Abstract
Yeast prions are self-propagating protein conformations that transmit heritable phenotypes in an epigenetic manner. The recently identified yeast prion [SWI+] is an alternative conformation of Swi1, a component of the evolutionarily conserved SWI/SNF chromatin-remodeling complex. Formation of the [SWI+] prion results in a partial loss-of-function phenotype for Swi1. The amino-terminal region of Swi1 is dispensable for its normal function but is required for [SWI+] formation and propagation; however, the precise prion domain (PrD) of Swi1 has not been elucidated. Here, we define the minimal Swi1 PrD as the first 37 amino acids of the protein. This region is extremely asparagine rich but, unexpectedly, contains no glutamine residues. This unusually small prion domain is sufficient for aggregation, propagation, and transmission of the [SWI+] prion. Because of its unusual size and composition, the Swi1 prion domain defined here has important implications for describing and identifying novel prions.
INTRODUCTION
Prion proteins are misfolded, infectious conformers of native host proteins. A unique feature of prions is their ability to self-perpetuate by recruiting soluble, correctly folded protein isomers and inducing them to adopt the prion conformation (38). The mammalian prion (PrPSc), the pathogen responsible for a group of transmissible neurodegenerative diseases known as prion diseases, is composed of misfolded conformers of the host protein PrPC (39, 56). Yeast prions are formed from native yeast proteins that adopt misfolded conformations and are transmitted as cytoplasmic traits during mating and budding (8, 52, 53, 58). Almost all yeast prions are composed of amyloid aggregates, which recruit soluble protein isomers into insoluble aggregates, which then assemble into fibrils (3, 42, 47, 57). Fragmentation of these fibrils produces prion “seeds,” which are transmitted in the cytoplasm, accounting for the heritable and infectious nature of yeast prions (11, 17, 19, 22).
While PrP is the only known mammalian prion protein, multiple yeast prions have been identified. [PSI+] (9) and [URE3] (23) were the first recognized prions in yeast and represent the prion forms of Sup35 and Ure2, respectively (56). Subsequent work has led to the identification of [PIN+], [β], [SWI+], [OCT+], [MOT3+], [GAR+], and, most recently, [NSI+] and [ISP+] (1, 5, 13, 15, 32, 40, 41, 44, 48). One of the common features shared by almost all yeast prion proteins is the modular nature of the protein. Most yeast prion proteins can be divided into two major regions: a functional domain, which is sufficient for native protein activity, and a transferrable prion domain (PrD), which is sufficient for formation and propagation of the prion (24, 48). Interestingly, two reports suggest that the PrDs of Sup35 and Ure2 may subtly affect protein activity (45, 54), although this phenomenon has not been described for other known yeast prions. In general, yeast PrDs are highly enriched in glutamine (Q) and asparagine (N) residues and have very few charged or hydrophobic residues (1, 10, 43). Several lines of evidence suggest that Q and N contribute similarly to the formation of amyloid fibers: aggregates formed from purified poly(Q), poly(N), or the Q/N-rich Sup35 PrD form analogous polar-zipper structures (34); both Q and N can form amide stacks, a strong stabilizing force for amyloid structure (30, 33); and compositional analysis of known yeast PrDs suggests similar propensities for Q and N in prion-forming domains (51). Furthermore, screening the yeast proteome for Q/N-rich proteins has directly resulted in the discovery of several known yeast prion proteins and has generated a list of multiple promising candidates (1, 26).
Swi1 was originally identified as a yeast prion candidate due to its high Q/N content, as well as its ability to promote [PSI+] formation (12, 15). Swi1 is a component of the multisubunit SWI/SNF chromatin-remodeling complex, which is responsible for the transcriptional regulation of approximately 6% of the yeast genome (36, 49). Deletion of SWI/SNF components results in defects in the mating type switch and in the metabolism of nonglucose carbon sources (28, 29). Formation of [SWI+] mimics knockdown of Swi1, which results in phenotypes that include poor sporulation and a reduced ability to grow on raffinose as a sole carbon source (15). Previous work from our laboratory has shown that these phenotypes are most likely attributable to the amino-terminal Q/N-rich region of Swi1 for several reasons: first, deletion of the first 523 amino acids of the protein has no significant effect on carbon source usage, a readily assayable trait regulated by Swi1 (15); second, a peptide comprising the first 327 amino acids of Swi1 forms fibrils in vitro that can induce [SWI+] in wild-type cells (14); and finally, deletion of the genomic sequence coding for the Swi1 amino terminus results in loss of [SWI+] (14). Based on these data, we hypothesized that the minimal Swi1 PrD lay within the first 327 amino acids of the Swi1 protein.
