Abstract
Neph proteins are evolutionarily conserved membrane proteins of the immunoglobulin superfamily that control the formation of specific intercellular contacts. Cell recognition through these proteins is essential in diverse cellular contexts such as patterning of the compound eye in Drosophila melanogaster, neuronal connectivity in Caenorhabditis elegans, and the formation of the kidney filtration barrier in mammals. Here we identify the PDZ and BAR domain protein PICK1 (protein interacting with C-kinase 1) as a Neph-interacting protein. Binding required dimerization of PICK1, was dependent on PDZ domain protein interactions, and mediated stabilization of Neph1 at the plasma membrane. Moreover, protein kinase C (PKCα) activity facilitated the interaction through releasing Neph proteins from their binding to the multidomain scaffolding protein zonula occludens 1 (ZO-1), another PDZ domain protein. In Drosophila, the Neph homologue Roughest is essential for sorting of interommatidial precursor cells and patterning of the compound eye. RNA interference-mediated knockdown of PICK1 in the Drosophila eye imaginal disc caused a Roughest destabilization at the plasma membrane and a phenotype that resembled rst mutation. These data indicate that Neph proteins and PICK1 synergistically regulate cell recognition and contact formation.
INTRODUCTION
Organization of cells into a specific spatial configuration is a prerequisite for the development of epithelial tissues and organs and requires selective cell adhesion and cell recognition. Multiple classes of cell surface molecules are involved in mediating cell recognition, many of which are highly conserved through evolution. Particularly important recognition molecules in humans are the Neph and nephrin proteins (15). Mutation of members of this class of adhesion proteins results in kidney failure and severe protein loss into urine due to the disruption of the kidney filtration barrier (9, 19). Neph proteins are characterized by an extracellular domain containing five immunoglobulin repeats, a transmembrane region, and an approximately 220-amino-acid cytoplasmic tail that mediates protein interactions and signaling (16). Whereas mammals contain three Neph proteins (Neph1 to -3), two Neph-like proteins can be found in Drosophila melanogaster (Roughest, Kirre) and at least one in Caenorhabditis elegans (SYG-1) (10). Neph proteins interact with the related proteins of the nephrin family. In the mammalian kidney it could be demonstrated that signaling through these junctional proteins directs formation, maturation, and maintenance of highly specialized, interdigitating secondary processes of kidney podocytes, the visceral epithelial cells of the glomerular filtration barrier (9, 19, 46). In Drosophila Neph-like Roughest and Kirre mediate cell recognition and morphogenesis in the development of the fly compound eye (1, 27). The adult compound eye of Drosophila is composed of about 750 unit eyes, or ommatidia, that are arranged in a highly ordered crystal-like pattern. This biological crystal develops from an unpatterned single-layered epithelium, the eye imaginal disc (5, 44). In the young pupa four lens-secreting cone cells and two primary pigment cells are separated by a pool of undifferentiated interommatidial precursor cells. During pupal development this interommatidial lattice is shaped into a very regular honeycomb-like pattern of secondary and tertiary pigment cells. This morphogenetic process, which requires cell sorting and extensive cell shaping, is controlled through Neph and nephrin proteins. Interestingly, these molecular mechanisms are evolutionarily highly conserved and have been shown to largely also apply to the development of the mammalian kidney filter, making the fly eye a perfect model system to address signaling processes at the kidney filter (6).
Although much has been learned about the role of Neph proteins in controlling morphogenetic processes, the integration of adhesion and cell signaling of the cell recognition module to orchestrate cell movement, cytoskeletal reorganization, and junction remodeling is poorly understood.
Here we show that Neph proteins interact with the BAR domain protein PICK1 (protein interacting with C kinase 1) and that this interaction stabilizes Neph proteins at the plasma membrane. PICK1 was first identified as an interactor of protein kinase C alpha (PKCα) (36). PICK1 harbors a PDZ domain (PSD95/DlgA/ZO-1) at the N terminus and a BAR domain (Bin/amphiphysin/Rvs) at the C terminus. BAR domains form crescent-shaped dimers and are involved in sensing and/or regulating membrane curvature, a prerequisite for vesicle formation (11, 17, 25). PICK1 has been shown to be involved in modulating the trafficking of neuronal receptors (14). We used the Drosophila eye imaginal disc to demonstrate that PICK1 and Neph proteins are involved in common pathways in vivo, suggesting a critical role for BAR domain proteins in controlling important morphogenetic processes.
MATERIALS AND METHODS
Reagents and plasmids.
Phorbol myristate acetate (PMA) (tetradecanoyl phorbol acetate [TPA], catalog no. 9905; Cell Signaling Technology) was used at a final concentration of 200 nM for 30 min. Full-length PICK1 cDNA was cloned from a human podocyte cDNA library using standard cloning techniques. Mouse Neph1, Neph2, and Neph3 cDNAs were cloned into a modified pcDNA6 expression vector coding for the CD5 signal peptide fused to the V5-tag sequence (sV5-tag) followed by restriction sites to insert the cDNA. ZO-1 plasmids have been described before (16). A membrane-bound fusion construct of the cytoplasmic domain of Neph1 (sIg.7.Neph1cyt) has been described previously (31). Antibodies were from Sigma (FLAG), Serotec (MAb V5), Millipore (polyclonal antibody [pAb] V5), and Santa Cruz (H-300 goat pAb anti-PICK1). A polyclonal anti-Neph2 antibody has been described previously (12). For Drosophila immunofluorescence the following antibodies were used: rat anti-DE-cadherin (Developmental Studies Hybridoma Bank), rabbit anti-Pyd (kind gift from R. Cagan), mouse anti-Rst (24A5; Fischbach lab), rabbit anti-Kirre (126intra; Fischbach lab), rabbit anti-Hbs (14intra; Fischbach lab), and rabbit anti-SNS (Fischbach lab).
Fly strains.
All crosses were kept at 25°C. The following fly lines were used: Rst3 (Fischbach lab), upstream activating sequence (UAS)-mCD8::GFP (Fischbach lab), GMR-GAL4 (Fischbach lab), GMR-GAL4, UAS-dicer2 and UAS-dicer2 (gift from R. Cagan), and UAS-PICK1-RNA interference (RNAi) (Vienna Drosophila RNAi Center, strains ID22268 and ID104486) flies.
Cell culture and coimmunoprecipitation.
HEK 293T cells were transfected with the calcium phosphate method and incubated for 24 h. Cells were then washed with phosphate-buffered saline (PBS), lysed in ice-cold IP buffer (1% Triton X-100, 20 mM Tris, pH 7.5, 25 mM NaCl, 50 mM NaF, 15 mM Na4P2O7, 1 mM EDTA, 0.25 mM phenylmethylsulfonyl fluoride [PMSF], 5 mM Na3VO4) on ice for 15 min and centrifuged (14,000 rpm, 4°C, 15 min). Supernatants containing equal amounts of total proteins were incubated for 1 h at 4°C with a polyclonal anti-V5 antibody (Chemicon) that was bound to protein G-Sepharose previously (1 h at 4°C) or with anti-FLAG antibody covalently coupled to agarose beads (catalog no. A2220; Sigma). The precipitates were washed three times with IP buffer, and bound proteins were resolved by 10% SDS-PAGE.
