA CpG methylation combining to mediate even more stringent silencing. Interestingly, H3K9 methylation has been recognized as a signal for DNA methylation (18, 46), and unmethylated H3K4 is believed to be required for de novo DNA methylation (37). As shown here, hypoxia can induce both H3K9 methylation and H3K4 demethylation at the BRCA1 promoter, so this could provide the foundation for subsequent DNA methylation. It is possible that the hypoxic conditions used here, by themselves, are not a strong enough stressor to induce cytosine methylation. Other tumor microenvironmental factors, such as low pH, nutrient deprivation, and cytokine exposure, might be needed to combine with hypoxia to drive eventual DNA methylation, but this remains to be determined.
Overall, our work demonstrates that hypoxia induces histone modifications at the BRCA1 and RAD51 promoters and can drive BRCA1 epigenetic silencing. Interestingly, RAD51 can be downregulated by hypoxia in a manner similar to that for BRCA1, but there is essentially no evidence for RAD51 silencing in human tumors. We speculate that this reflects the severe growth disadvantage that the absence of RAD51 would place on human cells, consistent with reports that full RAD51 knockout is lethal to cells (44). In contrast, BRCA1 knockout is not lethal to cells; in fact, cancer cells in which the BRCA1 promoter is silenced would lack the genome maintenance and tumor suppressor functions of the gene and so could in theory develop a growth advantage that would lead to expansion during tumor progression. This could explain the frequent observation of silenced BRCA1 genes in human cancers.
ACKNOWLEDGMENTS
This work was supported by NIH grants R01ES005775, R01CA148996, and P01CA129186 to P.M.G. and R01ES015191 to M.S.T. and DOD grant W81XWH-06-0579 to M.S.T. Y.L. was partially supported by NIH NRSA T32CA009259.
Footnotes
Published ahead of print on 13 June 2011.
REFERENCES
- 1. Banelli B., Casciano I., Romani M. 2000. Methylation-independent silencing of the p73 gene in neuroblastoma. Oncogene 19:4553–4556 [DOI] [PubMed] [Google Scholar]
- 2. Beyer S., Kristensen M. M., Jensen K. S., Johansen J. V., Staller P. 2008. The histone demethylases JMJD1A and JMJD2B are transcriptional targets of hypoxia-inducible factor HIF. J. Biol. Chem. 283:36542–36552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bindra R. S., et al. 2005. Hypoxia-induced down-regulation of BRCA1 expression by E2Fs. Cancer Res. 65:11597–11604 [DOI] [PubMed] [Google Scholar]
- 4. Bindra R. S., Glazer P. M. 2007. Co-repression of mismatch repair gene expression by hypoxia in cancer cells: role of the Myc/Max network. Cancer Lett. 252:93–103 [DOI] [PubMed] [Google Scholar]
- 5. Bindra R. S., Glazer P. M. 2005. Genetic instability and the tumor microenvironment: towards the concept of microenvironment-induced mutagenesis. Mutat. Res. 569:75–85 [DOI] [PubMed] [Google Scholar]
- 6. Bindra R. S., Glazer P. M. 2007. Repression of RAD51 gene expression by E2F4//p130 complexes in hypoxia. Oncogene 26:2048–2057 [DOI] [PubMed] [Google Scholar]
- 7. Bindra R. S., et al. 2004. Down-regulation of Rad51 and decreased homologous recombination in hypoxic cancer cells. Mol. Cell. Biol. 24:8504–8518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Catteau A., Harris W. H., Xu C. F., Solomon E. 1999. Methylation of the BRCA1 promoter region in sporadic breast and ovarian cancer: correlation with disease characteristics. Oncogene 18:1957–1965 [DOI] [PubMed] [Google Scholar]
- 9. Chen H., Yan Y., Davidson T. L., Shinkai Y., Costa M. 2006. Hypoxic stress induces dimethylated histone H3 lysine 9 through histone methyltransferase G9a in mammalian cells. Cancer Res. 66:9009–9016 [DOI] [PubMed] [Google Scholar]
- 10. Cloos P. A., Christensen J., Agger K., Helin K. 2008. Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev. 22:1115–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Coquelle A., Pipiras E., Toledo F., Buttin G., Debatisse M. 1997. Expression of fragile sites triggers intrachromosomal mammalian gene amplification and sets boundaries to early amplicons. Cell 89:215–225 [DOI] [PubMed] [Google Scholar]
- 12. Coquelle A., Toledo F., Stern S., Bieth A., Debatisse M. 1998. A new role for hypoxia in tumor progression: induction of fragile site triggering genomic rearrangements and formation of complex DMs and HSRs. Mol. Cell 2:259–265 [DOI] [PubMed] [Google Scholar]