We show here that a small region of 37 amino acid residues at the extreme amino terminus of Swi1 is sufficient for maintenance, propagation, and transmission of the [SWI+] prion. Swi11-37 decorates [SWI+] aggregates formed from full-length Swi1 and maintains its aggregated form when Swi1 is eliminated. Aggregated Swi11-37 can transmit the prion fold back to full-length Swi1, both in the same cell, by coexpression and to endogenous protein in a wild-type background by cytoplasmic mixing. Finally, Swi11-37-transmitted [SWI+] is curable by treatment with millimolar concentrations of guanidine hydrochloride (GdnHCl). These data strongly indicate that Swi11-37 represents the minimal Swi1 prion domain.
MATERIALS AND METHODS
Yeast strains and media.
All yeast strains used in this study were grown and maintained according to methods outlined in the Cold Spring Harbor manual (6). Strains were propagated on rich (yeast-peptone-dextrose [YPD]) or synthetic complete (SC) medium, dropping out certain amino acids to select for plasmid retention, and supplemented with 5 mM GdnHCl or 5-fluoroorotic acid (5-FOA) where indicated. For the raffinose growth phenotype, glucose was replaced with raffinose and the medium was supplemented with 1 μg/ml antimycin (Sigma). Plates were grown at 30°C for 3 days unless otherwise noted.
The BY4741 [SWI+] [PIN+], BY4741 swi1Δ, and c10B-49 [swi−] [pin−] MATα strains were described previously (14, 20). The BY4741 [SWI+] [pin−] strain was generated by transforming BY4741 [swi−] [pin−] cells with cell extract prepared from 74D-694 [SWI+] [pin−] TRP1::TEF1-RNQ1CFP using a procedure similar to what was described previously (14). To create 74D-694 [SWI+] [pin−] TRP1::TEF1-RNQ1CFP, 74D-694 [SWI+] [pin−] cells were transformed with a HindIII-linearized p304TEF-RNQ1CFP to integrate the TEF1-RNQ1CFP cassette into the TRP1 locus (RNQ1CFP was used as a marker for monitoring the prion status of Rnq1.
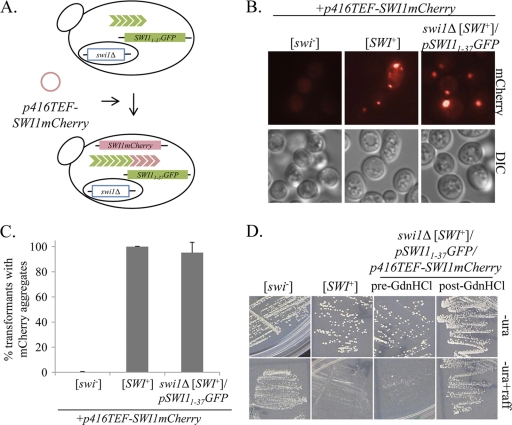
To construct the BY4741 swi1Δ [SWI+]/p416TEF-SWI1 strain, the [SWI+] prion was introduced to BY4741 swi1Δ by extract transformation from a c10B [SWI+] kar1-1 strain generated in our laboratory (14, 15). Log-phase c10B [SWI+] kar1-1 cells were suspended in 100 μl STC buffer (1 M sorbitol, 10 mM Tris-HCl, 10 mM CaCl2) and vortexed 4 times for 1 min each time in a bead beater (Biospecs) to lyse them. Fifty microliters of the crude lysates was transformed into the BY4741 swi1Δ/p416TEF-SWI1 strain using a standard yeast transformation protocol (see below), with p413TEF as a transformation marker. His+ Ura+ transformants were screened for the presence of [SWI+] by streaking them onto medium containing raffinose as a sole carbon source. Transformants showing reduced growth on raffinose were then transformed with p415TEF-SWI11-46YFP. Those isolates that exhibited Swi11-46-yellow fluorescent protein (YFP) aggregation (which forms aggregates in [SWI+] cells [see Fig. 3A]) were treated with 5 mM GdnHCl to cure the prion. Only isolates showing GdnHCl-curable aggregation and reduced growth on raffinose-containing media were considered [SWI+]. One of these isolates was designated swi1Δ [SWI+]/p416TEF-SWI1 and used for the experiments described here.