For endogenous coimmunoprecipitations, tissues were freshly prepared. Mouse brains and kidneys were homogenized using a glass potter, cleared by centrifugation, and solubilized in lysis buffer supplemented with 20 mM 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS) and 3 mM ATP. After centrifugation at 15,000 × g (15 min, 4°C) and centrifugation at 100,000 × g (30 min, 4°C), cell lysates containing equal amounts of total protein were precleared with protein A-Sepharose and then incubated for 1 h at 4°C with the appropriate antibody, followed by incubation with 30 μl of protein A-Sepharose beads for 3 h. The beads were washed extensively with lysis buffer, and bound proteins were resolved by 10% SDS-PAGE.
RNA interference experiments.
RNA interference experiments were done as described previously (29). In brief, short hairpin RNAs (shRNAs) were designed based on the prediction of publicly available prediction programs (RNAi Designer; Invitrogen). shRNAs were cloned into the transient microRNA expression vector pcDNA6.2-GW/emGFP/miR (Invitrogen). To monitor the efficiency of shRNA-mediated knockdown, we created a luciferase reporter construct using psicheck2 (Promega) in which the coding sequence of PICK1 was fused to the coding sequence of Renilla reniformis luciferase as an artificial 3′ untranslated region (UTR). In addition to R. reniformis luciferase, this construct expresses firefly luciferase for internal control. The reporter plasmid was cotransfected with the respective pcDNA6.2-GW/emGFP/miR shRNA construct into HEK 293T cells. R. reniformis luciferase and firefly luciferase activities were measured 24 h after transfection. Transfections and measurements were performed in triplicate. Selected hairpins (hp) (hp 1, TGCTGTCAAATACCTGGACGATATAGGTTTTGGCCACTGACTGACCTATATCGCAGGTATTTGA; hp 5, TGCTGACGACAGGTACTCAAACTTCAGTTTTGGCCACTGACTGACTGAAGTTTGTACCTGTCGT) were cloned into one cluster (chaining), and this cluster was Gateway cloned into pLenti6.2/V5-Dest (Invitrogen) for stable lentiviral expression in human podocytes cells.
Human podocyte culture.
Human immortalized podocytes (AB 8 cells) were cultivated as described previously (28). In brief, cells were grown in standard RPMI 1640 medium containing 10% fetal calf serum (FCS) and supplements either at the permissive temperature of 33°C (in 5% CO2) to promote cell propagation or at the nonpermissive temperature of 37°C (in 5% CO2) to allow terminal differentiation.
Pulldown assay.
MBP.Neph1cyt and GST.ZO-11–111 were expressed in Escherichia coli and affinity purified. Purified MBP.Neph1cyt was bound to amylose resin (New England BioLabs) and incubated at 4°C for 60 min with GST.ZO-11–111 that had been preincubated with buffer, recombinant PKCα (0.5 U; Invitrogen), or calf intestinal alkaline phosphatase (CIAP; 0.5 U; Invitrogen) for 30 min at 37°C. Bound proteins were eluted from the resin with Laemmli buffer and subjected to Western blot analysis.
Surface labeling.
At 24 h after transfection, HEK 293T cells were starved for 3 h with serum-free medium. Cells were washed twice with PBS and surface labeled with 0.6 mg/ml EZ Link Sulfo-NHS-SS-Biotin (Pierce) for 15 min at room temperature. Cells were incubated with quenching buffer (100 mM glycine in PBS) for 10 min at room temperature, washed twice with PBS, and lysed in IP buffer. Precleared lysates were incubated with NeutrAvidin UltraLink Resin (Pierce) for 8 h at 4°C. Bound proteins were eluted from the resin with Laemmli buffer and subjected to Western blot analysis.
In vitro phosphorylation.
His.Neph1cyt was expressed in E. coli, and 5 μg of His.Neph1cyt was bound to 30 μl of Ni-nitrilotriacetic acid agarose (Qiagen). In vitro phosphorylation of His.Neph1cyt was performed for 20 min at 30°C in a reaction in 60 μl of kinase reaction buffer containing 50 μM ATP (supplemented with 10 μCi of [γ-32P]ATP [Perkin Elmer]). Phosphorylation was initiated by the addition of 0.5 U of PKCα purified from rat brain (Sigma) in enzyme dilution buffer, or the His.Neph1cyt was incubated in enzyme dilution buffer alone (control) or with 0.5 U of CIAP (New England BioLabs). To monitor the incorporation of phosphate, radiolabeled His.Neph1cyt was visualized by SDS-PAGE and autoradiography.
Immunofluorescence.
HeLa cells were seeded on coverslips and transfected using Genejuice reagent (Novagen). After 24 to 48 h, cells were fixed for 10 min in 4% paraformaldehyde. Fixed cells were incubated for 30 min in blocking solution (5% normal donkey serum, 0.1% Triton X-100) and subsequently incubated with primary and secondary antibodies. After washing in PBS, the cells were mounted in Prolong Gold antifade (Invitrogen) and subjected to immunofluorescence microscopy with an Axiovert 200 M microscope/EC Plan-Neofluar 40×/1.30 oil objective equipped with a charge-coupled-device (CCD) camera (all from Carl Zeiss).
For labeling of surface/endocytosed sV5.Neph1, cells were incubated at 4°C with monoclonal antibody (MAb) anti-V5 (Serotec) diluted in full medium for 30 min. Cells were washed with medium three times and were allowed to recover at 37°C for 30 min. Cells were then washed with PBS, fixed, but not permeabilized, with 2% paraformaldehyde for 5 min, blocked with 5% normal donkey serum in PBS, and incubated with a donkey Alexa 488-coupled anti-mouse antibody (Invitrogen). After washing, cells and bound antibodies were fixed again for 5 min before cells were incubated with permeabilizing blocking solution (5% normal donkey serum, 0.1% Triton X-100). Then, a Cy3-coupled donkey anti-mouse secondary antibody (Jackson ImmunoResearch) was applied.
Drosophila retina immunohistology procedures were essentially as described previously (27). Briefly, staged pupal eye discs (18 to 19 h or 41 to 42 h after puparium formation [APF]) were dissected and fixed for 15 min in 4% paraformaldehyde, blocked with normal donkey serum in PBS with 0.1% Triton X-100, and incubated with primary antibody. A fluorescent-dye-coupled secondary antibody (Cy3, DyLight 549 or DyLight 488; Jackson ImmunoResearch) was used as the secondary antibody. Preparations were embedded in Vectashield (Vector), and image acquisition was done with a Zeiss LSM710/AxioObserver Z.1 equipped with an EC Plan-Neofluar 40×/1.30 oil objective (Carl Zeiss).
Adult eye fluorescence.
Adult flies were frozen at −80°C. The flies were glued on a slide, and autofluorescence was used to image the eye with a Zeiss LSM710/AxioObserver Z.1 equipped with an EC Plan Neofluar 10×/0.3 objective (Carl Zeiss).
Image acquisition and processing.
