Abstract
We report in vivo analysis of histone and RNA polymerase II (pol II) occupancy at the 601 element, which functions as a strong in vitro nucleosome-positioning element and transcriptional pause site. Surprisingly, nucleosomes were not strongly positioned over the 601 element inserted either within a yeast chromosomal open reading frame (ORF) (GAL1-YLR454W) or in an intergenic region. In fact 601 within GAL1-YLR454W was actually depleted of histones relative to flanking sequences and did not cause pol II pausing. Upstream of an inserted 601 element within GAL1-YLR454W, a positioned nucleosome was formed whose location depended on transcriptional history; it shifted after a round of activation and repression. Transcriptional activation caused histone eviction throughout the GAL1-YLR454W ORF, except at 601, where there was no loss and some net histone deposition. In contrast, a second round of activation after glucose shutoff caused histone eviction both at 601 and elsewhere in the ORF. We conclude that the intrinsic high-affinity histone-DNA interactions at 601 do not necessarily play a dominant role in establishing nucleosomes or pol II pause sites within a coding region in vivo and that transcriptional history can have an important influence on histone occupancy flanking this sequence.
INTRODUCTION
Nucleosome positioning controls the access of DNA sequences to the proteins that carry out genomic processes including replication, repair, and transcription. Two major factors determine nucleosome positioning, (i) intrinsic properties of the DNA sequence within a nucleosome that govern its flexibility and affinity for histones and (ii) extrinsic factors including ATP-dependent chromatin remodelers and boundary elements such as transcription start sites (5, 27, 47). There are conflicting results in the literature concerning the relative importance of an intrinsic “genomic code” for determining nucleosome positions based on DNA sequence alone in vivo in budding yeast (14, 17, 35, 41, 47). RNA polymerase II (pol II) transcription normally disrupts nucleosomes by eviction of H2A/H2B dimers (2, 40) or entire octamers (3, 20, 21, 34, 42, 43) followed by repair in which histones are restored by chaperones, including Spt16/FACT (facilitates chromatin transcription) (13, 34, 44). The chaperone activity of FACT can facilitate the removal of H2A/H2B dimers, nucleosome assembly (2, 28), and remodeling (45). FACT travels with elongating pol II and enhances histone deposition during transcription (26). Dynamic nucleosome eviction and replacement during transcription suggest that chromatin structure could change between successive rounds of activated transcription if nucleosomes were not replaced in their original locations; however, such an effect of transcriptional history has not been reported previously.
In vitro, many DNA sequences have been tested for the ability to position nucleosomes and the highest-affinity sequence so far identified is the 601 element isolated by SELEX (24). 601 has been used in numerous in vitro studies because it assembles into a very stable, highly positioned nucleosome (4, 6, 9, 12, 15, 30). The 601 nucleosome causes a strong orientation-dependent pause in transcription elongation by RNA pol II in vitro (4, 11, 15) because of particularly strong interactions that resist unwinding of the DNA from the histones at the boundary of the H3/H4 tetramer. The 601 nucleosome functions as a “fluctuating barrier” where pol II pauses while it waits for opportunities to elongate, which arise when DNA spontaneously unwraps from the histones (11). The RSC remodeling complex effectively counters pausing at a 601 nucleosome (6). Whether strongly positioned nucleosomes present a similar “fluctuating barrier” in vivo and how this may be affected by remodeling factors are important open questions. The effects of the 601 sequence on nucleosome positioning and transcription elongation in vivo have not been investigated extensively; however, two recent studies reported that 601 can position nucleosomes on plasmids in mammalian cells under some conditions (8, 38). Furthermore, pol II accumulation characteristic of pausing was observed when 601 was inserted within a reporter gene on a transiently transfected plasmid (38). In addition, a recent report showed that the nucleosome-positioning ability of a 133-bp sequence called the “superbinder” is predictable and depends on its location relative to activating sequences and on levels of transcription activation (41).
In this study, we investigated how transcription elongation and nucleosome positioning are affected when the 601 nucleosome-positioning element (NPE) was inserted into a chromosomal galactose-inducible gene or an intergenic region in budding yeast. Surprisingly, we found that in these locations, nucleosomes were not preferentially positioned over the 601 element and that 601 did not cause pol II pausing. However, 601 insertion did confer a change in nucleosome localization at adjacent sequences within an open reading frame (ORF) and this change depended on the transcriptional history of the gene.
MATERIALS AND METHODS
Yeast strains and plasmids.
The yeast strains (W303 background) used in this study are described in Table 1. A 280-bp-long PCR fragment containing 601 was amplified from pGEM3Z-601 (24) and inserted at +1242 or +5550 relative to the ATG codon of GAL1-YLR545W (FMP27) (37) by pop-in pop-out replacement of the Kluyveromyces lactis URA3 gene and selection with 5-fluoroorotic acid. This sequence has 72 bases upstream and 62 bases downstream of the 146-bp 601 core element. Previous nucleosome mapping studies have shown that position +1242 in YLR454W is within an internucleosomal region whereas +5550 is within a “fuzzy” nucleosome (22, 27). The sequences of the 601 insertions used in this study are shown in Table 2. Strains DBY1191 and DBY1193 were made by homologous recombination using a PCR fragment derived from DBY1109 that contained the 601 sequence, the GAL1 promoter, and the hygromycin resistance gene. The minimal 146-bp 601 sequence in the forward orientation was also inserted by pop-in pop-out replacement of the K. lactis URA3 gene at +1242 of GAL1-YLR454 (DBY1338) and in the intergenic region downstream of ADH4 at +2049 from the ATG codon 900 bases 3′ of the termination codon (DBY1343) (Table 2). A tetracycline-inducible ts degron (tet-degron) mutant form of STH1 was made as described previously (36), and the GAL1-YLR454W 601fw+1242::Hygro allele (DBY1229) was inserted by homologous recombination of a PCR product that contained the hygromycin resistance gene, the GAL1 promoter, and the 601 sequence. N-terminally myc-tagged H4 and H2B were expressed from plasmids pNOY436 and pNOY439 (18) transformed into DBY1109.
Table 1.