Fig. 3.
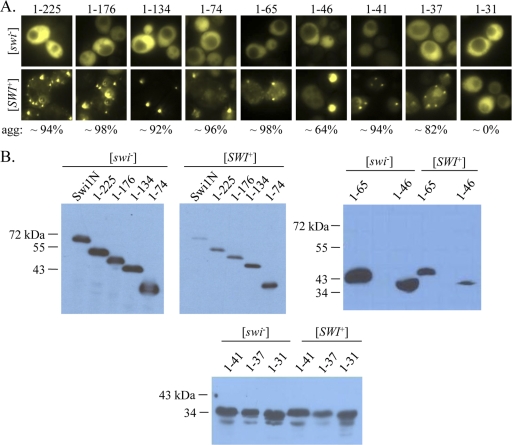
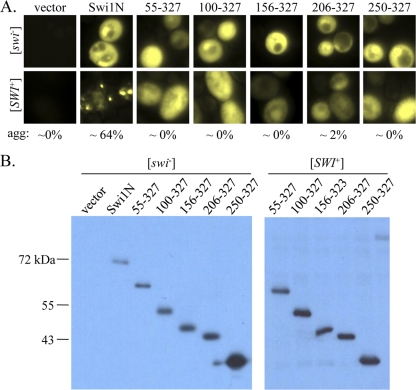
The extreme amino-terminus of Swi1 is sufficient for aggregation in [SWI+] cells. (A) Microscopy images (×100) of [swi−] and [SWI+] cells expressing Swi1N carboxyl-terminal truncation mutants fused to YFP. Ten to 20 individual transformants were imaged for each truncation; representative images are shown. The numbers indicate average percentages of [SWI+] cells with aggregates (agg). (B) Western blot of the Swi1N carboxyl-terminal-truncation YFP fusions. A polyclonal anti-GFP antibody was used to detect the fusion proteins.
Plasmid construction.
All constructs expressing truncation mutants were derived from p416TEF-NYFP, which was described previously (14). To construct the amino-terminal truncations, fragments of the SWI1 gene were amplified from genomic DNA using unique forward primers containing SpeI restriction sites and a common reverse primer containing a BamHI restriction site. Carboxyl-terminal truncations were amplified using a common forward primer containing an SpeI site and unique reverse primers containing BamHI sites. The amplified fragments were digested, purified, and ligated into the SpeI and BamHI sites of p416TEF-NYFP, replacing the Swi1N fragment with the truncated version. The correct insertion and sequence of each fragment were verified by sequencing.
To make p415TEF-SWI11-37GFP, the sequence encoding green fluorescent protein (GFP) was amplified from pFA6a-kanMX6-P3nmt1-GFP (2) using a forward primer containing a BamHI restriction site and a reverse primer containing an XhoI restriction site. The amplified fragment was digested, purified, and ligated into the BamHI and XhoI sites of p416TEF-SWI11-37YFP, yielding the plasmid p416TEF-SWI11-37GFP. The fragment encoding Swi11-37-GFP was cut out of p416TEF-SWI11-37GFP using SpeI and XhoI and ligated into the same sites in p415TEF (27).
To make p416TEF-SWI1mCherry, the sequence encoding mCherry was amplified from pmCherry-C1, a derivative of pEGFP-C1 (Clontech), using forward and reverse primers containing XhoI restriction sites. The amplified fragment was digested, purified, and ligated into the XhoI site of p416TEF-SWI1 (14). The correct orientation of the mCherry fragment was determined by restriction analysis. Finally, the SWI1 stop codon was mutated by site-directed mutagenesis to fuse in frame with mCherry. The correct insertion and sequence of mCherry was confirmed by sequencing.