Fluorescent images were acquired using Zeiss Axiovision 4.8 and Zeiss ZEN 2009 (Carl Zeiss) and assembled using Adobe Photoshop and Illustrator CS4 (Adobe Inc.) or ImageJ (26). For better visibility of membrane staining (see Fig. 2), gamma was set to 0.45 in Axiovision. ImageJ software was used for fluorescence intensity measurements (see Fig. 8) and intensity profiles (see Fig. 2) (RGB line profile plug-in).
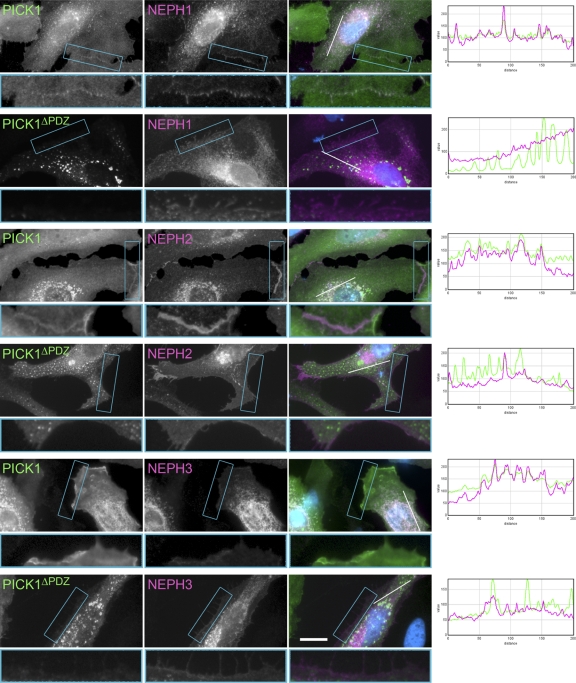
Fig. 2.
PICK1 colocalizes with Neph proteins. HeLa cells were transiently transfected with the constructs indicated (Neph constructs with sV5 tag; PICK1 constructs with FLAG tag) and stained with antibodies against the tags. The full-length Neph proteins colocalize with full-length PICK1 in cytoplasmic patches and at the plasma membrane. PICK1ΔPDZ does not colocalize with Neph proteins. The merged image shows the PICK1 channel in green and the Neph channel in magenta. Nuclei are stained with DAPI (blue). Boxed regions showing cell borders are shown in an enlarged view below each image. Intensity profiles for PICK1 (green) and Neph (magenta) along the white line in the merged image are shown to the right to assess the colocalization of PICK1 and Neph proteins and the random colocalization of PICK1ΔPDZ and Neph proteins.
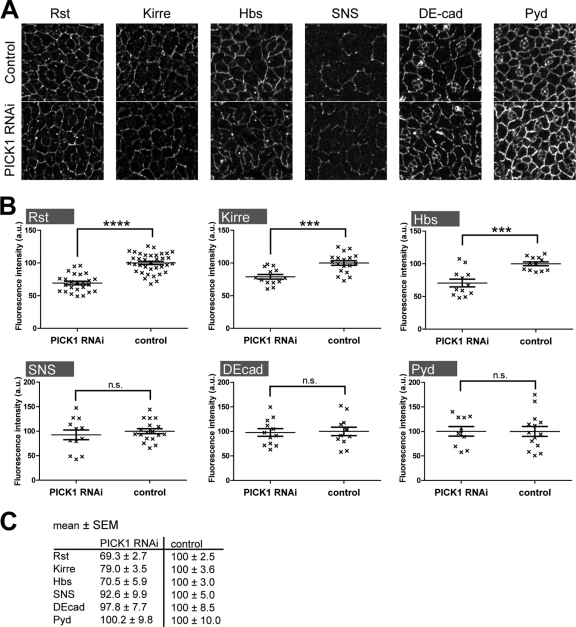
Fig. 8.
Knockdown of dPICK1 does not change the subcellular distribution but the total protein level of IRM proteins. (A) Pupal eye imaginal discs (18 to 19 h APF) stained with the antibodies indicated. The subcellular localization of the stained proteins is indistinguishable between dPICK1 knockdown (GMR-GAL4; UAS-dPICK1RNAi > UAS-dicer2) and control (GMR-GAL4 > UAS-dicer2). Brightness and contrast were adjusted for optimal visibility of the localization of the proteins (B and C) Quantitation of the fluorescence intensity. Genotypes as in panel A. Quantitation of the eye discs was performed by measuring the mean fluorescence intensity in dPICK1 knockdown eye discs compared with control eye discs. Graphs represent mean fluorescence intensities of individual eye discs (each data point is for one eye disc) ± standard errors of the means (SEM); ****, P < 0.0001; ***, P ≤ 0.0003 (unpaired Student's t test).
RESULTS AND DISCUSSION
Neph proteins bind PICK1 via PDZ domain-mediated interactions.
To identify critical regulators of the Neph/nephrin cell recognition module, we generated a cDNA library from human podocytes. The cDNA library was subjected to yeast two-hybrid screens using the intracellular domains of Neph2 and Neph3 as baits. These screens identified PICK1 as a major interactor of Neph2 and Neph3 (58% of hits in the Neph2 screen, 18% in the Neph3 screen). PICK1 is a 55-kDa cytosolic protein with several protein interaction domains (45).
To confirm the interaction, we cloned PICK1 from the human podocyte cDNA library and performed coimmunoprecipitation experiments from transiently transfected HEK 293T cells. These experiments indicated that PICK1 interacts with all three Neph proteins and confirmed the interaction with nephrin (Fig. 1 A) (42). The interaction could also be demonstrated for endogenous proteins precipitated from mouse kidney and mouse brain lysates (Fig. 1B and not shown), suggesting an in vivo complex formation of Neph and PICK1 proteins. PICK1 is expressed in podocytes, as demonstrated in knockdown experiments using a human podocyte cell line (Fig. 1C and D).
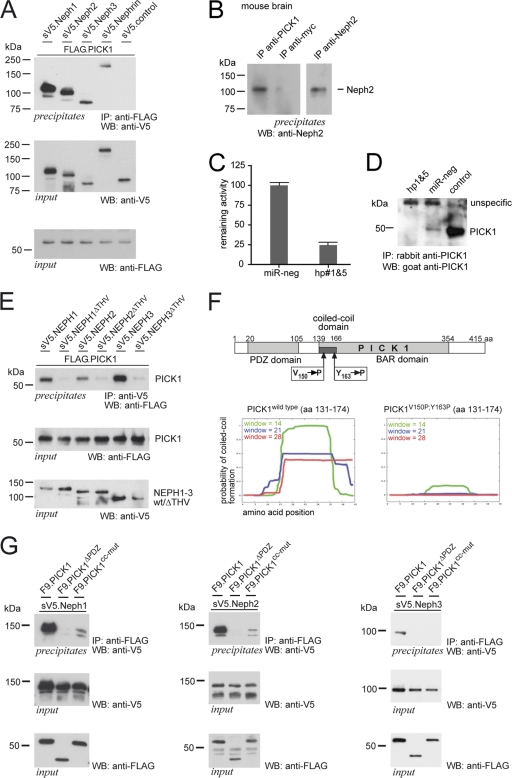
Fig. 1.