- 13. Ebert A., Lein S., Schotta G., Reuter G. 2006. Histone modification and the control of heterochromatic gene silencing in Drosophila. Chromosome Res. 14:377–392 [DOI] [PubMed] [Google Scholar]
- 14. Esteller M., Corn P. G., Baylin S. B., Herman J. G. 2001. A gene hypermethylation profile of human cancer. Cancer Res. 61:3225–3229 [PubMed] [Google Scholar]
- 15. Hegan D. C., et al. 2010. Inhibition of poly(ADP-ribose) polymerase down-regulates BRCA1 and RAD51 in a pathway mediated by E2F4 and p130. Proc. Natl. Acad. Sci. U. S. A. 107:2201–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Herman J. G., et al. 1998. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc. Natl. Acad. Sci. U. S. A. 95:6870–6875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hublitz P., Albert M., Peters A. H. 2009. Mechanisms of transcriptional repression by histone lysine methylation. Int. J. Dev. Biol. 53:335–354 [DOI] [PubMed] [Google Scholar]
- 18. Jackson J. P., Lindroth A. M., Cao X., Jacobsen S. E. 2002. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416:556–560 [DOI] [PubMed] [Google Scholar]
- 19. Jin W., et al. 2010. UHRF1 is associated with epigenetic silencing of BRCA1 in sporadic breast cancer. Breast Cancer Res. Treat. 123:359–373 [DOI] [PubMed] [Google Scholar]
- 20. Johnson A. B., Denko N., Barton M. C. 2008. Hypoxia induces a novel signature of chromatin modifications and global repression of transcription. Mutat. Res. 640:174–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Karray-Chouayekh S., et al. 2009. Clinical significance of epigenetic inactivation of hMLH1 and BRCA1 in Tunisian patients with invasive breast carcinoma. J. Biomed. Biotechnol. 2009:369129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kasper L. H., et al. 2005. Two transactivation mechanisms cooperate for the bulk of HIF-1-responsive gene expression. EMBO J. 24:3846–3858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kim J. K., Samaranayake M., Pradhan S. 2009. Epigenetic mechanisms in mammals. Cell. Mol. Life Sci. 66:596–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Klose R. J., et al. 2007. The retinoblastoma binding protein RBP2 is an H3K4 demethylase. Cell 128:889–900 [DOI] [PubMed] [Google Scholar]
- 25. Klose R. J., Zhang Y. 2007. Regulation of histone methylation by demethylimination and demethylation. Nat. Rev. Mol. Cell Biol. 8:307–318 [DOI] [PubMed] [Google Scholar]
- 26. Kondo Y., et al. 2008. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat. Genet. 40:741–750 [DOI] [PubMed] [Google Scholar]
- 27. Krieg A. J., et al. 2010. Regulation of the histone demethylase JMJD1A by hypoxia-inducible factor 1 alpha enhances hypoxic gene expression and tumor growth. Mol. Cell. Biol. 30:344–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Krishnan S., Horowitz S., Trievel R. C. 2011. Structure and function of histone H3 lysine 9 methyltransferases and demethylases. Chembiochem 12:254–263 [DOI] [PubMed] [Google Scholar]
- 29. Lee S. H., Kim J., Kim W. H., Lee Y. M. 2009. Hypoxic silencing of tumor suppressor RUNX3 by histone modification in gastric cancer cells. Oncogene 28:184–194 [DOI] [PubMed] [Google Scholar]
- 30. Li X., Liu J., Zhou R., Huang S., Chen X. M. 2010. Gene silencing of MIR22 in acute lymphoblastic leukaemia involves histone modifications independent of promoter DNA methylation. Br. J. Haematol. 148:69–79 [DOI] [PubMed] [Google Scholar]
- 31. Litt M. D., Simpson M., Gaszner M., Allis C. D., Felsenfeld G. 2001. Correlation between histone lysine methylation and developmental changes at the chicken beta-globin locus. Science 293:2453–2455 [DOI] [PubMed] [Google Scholar]
- 32. Maison C., et al. 2002. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat. Genet. 30:329–334 [DOI] [PubMed] [Google Scholar]
- 33. McGarvey K. M., et al. 2006. Silenced tumor suppressor genes reactivated by DNA demethylation do not return to a fully euchromatic chromatin state. Cancer Res. 66:3541–3549 [DOI] [PubMed] [Google Scholar]
- 34. Mihaylova V. T., et al. 2003. Decreased expression of the DNA mismatch repair gene Mlh1 under hypoxic stress in mammalian cells. Mol. Cell. Biol. 23:3265–3273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mirza S., et al. 2007. Promoter hypermethylation of TMS1, BRCA1, ERα and PRB in serum and tumor DNA of invasive ductal breast carcinoma patients. Life Sci. 81:280–287 [DOI] [PubMed] [Google Scholar]