To create the plasmid p304TEF-RNQ1CFP, a 1-kb fragment containing the GPD promoter and the CYC1 terminator cassette was digested from p426GPD (ATCC) and ligated into pRS304 to give rise to p304GPDCYC1 through the KpnI/SacI sites. The 1.3-kb CFP fragment from pECFP-N1 (Clontech) was used to replace the GFP fragment of p316CUP1-RNQ1GFP (26) through SacII/NaeI. The resulting plasmid, p316CUP1-RNQ1CFP, was partially digested with HpaI/BamHI to give rise to a 2.1-kb RNQ1CFP fragment, which was ligated into p304GPDCYC1 that was predigested with HincII/BamHI, yielding p304GPD-RNQ1CFP. Subsequently, the GPD promoter of p304GPD-RNQ1CFP was replaced by the TEF1 promoter from p416TEF-SWI1 (20) through the SacI/SpeI sites, resulting in p304TEF-RNQ1CFP.
Table 1 shows a list of primers used for this study.
Table 1.
Primers used in this study
| Primer name | Primer sequencea | Resulting plasmid |
|---|---|---|
| Swi1N rev | 5′-TGGATCCCGGGCAGAGAACA-3′ | Amino-terminal truncations |
| Swi1N fwd | 5′-ATCTAGAACTAGTATGGTTTCTTTAATTT-3′ | Carboxy-terminal truncations |
| 55 fwd | 5′-TACTACTAGTATGAACAATACTAATAACAACAATACGAATACC-3′ | p416TEF-SWI155-327YFP |
| 100 fwd | 5′-TACTACTAGTATGAATAACAGTAACACGGTAGCCTCC-3′ | p416TEF-SWI1100-327YFP |
| 156 fwd | 5′-TACTACTAGTATGAACAGCCTGTCTCCTCAGGC-3′ | p416TEF-SWI1156-327YFP |
| 206 fwd | 5′-TACTACTAGTATGATCACCAATGTTCAATCCATCAG-3′ | p416TEF-SWI1206-327YFP |
| 250 fwd | 5′-TACTACTAGTATGTCTAACCAGCTTATTTCTAATTACGC-3′ | p416TEF-SWI1250-327YFP |
| 225 rev | 5′-GGTGGATCCTTTGTGTTCGG-3′ | p416TEF-SWI11-225YFP |
| 176 rev | 5′-GGTGGATCCAAATTGGAAGAATC-3′ | p416TEF-SWI11-176YFP |
| 134 rev | 5′-GGTGGATCCTTAGCAGCTTTTC-3′ | p416TEF-SWI11-134YFP |
| 74 rev | 5′-GGTGGATCCTGAAAATCGTCTAC-3′ | p416TEF-SWI11-74YFP |
| 65 rev | 5′-GGTGGATCCTTGGTATTCGTATTGTTGTTATTAGTATTG-3′ | p416TEF-SWI11-65YFP |
| 46 rev | 5′-GTTGGATCCCTATTGTTGTTATTGGTATTATTAGCAGG-3′ | p416TEF-SWI11-46YFP |
| 41 rev | 5′-GTTGGATCCGTATTATTAGCAGGATTATTATTATT-3′ | p416TEF-SWI11-41YFP |
| 37 rev | 5′-GTTGGATCCGGATTATTATTATTATTAGTATTATTATT-3′ | p416TEF-SWI11-37YFP |
| 31 rev | 5′-GTTGGATCCGTATTATTATTATTAGTATTATTGTTATT-3′ | p416TEF-SWI11-31YFP |
| GFP fwd | 5′-TATGGATCCATCTTTAATTAACAGTAAAGGAGAAGAAC-3′ | p416TEF-SWI11-37GFP |
| GFP rev | 5′-ATACTCGAGCTATTTGTATAGTTCATCCATGCCATG-3′ | p416TEF-SWI11-37GFP |
| mCherry fwd | 5′-TATCTCGAGGTGAGCAAGGGCGAGGAG-3′ | p416TEF-SWI1mCherry |
| mCherry rev | 5′-TATCTCGAGCTACTTGTACAGCTCGTCCATG-3′ | p416TEF-SWI1mCherry |
The underlined nucleotides indicate restriction sites used for cloning.