Strains used in this study
| Strain | Derivation | Genotype | Reference or source |
|---|---|---|---|
| Strain | Derivation | Genotype | Reference or source |
| DBY446 | W303-1a | MATaura3his3leu2ade2trp1 (GAL1-YLR454W)::TRP1HPR1::myc13HIS3 | 37 |
| DBY1109 | DBY1109 | MATα ura3his3leu2ade2trp1can1lys2 (GAL1-YLR454W::601fw+1242)::Hygrospt16G132D | This study |
| DBY1111 | DBY1111 | MATα ura3his3leu2ade2trp1can1lys2 (GAL1-YLR454W::601rv+1242)::Hygrospt16G132D | This study |
| DBY1309 | DBY1109 | MATα ura3his3leu2ade2trp1can1lys2 (GAL1-YLR454W::601fw+1242)::Hygrospt16G132D/pNOY436(Myc-H4) | This study |
| DBY1311 | DBY1109 | MATα ura3his3leu2ade2trp1can1lys2 (GAL1-YLR454W::601fw+1242)::Hygrospt16G132D/pNOY439(Myc-H2B) | This study |
| DBY1191 | DBY446 | MATaura3his3leu2ade2trp1 (GAL1-YLR454W::601fw+1242)::HygroHPR1::myc13HIS3 | This study |
| DBY1193 | DBY446 | MATaura3his3leu2ade2trp1 (GAL1-YLR454W::601rv+1242)::HygroHPR1::myc13HIS3 | This study |
| DBY1197 | DBY446 | MATaura3his3leu2ade2trp1 (GAL1-YLR454W::601fw+5550)::TRP1HPR1::myc13HIS3 | This study |
| DBY1198 | DBY446 | MATaura3his3leu2ade2trp1 (GAL1-YLR454W::601rv+5550)::TRP1HPR1::myc13HIS3 | This study |
| DBY1229 | W303-1a | MATaura3-1his3-11,15leu2-3,112ade2-1trp1-1LEU2::pCM245 HIS3::pRS403-188sth1tet-degron::URA3 (GAL1-YLR454W::601fw-146bp+1242)::Hygro/pFL45S UBR1 | This study |
| DBY1333 | DBY1109 | MATα ura3his3leu2ade2trp1can1lys2 (GAL1-YLR454W::601fw+1242)::Hygrospt16G132D Δcbf1::Kan | This study |
| DBY1338 | DBY1109 | MATα ura3his3leu2ade2trp1can1lys2 (GAL1-YLR454W::601fw-146bp+1242)::Hygrospt16G132D | This study |
| DBY1343 | W303-1a | MATaade2-1ura3-1his3-11trp1-1leu2-3,112can1-100ADH4::601-146bp+2049 | This study |
Table 2.
Sequences of the 601 constructs used in this study
| Clone | Sequencea |
|---|---|
| YLR454W-601fw+1242 | ttgacatggaTCCTAATGACCAAGGAAAGCATGATTCTTCACACCGAGTTCATCCCTTATGTGATGGACCCTATACGCGGCCGCCCTGGAGAATCCCGGTGCCGAGGCCGCTCAATTGGTCGTAGACAGCTCTAGCACCGCTTAAACGCACGTACGCGCTGTCCCCCGCGTTTTAACCGCCAAGGGGATTACTCCCTAGTCTCCAGGCACGTGTCAGATATATACATCCTGTGCATGTATTGAACAGCGACCTTGCCGGTGCCAGTCGGATAGTGTTCCGAGCTCCCGGAtccagtaaat |
| YLR454W-601rv+1242 | ttgacatggaTCCGGGAGCTCGGAACACTATCCGACTGGCACCGGCAAGGTCGCTGTTCAATACATGCACAGGATGTATATATCTGACACGTGCCTGGAGACTAGGGAGTAATCCCCTTGGCGGTTAAAACGCGGGGGACAGCGCGTACGTGCGTTTAAGCGGTGCTAGAGCTGTCTACGACCAATTGAGCGGCCTCGGCACCGGGATTCTCCAGGGCGGCCGCGTATAGGGTCCATCACATAAGGGATGAACTCGGTGTGAAGAATCATGCTTTCCTTGGTCATTAGGAtccagtaaat |
| YLR454W-601fw+5550 | ggcacaatgtTCCTAATGACCAAGGAAAGCATGATTCTTCACACCGAGTTCATCCCTTATGTGATGGACCCTATACGCGGCCGCCCTGGAGAATCCCGGTGCCGAGGCCGCTCAATTGGTCGTAGACAGCTCTAGCACCGCTTAAACGCACGTACGCGCTGTCCCCCGCGTTTTAACCGCCAAGGGGATTACTCCCTAGTCTCCAGGCACGTGTCAGATATATACATCCTGTGCATGTATTGAACAGCGACCTTGCCGGTGCCAGTCGGATAGTGTTCCGAGCTCCCGGAttctttacgt |
| YLR454W-601rv+5550 | ggcacaatgtTCCGGGAGCTCGGAACACTATCCGACTGGCACCGGCAAGGTCGCTGTTCAATACATGCACAGGATGTATATATCTGACACGTGCCTGGAGACTAGGGAGTAATCCCCTTGGCGGTTAAAACGCGGGGGACAGCGCGTACGTGCGTTTAAGCGGTGCTAGAGCTGTCTACGACCAATTGAGCGGCCTCGGCACCGGGATTCTCCAGGGCGGCCGCGTATAGGGTCCATCACATAAGGGATGAACTCGGTGTGAAGAATCATGCTTTCCTTGGTCATTAGGAttctttacgt |
| YLR454W-601fw-146bp+1242 | ttgacatggaGCCCTGGAGAATCCCGGTGCCGAGGCCGCTCAATTGGTCGTAGACAGCTCTAGCACCGCTTAAACGCACGTACGCGCTGTCCCCCGCGTTTTAACCGCCAAGGGGATTACTCCCTAGTCTCCAGGCACGTGTCAGATATATACATCtccagtaaat |
| ADH4-601fw-146bp+2049 | gacgatatcaGCCCTGGAGAATCCCGGTGCCGAGGCCGCTCAATTGGTCGTAGACAGCTCTAGCACCGCTTAAACGCACGTACGCGCTGTCCCCCGCGTTTTAACCGCCAAGGGGATTACTCCCTAGTCTCCAGGCACGTGTCAGATATATACATCagcatgcctg |
Lowercase letters indicate flanking regions. Underlined sequences correspond to the minimal positioning sequence within the 280-bp 601 constructs. Putative binding sites for Abf1 (italic) and Cbf1 (boldface) are shown on separate sequences because they overlap.