PICK1 interacts with Neph/nephrin proteins via its PDZ domain. (A) Neph1-3 and nephrin coprecipitate with PICK1. FLAG-tagged PICK1 and sV5-tagged Neph/nephrin or control protein were expressed in HEK 293T cells and precipitated with anti-FLAG antibody. Western blot (WB) analysis was performed with a V5-specific antibody. Expression levels in the lysates are shown below. (B) Endogenous PICK1 interacts with Neph2 in the brain. Protein lysates from mouse brains were subjected to immunoprecipitation (IP) with a control antibody (anti-myc) or PICK1-specific antibody, washed extensively, and immunoblotted with specific anti-Neph2 antibody. The last lane depicts Neph2 expression in the brain, shown here after Neph2 precipitation due to low expression levels in the lysate. (C) To create an artificial 3′ UTR for luciferase, the coding sequence of human PICK1 was cloned into a bicistronic vector, fusing this sequence with the coding sequence of Renilla luciferase. Renilla luciferase activity, normalized to firefly luciferase activity that served as an expression control, was used to assay the efficiency of the shRNA-mediated knockdown of PICK1. The knockdown efficiencies of chained hairpins 1 and 5 are shown. Scrambled shRNA (miR-neg) was used as a control. Experiments were performed in triplicate. Error bars represent standard deviations (SD). (D) PICK1 is expressed in human podocytes. Human podocytes were stably transduced with PICK1 hairpins 1 and 5 or miReg-neg. PICK1 was precipitated from cell lysates with a rabbit anti-PICK1 antibody and immunoblotted with a goat anti-PICK1 antibody. A HEK293T cell lysate with overexpressed untagged human PICK1 served as a control. (E) PICK1 coprecipitation with Neph proteins is dependent on the PDZ domain binding motif. FLAG-tagged PICK1 and V5-tagged Neph1-3 full-length proteins or V5-tagged Neph proteins missing the PDZ-binding motif (ΔTHV) were expressed in HEK 293T cells and precipitated with anti-V5 antibody. Western blot analysis was performed with a FLAG-specific antibody. Expression levels in the lysates are shown below. (F) Schematic representation of PICK1 showing residues V150 and Y163 within the predicted coiled-coil domain. Predictions using the program COILS suggest that mutation of these residues to proline impairs the formation of a functional coiled-coil domain (http://www.ch.embnet.org/software/COILS_form.html). aa, amino acids. (G) Neph proteins coprecipitate with PICK1 in a PDZ domain- and coiled-coil domain-dependent manner. FLAG-tagged PICK1, PICK1 missing the PDZ domain (ΔPDZ), and PICK1V150P,Y163P (cc-mut) were coexpressed with V5-tagged Neph full-length proteins and precipitated with anti-FLAG antibody. Western blot analysis was performed with a V5-specific antibody. Expression levels in the lysates are shown below.
Neph proteins contain a cytoplasmic tail that harbors a type I PDZ binding motif at the very carboxyl terminus (16). Since PICK1 contains a PDZ domain, we tested whether the Neph-PICK1 interaction is mediated via a PDZ domain-mediated mechanism. Deletion of the last three amino acids representing the PDZ domain binding motif (ΔTHV) at the carboxyl termini of Neph1, Neph2, and Neph3 strongly abrogated binding to PICK1 (Fig. 1G). Moreover, deletion of the PDZ domain in PICK1 also abrogated the interaction (Fig. 1G), indicating that the Neph-PICK1 protein complex forms via PDZ domain-mediated interactions.
In PICK1 a short stretch at the N terminus of the BAR domain, identified as a coiled-coil motif, mediates self association (3) (Fig. 1F). To test whether dimerization of PICK1 may affect interaction with Neph proteins, we mutated valine at position 150 and tyrosine at position 163 into proline. These mutations were expected to interfere with the formation of a coiled-coil structure (Fig. 1F) and thus thought to abrogate dimerization. Coimmunoprecipitation experiments using wild-type PICK1, as well as the coiled-coil mutant PICK1V150P/Y163P (cc-mut), indicated that dimerization of PICK1 through the coiled-coil motif is required for the association of PICK1 with Neph proteins (Fig. 1G). Taken together, these data suggest that functional PICK1 dimers are able to bind Neph proteins and that the interaction is mediated through the PDZ domain of PICK1 and the PDZ-binding motif within the carboxyl terminus in Neph1 to -3.
PICK1 and Neph proteins colocalize at the plasma membrane and in intracellular vesicles derived from the plasma membrane.
PICK1 has recently emerged as a central mediator in regulating endocytosis and/or recycling of neuronal receptors such as AMPA-type glutamate receptors (GluR2) and metabotropic glutamate receptors (mGluR7) (7, 14, 24, 40). In these systems PICK1 can act in either direction: GluR2 is removed from the plasma membrane in a PICK1-dependent manner, while surface localization of mGluR7 is stabilized in a PICK1-dependent manner (21, 24, 40). Given the specific interaction between Neph proteins and PICK1, we speculated that PICK1 may be involved in recycling Neph adhesion proteins. To directly test this hypothesis, we performed immunofluorescence experiments. Coexpression of PICK1 and Neph proteins in HeLa cells identified a colocalization of these proteins. Besides the expected colocalization of the expressed proteins in the endoplasmic reticulum, we found a strong colocalization at the plasma membrane and in intracellular vesicles (Fig. 2). Strikingly, colocalization of Neph1 and PICK1, as well as plasma membrane staining of PICK1, was lost in a deletion mutant lacking the PDZ domain (Fig. 2 and data not shown). Recruitment of PICK1 to the membrane is a known prerequisite for the activation of the BAR domain (23).
We next went on to test whether PICK1 may influence vesicular trafficking, targeting, or stability of Neph1 at the plasma membrane. Endocytosis assays using a pulse-chase approach did not show gross differences of overall internalization of Neph1 in the presence or absence of PICK1 (not shown). However, surface biotinylation experiments revealed that expression of PICK1 significantly increased the amount of plasma membrane Neph1 whereas plasma membrane targeting of a control plasma membrane protein (transferrin receptor) was not affected (Fig. 3). These data indicated that PICK1 acts to stabilize Neph1 at the plasma membrane.
Fig. 3.