Yeast transformation.
Yeast cells were transformed according to a protocol adapted from the Cold Spring Harbor manual (6). Briefly, log-phase cells were collected by centrifugation, resuspended in LiPEG transformation solution (0.2 M lithium acetate, 40% polyethylene glycol [PEG] 3500), and supplemented with 2 μl of single-stranded salmon sperm carrier DNA (10 mg/ml; Sigma). One microliter of plasmid DNA (Qiagen miniprep) was then added, and the transformation mixture was vortexed briefly and incubated at 42°C for 15 to 45 min before being plated to selective medium. The plates were incubated at 30°C for 3 days until colonies appeared.
Microscopy.
Microscopy images were taken with a Zeiss Axiovert 200 epifluorescence microscope. Samples were viewed with a 100× objective and filters specific for differential interference contrast (DIC), YFP/GFP, or mCherry. Images were captured with an AxioCam HRm camera (Zeiss) using Axiovision AC (Zeiss).
Cytoduction.
Donor and recipient cells for cytoductions were grown separately in selective media overnight. Three hundred microliters of each overnight culture was then combined with 600 μl YPD rich medium, and the mixture was incubated at 30°C with agitation for 4 to 6 h. Aliquots of the cytoduction mixture were plated to selective medium and grown at 30°C for 3 days. Individual colonies were picked to selective medium to confirm correct nuclear markers.
Immunoblotting.
Samples for immunoblots were prepared by alkaline lysis as follows. One milliliter of an overnight yeast culture was pelleted, resuspended in 100 μl of 0.1 M NaOH, and incubated at room temperature for 5 min. The cells were then pelleted and resuspended in 100 μl 2× Laemmli sample buffer. The resuspended samples were boiled for 5 min and sonicated for 15 1-s pulses. The lysates were then spun down to pellet debris, and 5 μl of the supernatants was immediately loaded on a 7.5% (for larger peptides) or 15% (for smaller peptides) SDS-PAGE gel. The gels were transferred to polyvinylidene difluoride (PVDF) membranes and blotted with 1:2,500 mouse anti-GFP primary antibody (JL-8; Clontech) and 1:2,500 horseradish peroxidase (HRP)-conjugated rabbit anti-mouse secondary antibody (Thermo Scientific). The resulting chemiluminescence was detected using ECL Western Blotting Detection Reagents (Amersham).
Reverse transcription-PCR.
Total RNA was isolated from overnight cultures using the RNeasy Mini kit (Qiagen). cDNA was synthesized using the SuperScript III First-Strand Synthesis System (Invitrogen) according the manufacturer's instructions. Twenty nanograms of cDNA was used as the template for PCR amplification with primers specific for the Swi1 C terminus (Swi1 RT fwd, 5′-TCTTCGCGCCAGAGTTAGG-3′, and Swi1 RT rev, 5′-CCGAGTATTGCCAAGGAGTC-3′). The PCR products were run on a 2% agarose gel with the GeneRuler 100-bp Plus DNA ladder (Fermentas) and imaged on a Bio-Rad gel imager (Bio-Rad).
RESULTS
Swi11-37 decorates prion aggregates in [SWI+] cells.
We previously showed that the carboxyl-terminal portion of Swi1 is sufficient for readily assayable Swi1 functions, since it complements the carbon source usage defect of genomic SWI1 deletion (15). We also showed that in vitro-formed Swi11-327 fibrils can induce de novo [SWI+] and that chromosomal deletion of the coding region for Swi11-327 results in [SWI+] loss (14). Furthermore, Alberti et al. demonstrated that a peptide comprising a considerable portion of the Q/N-rich Swi1 N terminus (Swi11-385) forms SDS-resistant aggregates in vivo and amyloid fibers in vitro (1). Thus, we hypothesized that the minimal Swi1 PrD was located within the first 327 amino acids of Swi1 (here designated Swi1N). We reasoned that a domain that could propagate [SWI+] would also aggregate in [SWI+] cells; thus, we first aimed to identify the minimal region of Swi1N that could decorate [SWI+] aggregates.