Chromatin immunoprecipitations (ChIP) and antibodies.
ChIP and quantitative PCR (Q-PCR) were performed as previously described (16, 32). PCR primers are available upon request. For induction of GAL1-YLR454W, cells were grown at 30°C in lactate (Lac) medium (1× yeast extract-peptone-[YEP], 2% lactic acid, 3% glycerol, 0.05% glucose) to an optical density (OD) of 0.8, and then galactose was added to 2% for 30 min of incubation. For repression, cells were washed with phosphate-buffered saline (PBS) twice and resuspended in YEP plus 2% glucose for 2 h of incubation. For reactivation, cells were washed with PBS twice, resuspended in YEP plus 2% galactose, and incubated for 30 min. For the experiments using the spt16-G132D ts mutant (DBY1109 and DBY1111), cells were grown in Lac to an OD of 0.8 at 25°C, collected by centrifugation, resuspended in prewarmed medium, and incubated at 34°C for 30 min. Galactose was then added to a 2% final concentration, and cells were incubated at 34°C for 30 min. The Sth1 tet-degron (DBY1229) was inactivated by growing cells at 37°C in 2 μg/ml doxycycline for 2 h. Antibodies against the Pol II, H3, and H2B C termini were used as previously described (33, 46). Anti-c-Myc (Santa Cruz sc40-AC), anti-Abf1 (sc-25755), and anti-Htz1 (Abcam ab4626) were used at 2.5 μg/IP, 5 μg/IP, and 8 μg/IP, respectively.
MNase sensitivity assays.
Micrococcal nuclease (MNase) digestions were carried out by the method of Liu et al (23), with minor modifications. Cultures (50 ml) were grown in YEP plus 2% glucose to an OD at 600 nm of ∼0.9, and spheroplasts were prepared without cross-linking and digested in 3.9 ml of buffer Z (1 M sorbitol, 50 mM Tris-HCl [pH 7.4], 10 mM β-mercaptoethanol) with 150 U of lyticase (Sigma) for 30 min at 37°C while shaking at 120 rpm. Spheroplasts were resuspended in 600 μl of NP-S buffer (0.5 mM spermidine, 1 mM β-mercaptoethanol, 0.075% NP-40, 50 mM NaCl, 10 mM Tris [pH 7.4], 5 mM MgCl2, 1 mM CaCl2) and treated with 50 units of S7 MNase (Roche) for 30 min at 37°C. Mononucleosomal DNA was gel purified.
RESULTS
The 601 NPE does not induce Pol II transcriptional pausing in vivo.
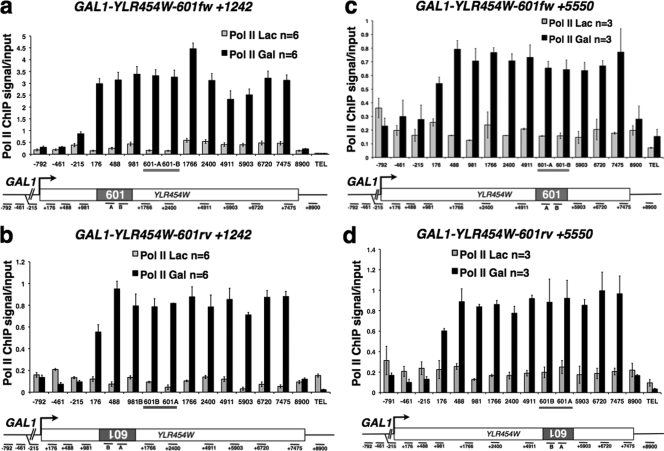
To test whether the 601 element causes pol II pausing in vivo, we inserted it into the yeast chromosomal GAL1-YLR454W gene in both orientations at position +1242 or +5550 relative to the start codon (Fig. 1a to d). We analyzed pol II occupancy by ChIP, and the results were quantified by Q-PCR of 15 amplicons in the YLR454W ORF, flanking sequences, and the 601 sequence itself (Fig. 1a to d). In uninduced cells grown in Lac, pol II occupancy was low throughout the gene, showing that the 601 insertion did not introduce a cryptic promoter (Fig. 1a to d). As expected, pol II density increased throughout the ORF upon galactose induction (Gal, Fig. 1a to d); however, it did not accumulate at 601 in either orientation at +1242 or +5550 (amplicons 601-A and 601-B, Fig. 1a to d). This result suggests that, contrary to in vitro results, the 601 sequence does not impose an orientation-dependent pause on elongating pol II in vivo.
Fig. 1.
The 601 sequence does not induce pol II pausing in vivo. Shown are relative pol II ChIP signals in the GAL1-YLR454W-601fw+1242 (a), GAL1-YLR454W-601rv+1242 (b), GAL1-YLR454W-601fw+5550 (c), and GAL1-YLR454W-601rv+5550 (d) genes with 601 in the forward (fw) and reverse (rv) orientations before (Lac) and after (galactose, Gal) transcription activation. Locations of Q-PCR amplicons relative to the ATG codon are indicated below the gene maps (dashes). The 601fw and 601rv amplicons are underlined. TEL indicates the telomere VIR control. Mean values from n PCRs from at least 3 IPs in panels a and b and 2 IPs in panels c and d with standard deviations are shown.
Lack of nucleosome occupancy over 601 in vivo.
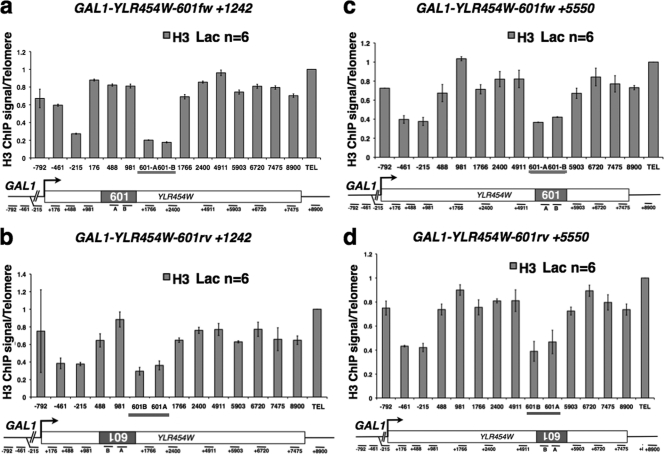
The unexpected lack of pol II pausing at 601 could be explained if a strongly positioned nucleosome within the ORF is not a real obstacle to pol II or if 601 does not, in fact, assemble a stable nucleosome. To test these possibilities, we monitored histone H3 occupancy by ChIP on the 601 sequence in both orientations at +1242 or +5550 in the uninduced GAL1-YLR454W gene. Surprisingly, this analysis showed that the 601 sequence had the lowest H3 ChIP signals of any region within the gene, regardless of its orientation or position (Fig. 2a to d). To rule out the possibility that the C terminus of H3 recognized by this antibody is specifically occluded on 601, we also performed ChIP analysis of N-terminally myc-tagged H2B and H4 expressed from plasmids (Fig. 3a and b). These experiments confirmed the H3 ChIP result showing that the 601 sequence has the lowest histone occupancy of any region we analyzed in the GAL1-YLR454W gene.