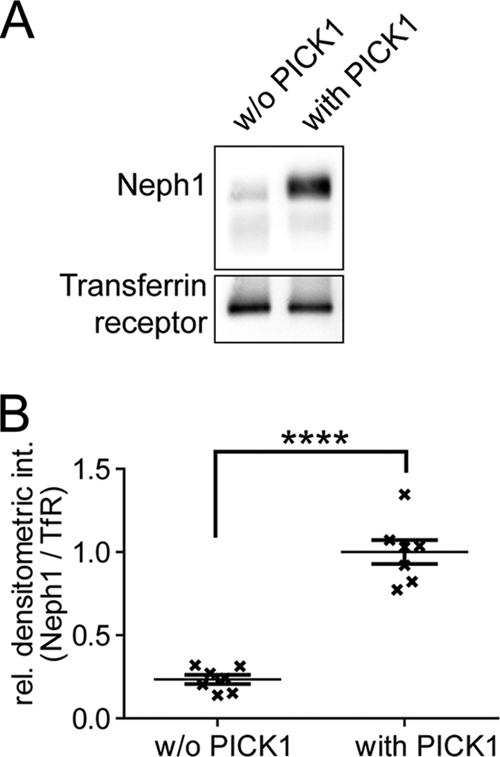
PICK1 stabilizes Neph1 at the plasma membrane. (A) sV5.Neph1 was expressed in HEK 293T cells either in the presence or in the absence of PICK1. Biotinylated surface-expressed proteins were precipitated with NeutrAvidin resin, and V5-tagged Neph1 was visualized by Western blotting using V5 antibody. Staining for the transferrin receptor served as a loading control. (B) Quantitation of the blots was performed by measuring the band intensity of Neph1 compared with the intensity of the band representing transferrin receptor. Graphs represent means ± standard errors of the means (SEM); ****, P < 0.0001 (unpaired Student's t test) (n = 7).
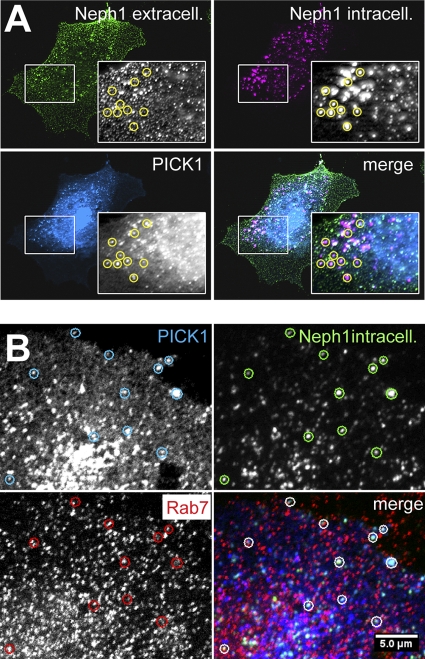
BAR domains act as sensors and/or regulators of membrane curvature (17). Since BAR domain dimers bind preferentially to liposomes of specific size (25), it is not unlikely that different BAR domains have slightly different geometries, which in turn determines the type of vesicle or invaginated membrane that is bound (14). PICK1 may therefore associate with a certain subset of vesicles. To test whether PICK1 specifically associates with Neph proteins in endocytic vesicles, we performed a fluorescence-based endocytosis assay (Fig. 4A). HeLa cells were cotransfected with sV5.Neph1 and with FLAG.PICK1. Living cells were pulse-labeled with a mouse anti-V5 antibody, washed, and transferred to 37°C for 30 min to allow internalization of protein antibody complexes. After fixation, the plasma membrane pool of Neph1 and the internalized Neph1 proteins were stained in different colors. PICK1 costaining revealed a perfect colocalization with endocytosed Neph1 originating from the plasma membrane (Fig. 4A). Interestingly, a subpopulation of the Neph1-PICK1 positive vesicles stained positive for the late endosome marker Rab7, which supported the concept that PICK1 may act to recycle Neph proteins from the late endosome and related compartments (Fig. 4B).
Fig. 4.
(A) PICK1 colocalizes with endocyosed Neph1. HeLa cells were cotransfected with sV5.Neph1 and with FLAG.PICK1. Living cells were labeled with mouse anti-V5 antibody for 30 min at 4°C. Cells were then washed and transferred to 37°C for an additional 30 min. Next, the cells were fixed and labeled with a secondary anti-mouse antibody–Alexa 488 (green fluorescent). After washing, the bound antibodies were briefly fixed. The cells were then permeabilized and labeled with an anti-mouse secondary antibody–Cy3 (red fluorescent) to mark internalized intracellular Neph1. PICK1 was stained with a rabbit anti-FLAG antibody. PICK1 clearly colocalizes with endocytosed Neph1 in intracellular vesicles (examples are marked with circles). (B) A subpopulation of PICK1/Neph1 endocytosed vesicles is positive for the late endosome marker Rab7. HeLa cells were cotransfected and treated as in panel A, except that extracellular V5 epitopes were not labeled but blocked with a anti-mouse antibody (nonfluorescent). In addition, Rab7 was stained with a rabbit anti-Rab7 antibody. A subpopulation of the vesicles is positive for endocytosed Neph1, PICK1, and Rab7.
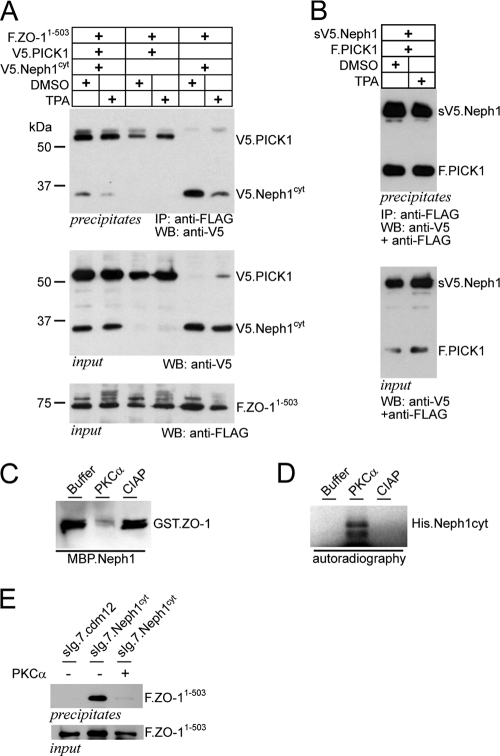
PKC activity controls preferential binding of PICK1 and release from the plasma membrane-associated multidomain protein zonula occludens 1 (ZO-1).
PICK1 has been shown to be important for aspects of neuronal biology such as hippocampal and cerebellar long-term depression, mainly by targeting neuronal receptors to endocytosis or retrieval through a PDZ domain interaction which is dependent on PKCα activity (18, 20, 37). Moreover, recent studies suggested that PICK1 and PKCα act together to stabilize mGluR7 at the plasma membrane. This stabilization required phosphorylation of mGluR7 by PKCα which in turn enhanced the binding of mGluR7 to PICK1 (40). Since phosphorylation of neuronal receptors disrupted their binding to the PDZ domain-containing scaffolding proteins, such as ABP/GRIP, but did not affect binding to PICK1 (22), we tested whether PKCα activity also influenced binding of Neph1 to the scaffolding protein zonula occludens 1 (ZO-1). We had previously demonstrated that Neph1 binds ZO-1, a large multidomain PDZ protein, similarly to the GluR2-ABP/GRIP interaction (16). ZO-1 interacts with Neph1 at the plasma membrane, providing a scaffold for Neph1 signaling activity (16, 32), and it has been suggested that Neph-dependent cell recognition requires constant remodeling of the cell junction (2, 15). Activation of PKCα with the phorbol ester PMA resulted in an inhibition of ZO-1 binding to Neph1 (Fig. 5A) without affecting the PICK1-Neph1 interaction (Fig. 5B). Of note, overexpressed PICK1 also coprecipitated with FLAG-tagged ZO-1 as shown in Fig. 5A. Repeated experiments revealed that this interaction was quite weak and occurred only with overexpressed proteins. To test whether ZO-1 is part of the endogenous PICK1 complex, we used a tandem affinity purification approach and characterized the interacting proteins (not shown). Although this study identified a series of specific partner proteins, including the published BAR domain protein ICA-69, ZO-1 was not among these candidates (not shown). Inhibition of ZO-1 binding to Neph1 was strongly enhanced in the presence of PICK1 (Fig. 5A). To test whether PKCα was able to phosphorylate the cytoplasmic tail of Neph1 to control ZO-1 binding, Neph1 and ZO-1 were recombinantly expressed and purified and subjected to PKCα phosphorylation followed by pulldown experiments. PKCα treatment resulted in direct phosphorylation of the Neph1 cytoplasmic tail (Fig. 5D) and disrupted the interaction of ZO-1 with Neph1 (Fig. 5C). Consistent with these findings, coexpression of PKCα with ZO-1 and Neph1 inhibited the interaction in HEK 293T cells (Fig. 5E). Taken together, these data show that the Neph1/ZO-1 interaction is dynamically regulated by PKCα and PICK1 and suggest that PICK1 may act in concert with PKC activity to control Neph1 recycling to the plasma membrane.