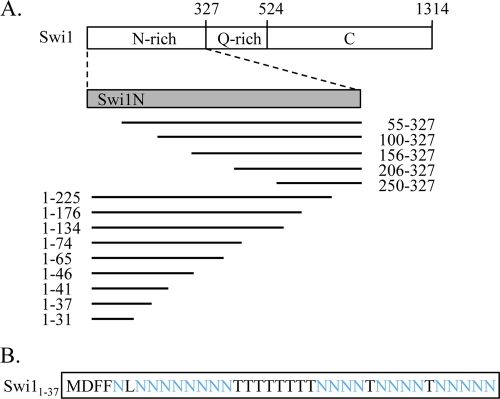
To address this question, we constructed a series of Swi1N truncations fused to a C-terminal YFP reporter (Fig. 1). These fusion proteins were then expressed in isogenic [swi−] [PIN+] and [SWI+] [PIN +] cells from a constitutive TEF1 promoter. All fusion proteins were expressed at the expected molecular weights in both [swi−] and [SWI+] cells (Fig. 2 B and 3 B). The Swi1N-YFP control and all of the truncation fusion proteins exhibited diffuse fluorescence in [swi−] cells (Fig. 2A and 3A), indicating that no aggregation occurred as a result of overexpression. The positive control, Swi1N-YFP, formed fluorescent puncta in [SWI+] cells, as expected (Fig. 2A). Deletion of 55 or more amino acids from the Swi1N amino terminus, however, resulted in loss of aggregation: all fusion proteins representing Swi1N amino-terminal truncation exhibited diffuse fluorescence in [SWI+] cells (Fig. 2A). This suggests that the extreme amino terminus of the protein is important for aggregation. Indeed, truncation of Swi1N from the carboxyl terminus revealed that only the extreme amino terminus is required for aggregation, as a fusion protein representing only the first 37 amino acids of Swi1 exhibited robust aggregation in [SWI+] cells (Fig. 3A). Further truncation to 31 amino acids resulted in complete loss of aggregation (Fig. 3A). Taken together, these data indicate that the extreme amino terminus of Swi1, a region of 37 amino acids, is sufficient for decoration of Swi1 aggregates in [SWI+] cells.
Fig. 1.
Truncation mutants used in this study. (A) Schematic of the Swi1 protein and truncation peptides used in the aggregation study (not to scale). Swi1N was previously shown to decorate [SWI+] aggregates (14) and is used as a positive control. The numbers indicate amino acids. All truncation mutants are fused with a C-terminal YFP reporter. (B) Amino acid sequence of Swi11-37, the region identified as the minimal Swi1 PrD in this study. Asparagine residues are highlighted in blue.
Fig. 2.
Deletion of the extreme amino terminus of Swi1 abolishes aggregation in [SWI+] cells. (A) Microscopy images (×100) of [swi−] and [SWI+] cells expressing Swi1N amino-terminal truncation mutants fused to YFP. Ten to 20 individual transformants were imaged for each truncation; representative images are shown. The numbers indicate the average percentages of [SWI+] cells with aggregates (agg). (B) Western blot of the Swi1N amino-terminal-truncation YFP fusions. A polyclonal anti-GFP antibody was used to detect the fusion proteins.
Swi11-37 maintains the prion conformation in the absence of Swi1.