Fig. 2.
Low histone occupancy on the 601 sequence in vivo. Shown are relative histone H3 ChIP signals in the uninduced GAL1-YLR454W-601fw+1242 (a), GAL1-YLR454W-601rv+1242 (b), GAL1-YLR454W-601fw+5550 (c), and GAL1-YLR454W-601rv+5550 (d) genes in Lac-grown cells. Q-PCR amplicons are indicated as in Fig. 1. Mean values from n PCRs from at least 3 IPs with standard deviations are shown.
Fig. 3.
Low histone occupancy at both 601 and its minimal positioning element. Shown are relative ChIP signals for N-terminally myc-tagged H2B (a) and myc-tagged H4 (b) at the uninduced GAL1-YLR454W-601fw+1242 gene in Lac, histone H3 occupancy in the uninduced GAL1-YLR454W-601fw-146bp+1242 gene (601 minimal positioning element) (c), and histone H3 occupancy at the 601 minimal positioning element inserted into the 3′ flanking region of the ADH4 gene (ADH4-601fw-146bp+2049) (d). A map of the amplicons is also shown. Mean values from n PCRs from at least 3 IPs with standard deviations are shown.
These surprising results were obtained using the core 601 element flanked by 62 to 72 bases of 5′ and 3′ vector sequence present in the original SELEX insert (24) (Table 2). To rule out a possible effect of these flanking sequences on nucleosome positioning in vivo, we inserted the core 146-bp 601 sequence in the forward orientation at position +1242 in GAL1-YLR454W. ChIP analysis of H3 occupancy over the minimal 601 element at this position showed the same depletion observed when it was flanked by additional vector sequences (Fig. 3c). We conclude that nucleosome depletion at 601 within the YLR454W ORF is a property of the core 146-bp positioning sequence.
To determine whether failure to position a nucleosome over 601 was specific to insertion within an ORF, we inserted the minimal 146-bp element into an intergenic region downstream of ADH4 (ADH4-601fw-146bp+2049) and monitored histone H3 occupancy. H3 occupancy at 601 in this intergenic region was neither elevated nor depleted relative to flanking sequences (Fig. 3d). Together, the results in Fig. 2 and 3 therefore demonstrate that the 601 sequence does not necessarily position a stable nucleosome in vivo as it does in vitro and that its effects on local histone occupancy vary, depending on the sequence context.
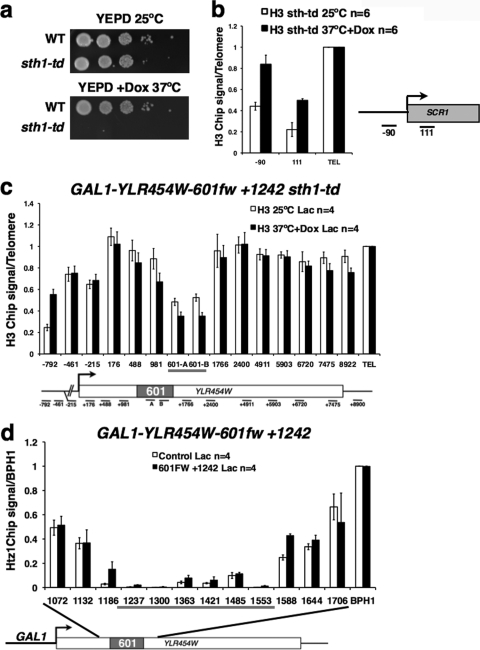
The experiments in Fig. 2 and 3 suggest that 601 insertion into the GAL1-YLR454W ORF, but not into an intergenic region, establishes chromatin that resembles nucleosome-depleted regions (NDRs) previously described at promoters (10, 27, 31). We compared the 601-dependent region of low nucleosome occupancy at +1242 in GAL1-YLR454W with those at promoters by examining several features characteristic of NDRs, including sensitivity to the Sth1 ATPase in the RSC complex, enrichment of Htz1 (H2AZ) in flanking nucleosomes, and Abf1 binding (10, 31). We measured histone occupancy on the GAL1-454W-601fw+1242 gene before and after Sth1 inactivation using a doxycycline-inducible degron (sth1-td) (DBY1229, Fig. 4a and c). Functional inactivation of Sth1 under nonpermissive conditions (37°C for 2 h with doxycycline) was confirmed by the increase in H3 occupancy at the pol III-transcribed SCR1 locus as previously reported (29) (Fig. 4b). In contrast, Sth1 inactivation did not markedly increase histone occupancy at 601 in the YLR454W ORF(Fig. 4c), showing that remodeling by RSC is not required for histone depletion at this location.
Fig. 4.
The region of low histone occupancy induced by 601 differs from NDRs at promoters. (a) Growth defect of the sth1-td strain (DBY1229) at 37°C with 2 μg/ml doxycycline. WT, wild type. (b, c) Histone H3 occupancy at the control SCR1 (29) locus and in GAL1-YLR454W-601fw+1242 before and after Sth1 depletion for 2 h at 37°C with 2 μg/ml doxycycline. Note that Sth1 inactivation increases histone occupancy at SCR1 as expected but not around the 601 element in GAL1-YLR454W. Mean values normalized to TELVIR (TEL) with standard deviations are shown. Dox, doxycycline. (d) Histone Htz1 ChIP using tiled Q-PCR amplicons in DBY1191 GAL1-YLR454W-601fw+1242 and in the control strain without a 601 insertion relative to the BPH1 promoter, which is a positive control for Htz1 (31). Low signals for 601 amplicons (underlined) in the control strain are background. Mean values from n PCRs from at least 2 IPs with standard deviations are shown.