- 36. Nakayama J.-i., Rice J. C., Strahl B. D., Allis C. D., Grewal S. I. S. 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292:110–113 [DOI] [PubMed] [Google Scholar]
- 37. Ooi S. K. T., et al. 2007. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448:714–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Oyer J. A., Chu A., Brar S., Turker M. S. 2009. Aberrant epigenetic silencing is triggered by a transient reduction in gene expression. PLoS One 4:e4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Peters A. H. F. M., et al. 2002. Histone H3 lysine 9 methylation is an epigenetic imprint of facultative heterochromatin. Nat. Genet. 30:77–80 [DOI] [PubMed] [Google Scholar]
- 40. Reynolds T. Y., Rockwell S., Glazer P. M. 1996. Genetic instability induced by the tumor microenvironment. Cancer Res. 56:5754–5757 [PubMed] [Google Scholar]
- 41. Shi Y. 2007. Histone lysine demethylases: emerging roles in development, physiology and disease. Nat. Rev. Genet. 8:829–833 [DOI] [PubMed] [Google Scholar]
- 42. Shi Y., et al. 2004. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119:941–953 [DOI] [PubMed] [Google Scholar]
- 43. Shi Y., Whetstine J. R. 2007. Dynamic regulation of histone lysine methylation by demethylases. Mol. Cell 25:1–14 [DOI] [PubMed] [Google Scholar]
- 44. Sonoda E., et al. 1998. Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J. 17:598–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stirzaker C., Song J. Z., Davidson B., Clark S. J. 2004. Transcriptional gene silencing promotes DNA hypermethylation through a sequential change in chromatin modifications in cancer cells. Cancer Res. 64:3871–3877 [DOI] [PubMed] [Google Scholar]
- 46. Tamaru H., et al. 2003. Trimethylated lysine 9 of histone H3 is a mark for DNA methylation in Neurospora crassa. Nat. Genet. 34:75–79 [DOI] [PubMed] [Google Scholar]
- 47. Turker M. S., Mummaneni P., Bishop P. L. 1991. Region- and cell type-specific de novo DNA methylation in cultured mammalian cells. Somat. Cell Mol. Genet. 17:151–157 [DOI] [PubMed] [Google Scholar]
- 48. Vakoc C. R., Sachdeva M. M., Wang H., Blobel G. A. 2006. Profile of histone lysine methylation across transcribed mammalian chromatin. Mol. Cell. Biol. 26:9185–9195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. van Zeeland A. A., Simons J. U. 1976. The use of correction factors in the determination of mutant frequencies in populations of human diploid skin fibroblasts. Mutat. Res. 34:149–158 [DOI] [PubMed] [Google Scholar]
- 50. Vasilatos S. N., et al. 2009. CpG island tumor suppressor promoter methylation in non-BRCA-associated early mammary carcinogenesis. Cancer Epidemiol. Biomarkers Prev. 18:901–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wei M., et al. 2005. BRCA1 promoter methylation in sporadic breast cancer is associated with reduced BRCA1 copy number and chromosome 17 aneusomy. Cancer Res. 65:10692–10699 [DOI] [PubMed] [Google Scholar]
- 52. Wei M., et al. 2008. Estrogen receptor alpha, BRCA1, and FANCF promoter methylation occur in distinct subsets of sporadic breast cancers. Breast Cancer Res. Treat. 111:113–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Xia X., et al. 2009. Integrative analysis of HIF binding and transactivation reveals its role in maintaining histone methylation homeostasis. Proc. Natl. Acad. Sci. U. S. A. 106:4260–4265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Xiong Y., Dowdy S. C., Eberhardt N. L., Podratz K. C., Jiang S.-W. 2006. hMLH1 promoter methylation and silencing in primary endometrial cancers are associated with specific alterations in MBDs occupancy and histone modifications. Gynecol. Oncol. 103:321–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yamane K., et al. 2007. PLU-1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol. Cell 25:801–812 [DOI] [PubMed] [Google Scholar]
- 56. Yang J., et al. 2009. Role of hypoxia-inducible factors in epigenetic regulation via histone demethylases. Ann. N. Y. Acad. Sci. 1177:185–197 [DOI] [PubMed] [Google Scholar]
- 57. Yang X. J., Seto E. 2007. HATs and HDACs: from structure, function and regulation to novel strategies for therapy and prevention. Oncogene 26:5310–5318 [DOI] [PubMed] [Google Scholar]
- 58. Young S. D., Marshall R. S., Hill R. P. 1988. Hypoxia induces DNA overreplication and enhances metastatic potential of murine tumor cells. Proc. Natl. Acad. Sci. U. S. A. 85:9533–9537 [DOI] [PMC free article] [PubMed] [Google Scholar]