Fig. 5.
PKCα and PICK1 cooperatively regulate the Neph1/ZO-1 plasma membrane complex. (A) FLAG-tagged ZO-11–503 and V5-tagged Neph1cyt were coexpressed with PICK1 or empty vector. ZO-1 was precipitated with anti-FLAG antibody. Western blot analysis was performed with a V5-specific antibody to detect coprecipitated Neph protein. Activation of protein kinase C (PKCα) with 200 nM PMA inhibits the interaction (lane 5 and 6). Coexpression of PICK1 further augments the PKC effect on the Neph1/ZO-1 interaction (lane 1 and 2) without affecting protein levels (middle and lower panels). (B) PMA does not influence the PICK1/NEHP1 interaction. Experimental setting as in panel A with the constructs indicated. (C and D) The interaction Neph1/ZO-1 is regulated through PKCα activity. (C) Pulldown assay of bacterially expressed GST.ZO-11–111 and MBP.Neph1cyt in the presence of buffer, PKCα, or calf intestinal alkaline phosphatase (CIAP). Addition of PKCα abrogates the Neph1/ZO-1 interaction, whereas CIAP did not affect the interaction. (D) Kinase assay of bacterially expressed His.Neph1cyt in the presence of radioactively labeled ATP and buffer, PKCα or CIAP. Addition of PKCα led to phosphorylation of Neph1cyt. Autoradiograph after in vitro phosphorylation of His.Neph1cyt (E) Membrane-bound Neph1cyt (sIg7.Neph1cyt) or a control plasmid (sIg.7.cdm12) and FLAG-tagged ZO-11–503 were expressed in HEK 293T cells and precipitated with protein G. Western blot analysis was performed with a FLAG-specific antibody. Overexpression of PCKα abrograted the Neph1/ZO-1 interaction.
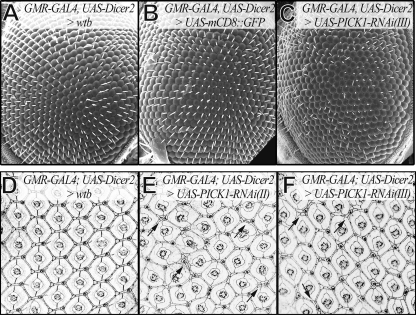
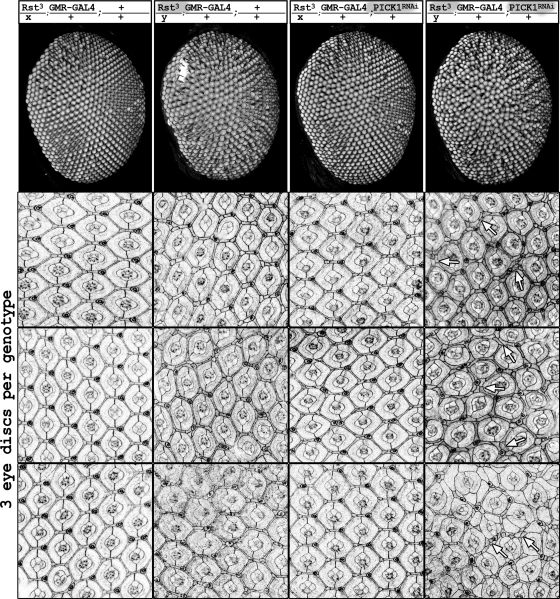
dPICK1 and Roughest, a Drosophila homolog of Neph1, cooperatively control patterning of the Drosophila compound eye.
Drosophila eye development involves cell sorting, cell death, and remodeling of cell contacts and shapes—processes that have been shown to be controlled by Neph- and nephrin-mediated adhesion and signaling (1, 10, 10, 13, 27). The primary pigment cells express nephrin proteins, Hibris (Hbs) and Sticks-and-stones (Sns), that interact with the Neph-like proteins, Roughest (Rst) and Kin-of-irre (kirre), present on neighboring cells. Heterophilic adhesion between nephrin- and Neph-like proteins results in remodeling contacts between cells to favor their contact with the pigment cells (1, 10). This sorting and culling process results in a pattern with a precise number and position of interommatidial cells (44). Double-stranded RNAs corresponding to PICK1 mRNA were expressed under the control of the eye-specific driver GMR-GAL4, together with the RNA interference (RNAi) enhancer dicer2 (8). Interestingly, knockdown of dPICK1 resulted in a rough eye phenotype closely resembling the rst phenotype (Fig. 6). The phenotype was confirmed using two independent RNAi lines (Fig. 6E and F). To demonstrate that the PICK1 phenotype was dependent on the Neph function, we tested for genetic interaction and made use of the weak allele rst3 (41). The presence of one rst3 allele in females, which have one intact copy of the rst gene on the X chromosome and one copy of the GMR-GAL4 transgene on the second chromosome, leads to a very mild phenotype. These flies demonstrated almost wild-type-patterned eyes (Fig. 7, first column). In males, however, the weak rst3 phenotype was displayed (Fig. 7, second column). The addition of RNAi-mediated knockdown of dPICK1 using GMR-GAL4, without the RNAi enhancer dicer2, did not show any dramatic effect in females carrying one rst3 allele (Fig. 7, third column). However, adding the PICK1 knockdown to the rst3 allele in males (still in the absence of dicer2) resulted in a dramatic enhancement of the phenotype, clearly confirming the genetic interaction (Fig. 7, last column). Taken together, these data indicated that PICK1, very similarly to its action in mammals, acts to support Neph/Roughest function in the fly.
Fig. 6.