We compared Htz1 localization around the 601 element in GAL1-YLR454W (+1242fw) with that in the BPH1 3′ end, which has a positioned Htz1 nucleosome (31). ChIP analysis was performed with a set of tiled Q-PCR primer pairs that span 601. Only low levels of Htz1 were observed in the regions flanking 601 compared to those in the BPH1 positive control (Fig. 4d). The region of low histone occupancy at 601 therefore differs from promoter-associated NDRs that are flanked by positioned, Htz1-containing nucleosomes.
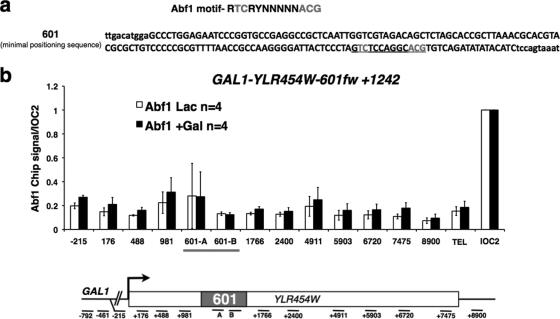
Abf1 binding contributes to the formation of NDRs at promoters, and a putative Abf1 binding site occurs within the 601 sequence (Table 2; Fig. 5a); however, anti-Abf1 ChIP revealed no significant binding at 601 relative to the IOC2 positive control (10, 25) (Fig. 5b). Abf1 therefore appears not to be involved in nucleosome depletion at 601. There is also a consensus Cbf1 binding site within 601 (Table 2), but CBF1 deletion did not reverse the relative depletion of histones at 601 in the GAL1-YLR454W-601fw+1242 gene (data not shown). In summary, neither Abf1 nor Cbf1 binding within 601 is a major cause of histone depletion at this sequence. We conclude that although the 601 element induces a region of low histone occupancy within the YLR454W ORF, it does not mimic other properties of NDRs at promoters.
Fig. 5.
Abf1 does not bind 601 in vivo. (a) Putative Abf1 binding site within 601 (see Table 2). (b) Relative occupancy of Abf1 in the GAL1-YLR454W-601fw+1242 gene before (Lac) and after (+Gal [galactose]) transcription activation and normalized to the previously characterized Abf1 binding site in the IOC2 promoter (10, 25). Means and standard deviations for n PCRs are normalized to TELVIR.
Transcriptional history determines nucleosome occupancy near 601.
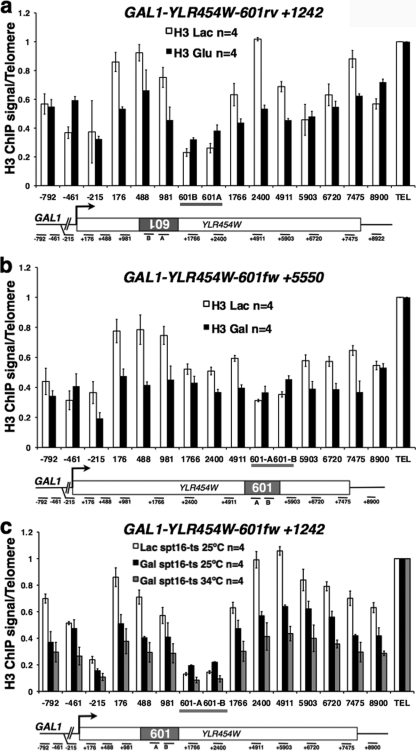
We asked how transcription would affect histone occupancy on 601 at position +1242 in GAL1-YLR454W during the course of galactose activation, glucose repression, and galactose reactivation. As expected, pol II density increased with galactose induction, fell to background levels in glucose, and increased once more with reactivation (Fig. 6a). Histone H3 and H2B levels were reduced at most positions within the ORF following transcriptional activation as previously reported (21, 34) (Fig. 6b, white versus green bars, and c, white versus red bars). Remarkably, the 601 sequence is an exception to this rule, where occupancy of H3 and H2B actually increased upon galactose induction (Fig. 6b and c, amplicons 601-A and 601-B). Following glucose shutoff, histone occupancy further increased at 601 and throughout the ORF, consistent with nucleosome replacement (Fig. 6b and c, gray bars). Consequently, after a round of activation and repression, the chromatin had been remodeled to achieve approximately equivalent nucleosome occupancy across 601 and its flanking sequences. In contrast to the first round of transcription, histones H3 and H2B were evicted from the 601 sequence when it was challenged by a second round of galactose-induced transcription (Fig. 6b and c, gray versus blue bars). In common with the first round of activation, we found no evidence of pol II pausing at 601 during a second galactose activation (Fig. 6a, blue bars).
Fig. 6.
Effects of transcriptional history on nucleosome occupancy at 601. Shown are relative pol II (a), histone H3 (b), and H2B (c) ChIP signals at the GAL1-YLR454W-601fw+1242 gene in cells grown in Lac and during a cycle of galactose activation (30 min, +Gal-1), glucose repression (120 min, +Glu), and galactose reactivation (30 min, +Gal-2). Note that histone occupancy at 601 increased on the first activation (white versus green bars in panel b and white versus red bars in panel c) and decreased on the second activation (gray versus blue bars in panels b and c). Means and standard deviations for n PCRs are normalized to the input in panel a and to TELVIR in panels b and c.
We investigated whether the surprising increase in histone occupancy at 601 following galactose induction is peculiar to one construct or is a more general phenomenon. Histone H3 occupancy was analyzed before and after galactose induction at 601 in the reverse orientation at +1242 and in the forward orientation at +5550 (Fig. 7a and b). In agreement with the results in Fig. 6, H3 levels increased within 601 in both of these constructs after galactose induction and decreased elsewhere throughout the YLR454W ORF. These results show that 601 elements inserted in multiple sequence contexts within a galactose-inducible gene interfere with the histone eviction that normally accompanies transcriptional activation.
Fig. 7.
Spt16/FACT-dependent histone deposition at 601. (a, b) Anti-H3 ChIP of the GAL1-YLR454W-601rev+1242 and GAL1-YLR454W-601fw+5550 genes in Lac and induced with galactose (Gal) for 30 min. Note that, as in Fig. 6b, H3 occupancy decreased at most locations on Gal induction, but in contrast, it slightly increased at 601. (c) Anti-H3 ChIP of the GAL1-YLR454W-601fw+1242 gene (DBY1109 spt16ts) in Lac or galactose (Gal) at 25°C or 34°C. Note histone deposition at 601 when the gene was activated at 25°C and eviction at 34°C. Means and standard deviations for n PCRs are normalized to TELVIR.
The histone chaperone Spt16/FACT helps to repair chromatin by depositing histones in the wake of elongating transcription complexes (13, 34, 44). We asked whether Spt16/FACT is also required for the unexpected net histone deposition around 601 during activated transcription. For these experiments, we monitored H3 occupancy at GAL1-YLR454W genes with a 601 insertion during galactose activation in the spt16-G132D ts mutant (7). The results in Fig. 7c show that Spt16 inactivation reversed the galactose-induced increase in H3 occupancy at 601. As expected, Spt16 inactivation also reduced histone occupancy throughout the rest of the ORF during galactose induction. We conclude that Spt16/FACT is required for the unusual net histone deposition at 601 under galactose-induced conditions.
Transcriptional history reorganizes the chromatin flanking 601.
To investigate nucleosome positioning at 601 and its flanking sequences by an independent method at higher resolution, we performed MNase sensitivity assays. MNase-digested mononucleosomal DNA from uninduced cells and glucose-repressed cells following galactose induction was analyzed by Q-PCR using 27 tiled amplicons spanning 900 bases around the 601 sequence (Fig. 8a). Signals were quantified relative to the well-characterized PHO5 −1 nucleosome (1). Consistent with anti-histone ChIP, the 601 sequence was more sensitive to MNase than flanking regions and the dyad (arrow) was never strongly protected (Fig. 8c and d). 601 in the forward, but not the reverse, orientation caused pronounced positioning of an upstream nucleosome relative to the parent gene without a 601 insertion (compare amplicons 1132 and 1158 in Fig. 8b and c). Notably, after a round of transcription activation and glucose shutoff, there is a marked reorganization of chromatin flanking 601 in the forward orientation (Fig. 8c). A downstream nucleosome becomes better positioned (Fig. 8c, gray bars, amplicons 1456 to 1588) and the upstream nucleosome shifts about 80 bp toward the 5′ end of 601 (Fig. 8c, amplicons 1132 to 1216). Histone deposition at sequences flanking 601 in the glucose repressed cells (Fig. 8c, gray bars) is consistent with the results of the lower-resolution histone ChIP assays in Fig. 6b and c. We conclude that chromatin flanking 601 within a gene can be restructured in a way that depends on 601 orientation during the course of a cycle of transcriptional activation and repression.
Fig. 8.
Transcriptional history determines nucleosome occupancy flanking 601. (a) Experimental design and map of Q-PCR primer pairs used for MNase sensitivity mapping. Position numbers represent the middle of each amplicon relative to the ATG. Primer pairs overlapping 601 are in red. MNase sensitivity of chromatin at GAL1-YLR454W (b) in DBY446 and in GAL1-YLR454W-601fw+1242 (c) and GAL1-YLR454W-601rv+1242 (d) in cells grown in Lac or repressed with glucose (Glu) after galactose activation. Dashed lines represent the moving average of the values at each amplicon. Note the MNase sensitivity across the 601 dyad at position 1395 (red arrow in panel c and blue arrow in panel d). Amplicons in inverted 601 are numbered relative to the AIG. Mean values from 2 IPs and 4 PCRs per IP with standard deviations are normalized to the signals at the PHO5 −1 nucleosome.
DISCUSSION
In this study, we analyzed the effects of inserting the strong NPE 601 within a transcribed chromosomal gene in budding yeast. In vitro, very stable nucleosomes assemble on this sequence at a unique position. Furthermore, in a purified system, the 601 nucleosome causes strong, orientation-dependent pausing by RNA pol II before it reaches the nucleosomal dyad (4). Surprisingly, we found that 601 insertion in either orientation at two positions within the GAL1-YLR454W gene caused no detectable polymerase pausing, as detected by anti-pol II ChIP. Furthermore, nucleosomes were not preferentially localized over the 601 element when it was inserted either within the YLR454W ORF or in an intergenic region (Fig. 2 and 3). On the contrary, we observed that in both orientations at both positions within GAL1-YLR454W, 601 actually induced the formation of a chromatin region with the lowest histone occupancy of any sequence within the gene (Fig. 2, 3, 6, and 7). Furthermore, high-resolution MNase mapping showed no evidence of nucleosomes that preferentially localized over the dyad, where nucleosomes assemble on 601 in vitro (Fig. 8c and d). How nucleosomes are excluded from 601 within the YLR454W ORF is not known, but it appears to differ significantly from NDRs at promoters (10, 31) because it does not require Sth1 and it is not associated with accumulation of high levels of Htz1 in flanking nucleosomes or with Abf1 binding (Fig. 4 and 5). We conclude that at the locations we examined in the context of the chromosomal YLR454W ORF and the ADH4 3′ flank, the innate affinity of the 601 sequence for histones is not the dominant determinant of nucleosome positioning. Our results are consistent with previous observations showing that another in vitro nucleosome-positioning sequence, the TG5 element designed to bend around nucleosomes with a strong rotational setting, also failed to assemble positioned nucleosomes in yeast (39). These results suggest that in vivo chromatin remodelers and assembly factors are able to overcome the intrinsic affinity of a sequence for nucleosomes, with the result that a nucleosome can actually be partially excluded from a strong positioning element. In summary, these experiments suggest that intrinsic DNA-histone interactions are not the main determinant of nucleosome positioning in vivo at many locations in the yeast genome, consistent with previous conclusions (47).