Knockdown of dPICK1 causes a rst mutant-like phenotype. (A to C) Scanning electron micrographs of adult eyes. (A and B) Controls have a wild-type patterned eye. (C) GMR-GAL4-driven knockdown of dPICK1 (RNAi transgene on the third chromosome) leads to a rough eye phenotype. The RNAi enhancer dicer2 was coexpressed. (D and E) Midpupal eye imaginal discs stained for DE-cadherin. GMR-GAL4-driven dPICK1 knockdown with two independent RNAi transgenes leads to the same Rst-like phenotype. Many ommatidia are separated not by a single row but by a double row of interommatidial cells (arrows). The RNAi enhancer dicer2 was coexpressed. (D) Driver line control. (E) dPICK1 RNAi transgene on the second chromosome. (F) dPICK1 RNAi transgene on the third chromosome.
Fig. 7.
Genetic interaction between Rst and dPICK1. The genotype for each column is indicated on top. First row, images of adult eyes. The weak phenotypes of the Rst3 allele (2nd column) and dPICK1 RNAi knockdown (3rd column) enhance each other to a severe phenotype if both lesions are present in males (rightmost column). Pupal eye imaginal discs (3 individual discs per genotype, 42 h APF) stained with anti-Pyd illustrate the same effect. Clear sorting defects are visible in the eye imaginal discs of these animals. Many ommatidia are not separated by a single row but by a double row of interommatidial cells (arrows).
To examine the PICK1 function in the fly eye in greater detail, we analyzed Neph- and nephrin-like proteins, collectively known as IRM proteins (10), during eye development and stained pupal eye discs at a morphogenetically highly active stage (18 to 19 h APF). Interestingly, dPICK1 knockdown did not result in an obvious subcellular redistribution of IRM proteins in cells of the eye discs (Fig. 8). However, we realized that the fluorescence intensity was generally weaker in the dPICK1 knockdown eye discs than in controls. Quantification of the fluorescence intensity confirmed this effect (Fig. 8B and C). Knockdown of dPICK1 in the fly eye resulted in a significant decrease of Rst and Kirre expression at the plasma membrane but did not affect the expression of other proteins (E-cadherin, Pyd, SNS) (Fig. 8B and C). Since the IRM proteins are required for cell sorting at this stage of development (27), this reduction could well explain the rough eye phenotype.
Taken together, this work highlights the important role of PICK1 in regulating the Neph-dependent cell recognition module and Neph-mediated steps in morphogenesis. Members of the Neph/nephrin cell recognition module are conserved across species (10). They are involved in the recognition of different cell types and are mediating cell contacts of opposing cell membranes. Thus, it is highly conceivable that similar mechanisms may regulate the dynamic function of the Neph/nephrin cell recognition module in many species. We observed an increased Neph level at the plasma membrane in the presence of PICK1 in a cell culture biotinylation assay and a decreased Neph level in dPICK1 knockdown Drosophila eye discs. Thus, PICK1 seems to affect the rate of turnover and recycling of Neph adaptor proteins to affect their signaling abilities. In light of this hypothesis, it is important to consider that Neph proteins do not only function as adhesion molecules. They are involved in signaling for the regulation of cellular processes such as remodeling of cell contacts, organizing cell shape changes and cell sorting during Drosophila eye development (1, 10, 27), cell fusion in Drosophila muscle development (4, 38), correct wiring of the Drosophila brain (30, 39), localization and stabilization of presynaptic sites in C. elegans (34, 35), the development and maintenance of the kidney podocyte (9, 19), and the wiring of olfactory sensory neurons in the olfactory bulb of the mouse (33). It is obvious that these processes require the tightly regulated and timed expression of the Neph/nephrin proteins at the cell surface. The PDZ/BAR domain protein PICK1 has been shown to regulate the timed expression and/or recycling of associated proteins at the plasma membrane (22, 43). In conclusion, PICK1 was identified as an essential novel regulator of Neph/nephrin proteins, most likely by influencing their availability to mediate signal transduction at the plasma membrane.
ACKNOWLEDGMENTS
We thank Stefanie Keller, Bettina Maar, Ruth Herzog, Manuela Hochberger, Christina Engel, and Charlotte Meyer for excellent technical assistance and members of the laboratories for helpful discussions. We also thank Ross Cagan, the Vienna Drosophila RNAi Center, and Developmental Studies Hybridoma Bank, University of Iowa, for flies and antibodies.
H.C.R. received funding through the Deutsche Forschungsgemeinschaft (RE2246/1-1, RE2246/2-1, and SFB832), the Deutsche Nierenstiftung, and the Köln Fortune Program. T.B.H. received funding through the Deutsche Forschungsgemeinschaft and through exc 294. T.B. received funding through the Deutsche Forschungsgemeinschaft (BE2212, SFB572, and SFB635).
Footnotes
Published ahead of print on 20 June 2011.
REFERENCES
- 1. Bao S., Cagan R. L. 2005. Preferential adhesion mediated by Hibris and Roughest regulates morphogenesis and patterning in the Drosophila eye. Dev. Cell 8:925–935 [DOI] [PubMed] [Google Scholar]
- 2. Benzing T. 2004. Signaling at the slit diaphragm. J. Am. Soc. Nephrol. 15:1382–1391 [DOI] [PubMed] [Google Scholar]
- 3. Boudin H., Craig A. M. 2001. Molecular determinants for PICK1 synaptic aggregation and mGluR7a receptor coclustering. J. Biol. Chem. 276:30270–30276 [DOI] [PubMed] [Google Scholar]
- 4. Bour B. A., Chakravarti M., West J. M., Abmayr S. M. 2000. Drosophila SNS, a member of the immunoglobulin superfamily that is essential for myoblast fusion. Genes Dev. 14:1498–1511 [PMC free article] [PubMed] [Google Scholar]
- 5. Cagan R. L. 2009. Principles of Drosophila eye differentiation. Curr. Top. Dev. Biol. 89:115–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cagan R. L. 2003. The signals that drive kidney development: a view from the fly eye. Curr. Opin. Nephrol. Hypertens. 12:11–17 [DOI] [PubMed] [Google Scholar]
- 7. Dev K. K., et al. 2000. PICK1 interacts with and regulates PKC phosphorylation of mGLUR7. J. Neurosci. 20:7252–7257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dietzl G., et al. 2007. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448:151–156 [DOI] [PubMed] [Google Scholar]
- 9. Donoviel D. B., et al. 2001. Proteinuria and perinatal lethality in mice lacking NEPH1, a novel protein with homology to NEPHRIN. Mol. Cell. Biol. 21:4829–4836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fischbach K.-F., et al. 2009. The irre cell recognition module (IRM) proteins. J. Neurogenet. 23:48–67 [DOI] [PubMed] [Google Scholar]
- 11. Frost A., Unger V. M., Camilli P. D. 2009. The BAR domain superfamily: membrane-molding macromolecules. Cell 137:191–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gerke P., et al. 2006. Neuronal expression and interaction with the synaptic protein CASK suggest a role for Neph1 and Neph2 in synaptogenesis. J. Comp. Neurol. 498:466–475 [DOI] [PubMed] [Google Scholar]