Our results do not exclude the possibility that at other locations in the genome, 601 may be able to exert a dominant nucleosome-positioning effect. Consistent with this possibility, 601 was found to induce nucleosome positioning under some conditions on plasmids introduced into mammalian cells. A positioned nucleosome assembled on a variant of 601 inserted into the EF-1a promoter on plasmids in mouse liver cells at early time points when the gene was expressed but not at later times after it had been silenced (8). Note that the sequence used in this study differed from the original 601 element we used (24) at 4 positions within the 146-bp core. Furthermore, pol II pausing occurred upstream of a positioned 601 nucleosome within a luciferase reporter gene on a plasmid transiently transfected into 293T cells (38); however, it is not clear whether this is an orientation-dependent effect like that found in vitro (4). The differences between 601 positioning behavior in yeast and that in mammalian cells could be due to inherent differences between these systems, as well as differences between chromatin assembly on episomal plasmids and that on chromosomes. Sequence context also influences how 601 affects nucleosome occupancy in vivo. This point is illustrated by the fact that the histone occupancy at 601 differed from that at its flanking regions, depending on whether it was inserted within the YLR454W ORF or in the ADH4 3′ flanking region (Fig. 3). Similarly, histone occupancy at a 601-related positioning element differed between two positions where it was inserted in the GAL1 promoter (41).
Introduction of 601 in the forward orientation at +1242 in the YLR454W ORF localized a nucleosome upstream prior to transcriptional activation (see positions 1132 and 1158 in Fig. 8c). This effect may be in part due to statistical positioning exerted by a boundary effect caused by nucleosome depletion over 601 (19, 27). Interestingly, this effect is polar, as 601 in the reverse orientation at the same location does not cause strong positioning of an upstream nucleosome (Fig. 8d).
Our investigation of how transcriptional activation and repression and the chromatin repair/assembly factor Spt16/FACT affect histone occupancy at 601 revealed two unusual properties of this element. (i) It conferred net histone deposition around it during galactose-induced transcriptional activation, and (ii) it conferred a stable change in local chromatin structure that is dependent on transcriptional history. Normally galactose-induced transcription causes histone eviction within ORFs (20, 34), as we observed at most positions in the activated GAL1-YLR454W ORF (Fig. 6b and c and 7a and b). The region around 601 is an exception where histone occupancy reproducibly increased during an initial round of galactose activation. This effect was observed at 601 in both orientations and at two different positions within the GAL-YLR454W gene (Fig. 6b and c and 7a and b). The unexpected net histone deposition at 601 during galactose activation required the histone chaperone Spt16/FACT (Fig. 7). We speculate that the nucleosome-depleted 601 sequence may be a preferred sink for Spt16-mediated nucleosome replacement during active transcription. Additional histone deposition occurred at 601 (and elsewhere in the gene) when transcription was subsequently repressed by glucose. In a second round of galactose-induced transcription, these histones are evicted from 601. Thus, histone occupancy around 601 is modulated downward and then upward in successive rounds of transcriptional activation.
There is a marked difference in where nucleosomes are situated around 601 before and after a cycle of transcriptional activation and repression. Before activation, in lactose-grown cells with 601 in the forward orientation (at +1242), a nucleosome is positioned approximately 250 bases upstream of the 601 dyad and no highly positioned nucleosome was detected immediately downstream (Fig. 8c, blue bars). After a round of galactose activation and glucose repression, however, the chromatin did not return to its original state but instead was rearranged in a way that established at least three newly positioned nucleosomes that flank both sides of 601 (Fig. 8c, gray bars). Histones are therefore not necessarily accurately replaced at their original positions after a round of transcription. This rearrangement appears to result from sliding of the upstream nucleosome closer to 601 and deposition of two positioned nucleosomes downstream of 601. Based on the evidence in Fig. 7c, we propose that this transcription-dependent chromatin rearrangement around 601 involves Spt16/FACT and possibly also remodelers that can slide nucleosomes. Although the nucleosomes that come to flank both sides of 601 after a cycle of activation and repression are well positioned, they did not cause detectable pol II pausing when a second round of activated transcription was induced by galactose (Fig. 6a, blue bars). It remains possible, however, that subtle differences in transcriptional elongation between successive rounds of activation would be missed at the resolution of our ChIP analysis.
In summary, these results show that chromatin structure within a gene can be stably remodeled in a way that depends on the recent transcriptional history of the gene. A potential implication of this observation is that over the course of successive cycles of activation of a particular gene, RNA pol II may confront different chromatin landscapes that could influence transcriptional elongation and coupled cotranscriptional events such as pre-mRNA processing. An important unresolved question is whether the ability to affect local chromatin in a manner that depends on transcriptional history is unique to the synthetic 601 element. In the future, it will be of interest to determine whether naturally occurring sequences have similar properties.
ACKNOWLEDGEMENTS
We thank J. Widom (Northwestern University) and M. Nomura (University of California, Irvine) for plasmids and K. Struhl and T. Formosa for yeast strains. We thank C. Wu (NIH), J. Tyler, P. Megee, J. Hesselberth, S. Johnson, and S. Kim (University of Colorado, Denver) for helpful discussions and comments on the manuscript.
This work was supported by NIH grant GM063873 and a Research Supplement to Promote Diversity (GM58613) to R.P.
Footnotes
Published ahead of print on 20 June 2011.
REFERENCES
- 1. Almer A., Rudolph H., Hinnen A., Horz W. 1986. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 5:2689–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Belotserkovskaya R., et al. 2003. FACT facilitates transcription-dependent nucleosome alteration. Science 301:1090–1093 [DOI] [PubMed] [Google Scholar]
- 3. Boeger H., Griesenbeck J., Strattan J., Kornberg R. 2004. Removal of promoter nucleosomes by disassembly rather than sliding in vivo. Mol. Cell 14:667–673 [DOI] [PubMed] [Google Scholar]
- 4. Bondarenko V., et al. 2006. Nucleosomes can form a polar barrier to transcript elongation by RNA polymerase II. Mol. Cell 24:469–479 [DOI] [PubMed] [Google Scholar]
- 5. Cairns B. 2009. The logic of chromatin architecture and remodelling at promoters. Nature 461:193–198 [DOI] [PubMed] [Google Scholar]
- 6. Carey M., Li B., Workman J. L. 2006. RSC exploits histone acetylation to abrogate the nucleosomal block to RNA polymerase II elongation. Mol. Cell 24:481–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Formosa T., et al. 2001. Spt16-Pob3 and the HMG protein Nhp6 combine to form the nucleosome-binding factor SPN. EMBO J. 20:3506–3517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gracey L. E., et al. 2010. An in vitro-identified high-affinity nucleosome-positioning signal is capable of transiently positioning a nucleosome in vivo. Epigenetics Chromatin 3:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hall M. A., et al. 2009. High-resolution dynamic mapping of histone-DNA interactions in a nucleosome. Nat. Struct. Mol. Biol. 16:124–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hartley P. D., Madhani H. D. 2009. Mechanisms that specify promoter nucleosome location and identity. Cell 137:445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hodges C., Bintu L., Lubkowska L., Kashlev M., Bustamante C. 2009. Nucleosomal fluctuations govern the transcription dynamics of RNA polymerase II. Science 325:626–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huynh V. A. T., Robinson P. J. J., Rhodes D. 2005. A method for the in vitro reconstitution of a defined “30 nm” chromatin fibre containing stoichiometric amounts of the linker histone. J. Mol. Biol. 345:957–968 [DOI] [PubMed] [Google Scholar]
- 13. Jamai A., Puglisi A., Strubin M. 2009. Histone chaperone spt16 promotes redeposition of the original h3-h4 histones evicted by elongating RNA polymerase. Mol. Cell 35:377–383 [DOI] [PubMed] [Google Scholar]
- 14. Jiang C., Pugh B. F. 2009. Nucleosome positioning and gene regulation: advances through genomics. Nat. Rev. Genet. 10:161–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jin J., et al. 2010. Synergistic action of RNA polymerases in overcoming the nucleosomal barrier. Nat. Struct. Mol. Biol. 17:745–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Johnson S., Cubberley G., Bentley D. 2009. Cotranscriptional recruitment of the mRNA export factor Yra1 by direct interaction with the 3′ end processing factor Pcf11. Mol. Cell 33:215–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kaplan N., et al. 2009. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458:362–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Keener J., Dodd J. A., Lalo D., Nomura M. 1997. Histones H3 and H4 are components of upstream activation factor required for the high-level transcription of yeast rDNA by RNA polymerase I. Proc. Natl. Acad. Sci. U. S. A. 94:13458–13462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kornberg R. D., Stryer L. 1988. Statistical distributions of nucleosomes: nonrandom locations by a stochastic mechanism. Nucleic Acids Res. 16:6677–6690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kristjuhan A., Svejstrup J. Q. 2004. Evidence for distinct mechanisms facilitating transcript elongation through chromatin in vivo. EMBO J. 23:4243–4252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee C.-K., Shibata Y., Rao B., Strahl B. D., Lieb J. D. 2004. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 36:900–905 [DOI] [PubMed] [Google Scholar]
- 22. Lee W., et al. 2007. A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 39:1235–1244 [DOI] [PubMed] [Google Scholar]
- 23. Liu C. L., et al. 2005. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 3:e328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lowary P., Widom J. 1998. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 276:19–42 [DOI] [PubMed] [Google Scholar]
- 25. MacIsaac K. D., et al. 2006. An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics 7:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mason P., Struhl K. 2003. The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol. Cell. Biol. 23:8323–8333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mavrich T., et al. 2008. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 18:1073–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Orphanides G., LeRoy G., Chang C., Luse D., Reinberg D. 1998. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell 92:105–116 [DOI] [PubMed] [Google Scholar]
- 29. Parnell T. J., Huff J. T., Cairns B. R. 2008. RSC regulates nucleosome positioning at Pol II genes and density at Pol III genes. EMBO J. 27:100–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Partensky P. D., Narlikar G. J. 2009. Chromatin remodelers act globally, sequence positions nucleosomes locally. J. Mol. Biol. 391:12–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Raisner R. M., et al. 2005. Histone variant H2A. Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell 123:233–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schroeder S., Schwer B., Shuman S., Bentley D. 2000. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 14:2435–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schroeder S., Zorio D., Schwer B., Shuman S., Bentley D. 2004. A function of yeast mRNA cap methyltransferase, Abd1, in transcription by RNA polymerase II. Mol. Cell 13:377–387 [DOI] [PubMed] [Google Scholar]
- 34. Schwabish M. A., Struhl K. 2004. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 24:10111–10117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Segal E., et al. 2006. A genomic code for nucleosome positioning. Nature 442:772–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Seward D., et al. 2007. Demethylation of trimethylated histone H3 Lys4 in vivo by JARID1 JmjC proteins. Nat. Struct. Mol. Biol. 14:240–242 [DOI] [PubMed] [Google Scholar]
- 37. Strässer K., et al. 2002. TREX is a conserved complex coupling transcription with mRNA export. Nature 417:304–308 [DOI] [PubMed] [Google Scholar]
- 38. Subtil-Rodríguez A., Reyes J. C. 2010. BRG1 helps RNA polymerase II to overcome a nucleosomal barrier during elongation, in vivo. EMBO Rep. 11:751–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tanaka S., Zatchej M., Thoma F. 1992. Artificial nucleosome positioning sequences tested in yeast minichromosomes: a strong rotational setting is not sufficient to position nucleosomes in vivo. EMBO J. 11:1187–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Thiriet C., Hayes J. 2005. Replication-independent core histone dynamics at transcriptionally active loci in vivo. Genes Dev. 19:677–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang X., Bryant G. O., Floer M., Spagna D., Ptashne M. 2011. An effect of DNA sequence on nucleosome occupancy and removal. Nat. Struct. Mol. Biol. 18:507–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Weiner A., Hughes A., Yassour M., Rando O. J., Friedman N. 2010. High-resolution nucleosome mapping reveals transcription-dependent promoter packaging. Genome Res. 20:90–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Williams S. K., Truong D., Tyler J. K. 2008. Acetylation in the globular core of histone H3 on lysine-56 promotes chromatin disassembly during transcriptional activation. Proc. Natl. Acad. Sci. U. S. A. 105:9000–9005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Workman J. 2006. Nucleosome displacement in transcription. Genes Dev. 20:2009–2017 [DOI] [PubMed] [Google Scholar]
- 45. Xin H., et al. 2009. yFACT induces global accessibility of nucleosomal DNA without H2A-H2B displacement. Mol. Cell 35:365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang L., Schroeder S., Fong N., Bentley D. 2005. Altered nucleosome occupancy and histone H3K4 methylation in response to ‘transcriptional stress’. EMBO J. 24:2379–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang Y., et al. 2009. Intrinsic histone-DNA interactions are not the major determinant of nucleosome positions in vivo. Nat. Struct. Mol. Biol. 16:847–852 [DOI] [PMC free article] [PubMed] [Google Scholar]