- 13. Grillo-Hill B. K., Wolff T. 2009. Dynamic cell shapes and contacts in the developing Drosophila retina are regulated by the Ig cell adhesion protein hibris. Dev. Dyn. 238:2223–2234 [DOI] [PubMed] [Google Scholar]
- 14. Hanley J. G. 2008. PICK1: a multi-talented modulator of AMPA receptor trafficking. Pharmacol. Ther. 118:152–160 [DOI] [PubMed] [Google Scholar]
- 15. Huber T. B., Benzing T. 2005. The slit diaphragm: a signaling platform to regulate podocyte function. Curr. Opin. Nephrol. Hypertens. 14:211–216 [DOI] [PubMed] [Google Scholar]
- 16. Huber T. B., et al. 2003. The carboxyl terminus of Neph family members binds to the PDZ domain protein zonula occludens-1. J. Biol. Chem. 278:13417–13421 [DOI] [PubMed] [Google Scholar]
- 17. Itoh T., De Camilli P. 2006. BAR, F-BAR (EFC) and ENTH/ANTH domains in the regulation of membrane-cytosol interfaces and membrane curvature. Biochim. Biophys. Acta 1761:897–912 [DOI] [PubMed] [Google Scholar]
- 18. Jo J., et al. 2008. Metabotropic glutamate receptor-mediated LTD involves two interacting Ca2+ sensors, NCS-1 and PICK1. Neuron. 60:1095–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kestilä M., et al. 1998. Positionally cloned gene for a novel glomerular protein-nephrin-is mutated in congenital nephrotic syndrome. Mol. Cell 1:575–582 [DOI] [PubMed] [Google Scholar]
- 20. Kim C.-H., Chung H. J., Lee H.-K., Huganir R. L. 2001. Interaction of the AMPA receptor subunit GluR2/3 with PDZ domains regulates hippocampal long-term depression. Proc. Natl. Acad. Sci. U. S. A. 98:11725–11730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lin D.-T., Huganir R. L. 2007. PICK1 and Phosphorylation of the glutamate receptor 2 (GluR2) AMPA receptor subunit regulates GluR2 recycling after NMDA receptor-induced internalization. J. Neurosci. 27:13903–13908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lu W., Ziff E. B. 2005. PICK1 interacts with ABP/GRIP to regulate AMPA receptor trafficking. Neuron 47:407–421 [DOI] [PubMed] [Google Scholar]
- 23. Madsen K. L., et al. 2008. Membrane localization is critical for activation of the Pick1 bar domain. Traffic 9:1327–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Perez J. L., et al. 2001. PICK1 targets activated protein kinase Cα to AMPA receptor clusters in spines of hippocampal neurons and reduces surface levels of the AMPA-type glutamate receptor subunit 2. J. Neurosci. 21:5417–5428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Peter B. J., et al. 2004. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science 303:495–499 [DOI] [PubMed] [Google Scholar]
- 26. Rasband W. 1997. ImageJ. U.S. National Institutes of Health, Bethesda, MD: http://imagej.nih.gov/ij/ [Google Scholar]
- 27. Reiter C., Schimansky T., Nie Z., Fischbach K.-F. 1996. Reorganization of membrane contacts prior to apoptosis in the Drosophila retina: the role of the IrreC-rst protein. Development 122:1931–1940 [DOI] [PubMed] [Google Scholar]
- 28. Saleem M. A., et al. 2002. A conditionally immortalized human podocyte cell line demonstrating nephrin and podocin expression. J. Am. Soc. Nephrol. 13:630–638 [DOI] [PubMed] [Google Scholar]
- 29. Schermer B., et al. 2006. The von Hippel-Lindau tumor suppressor protein controls ciliogenesis by orienting microtubule growth. J. Cell Biol. 175:547–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schneider T., et al. 1995. Restricted expression of the irreC-rst protein is required for normal axonal projections of columnar visual neurons. Neuron 15:259–271 [DOI] [PubMed] [Google Scholar]
- 31. Sellin L., et al. 2003. NEPH1 defines a novel family of podocin interacting proteins. FASEB J. 17:115–117 [DOI] [PubMed] [Google Scholar]
- 32. Seppa M. J., Johnson R. I., Bao S., Cagan R. L. 2008. Polychaetoid controls patterning by modulating adhesion in the Drosophila pupal retina. Dev. Biol. 318:1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Serizawa S., et al. 2006. A neuronal identity code for the odorant receptor-specific and activity-dependent axon sorting. Cell 127:1057–1069 [DOI] [PubMed] [Google Scholar]
- 34. Shen K., Bargmann C. I. 2003. The immunoglobulin superfamily protein SYG-1 determines the location of specific synapses in C. elegans. Cell 112:619–630 [DOI] [PubMed] [Google Scholar]
- 35. Shen K., Fetter R. D., Bargmann C. I. 2004. Synaptic specificity is generated by the synaptic guidepost protein SYG-2 and its receptor, SYG-1. Cell 116:869–881 [DOI] [PubMed] [Google Scholar]
- 36. Staudinger J., Zhou J., Burgess R., Elledge S. J., Olson E. N. 1995. PICK1: a perinuclear binding protein and substrate for protein kinase C isolated by the yeast two-hybrid system. J. Cell Biol. 128:263–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Steinberg J. P., et al. 2006. Targeted in vivo mutations of the AMPA receptor subunit GluR2 and its interacting protein PICK1 eliminate cerebellar long-term depression. Neuron 49:845–860 [DOI] [PubMed] [Google Scholar]
- 38. Strünkelnberg M., et al. 2001. rst and its paralogue kirre act redundantly during embryonic muscle development in Drosophila. Development 128:4229–4239 [DOI] [PubMed] [Google Scholar]
- 39. Sugie A., Umetsu D., Yasugi T., Fischbach K.-F., Tabata T. 2010. Recognition of pre- and postsynaptic neurons via nephrin/NEPH1 homologs is a basis for the formation of the Drosophila retinotopic map. Development 137:3303–3313 [DOI] [PubMed] [Google Scholar]
- 40. Suh Y. H., et al. 2008. Corequirement of PICK1 binding and PKC phosphorylation for stable surface expression of the metabotropic glutamate receptor mGluR7. Neuron 58:736–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tanenbaum S. B., Gorski S. M., Rusconi J. C., Cagan R. L. 2000. A screen for dominant modifiers of the irreC-rst cell death phenotype in the developing Drosophila retina. Genetics 156:205–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tossidou I., et al. 2010. Podocytic PKC-alpha is regulated in murine and human diabetes and mediates nephrin endocytosis. PLoS One 5:e10185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Williams M. E., Wu S. C.-Y., McKenna W. L., Hinck L. 2003. Surface expression of the netrin receptor UNC5H1 is regulated through a protein kinase C-interacting protein/protein kinase-dependent mechanism. J. Neurosci. 23:11279–11288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wolff T., Ready D. F. 1993. Pattern formation in the Drosophila retina, p. 1277–1325 In Bate M., Arias A. M. (ed.), The development of Drosophila melanogaster. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 45. Xu J., Xia J. 2007. Structure and function of PICK1. Neurosignals 15:190–201 [DOI] [PubMed] [Google Scholar]
- 46. Zenker M., Machuca E., Antignac C. 2009. Genetics of nephrotic syndrome: new insights into molecules acting at the glomerular filtration barrier. J. Mol. Med. 87:849–857 [DOI] [PubMed] [Google Scholar]