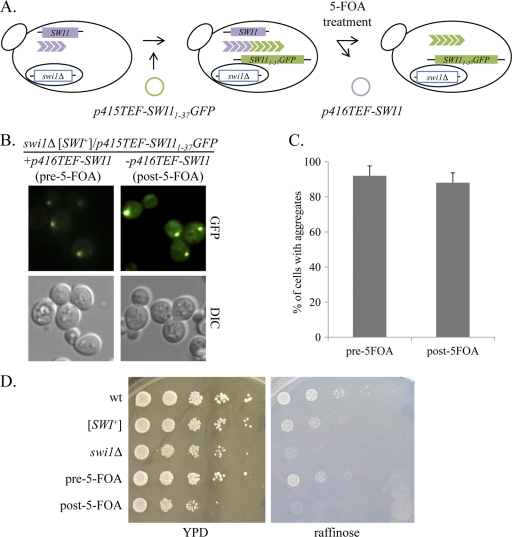
Given that Swi11-37 decorates [SWI+] aggregates in cells expressing Swi1, we next asked whether Swi11-37 was able to maintain the aggregated prion conformation by itself (in the absence of full-length Swi1). To test this, we needed to construct a [SWI+] strain coexpressing Swi1 and Swi11-37 in which Swi1 expression could be “turned off,” since maintenance of a prion requires a continuous supply of soluble prion protein. We started by transforming a swi1Δ strain with p416TEF-SWI1. The p416TEF-SWI1 plasmid is maintained by Ura+ selection, so it can be counterselected for by treatment with 5-FOA (4). Next, we introduced the [SWI+] prion by extract transformation (see Materials and Methods for details), resulting in the strain swi1Δ [SWI+] [pin−]/p416TEF-SWI1 (Fig. 4 A). This strain was then transformed with p415TEF-SWI11-37GFP (Fig. 4A). As expected, the fusion protein formed fluorescent foci in these cells, due to Swi11-37-GFP decoration of preexisting [SWI+] aggregates formed from full-length Swi1 (Fig. 4B and C). These cells exhibited an intermediate or slow growth phenotype on raffinose as a sole carbon source (defined as Raf+/−), due to the presence of the prion (Fig. 4D, pre-5-FOA). The swi1Δ [SWI+] [pin−]/p416TEF-SWI1/p415TEF-SWI11-37GFP cells were then treated with 5-FOA to select for cells that had lost the URA3-based plasmid p416TEF1-SWI1 (Fig. 4A). After 5-FOA treatment, Swi11-37-GFP still formed fluorescent foci (Fig. 4B and C), suggesting that it is able to maintain the [SWI+] conformation in the absence of full-length Swi1. To confirm that these cells were in fact cured of the p416TEF-SWI1 plasmid, they were spotted to medium containing raffinose as the sole source of carbon before and after treatment with 5-FOA. If p416TEF-SWI1 was successfully eliminated by 5-FOA treatment, then the lack of Swi1 should result in no growth at all on raffinose-containing medium (Raf−). Thus, 5-FOA treatment should result in a transition from a Raf+/− phenotype to a Raf− phenotype. This is indeed what we saw: before 5-FOA treatment, the swi1Δ [SWI+] [pin−]/p415TEF-SWI11-37GFP/p416TEF-SWI1 strain was Raf+/−, and after 5-FOA treatment, the resulting swi1Δ [SWI+] [pin−]/p415TEF-SWI11-37GFP strain was Raf− (Fig. 4D). The absence of SWI1 expression was also confirmed by reverse transcription-PCR (data not shown). Thus, we conclude that Swi11-37 maintains the [SWI+] prion fold on its own in the absence of full-length Swi1 protein.
Fig. 4.
Swi11-37 maintains the [SWI+] phenotype in the absence of Swi1. (A) Schematic of the experimental design. A swi1Δ [SWI+]/p416TEF-SWI1 strain was transformed with p415TEF-SWI11-37GFP. The cells were treated with 5-FOA to select for those that had lost p416TEF-SWI1. (B) Microscopy images (×100) of cells before and after treatment with 5-FOA. Ten colonies were imaged for both the pre-5-FOA and post-5-FOA groups; representative images are shown. (C) Quantification of cells with aggregates. Fifty individual cells were counted from one representative colony. The error bars indicate standard deviations. (D) Loss of SWI1 was confirmed by the growth phenotype on medium containing raffinose as the sole carbon source. Log-phase cells were serially diluted 1:10 and spotted to rich medium (YPD) or raffinose medium (raffinose). Ten colonies were tested; representative images are shown. Wild-type (wt), [SWI+], and swi1Δ controls are included.
Swi11-37 can transmit the [SWI+] prion conformation to soluble Swi1.
Since Swi11-37 is able to maintain the [SWI+] aggregation phenotype by itself, we next asked whether the peptide can transmit the prion fold back to full-length Swi1. To test this, the swi1Δ [SWI+]/p415TEF-SWI11-37GFP strain generated as described above was transformed with p416TEF-SWI1mCherry (Fig. 5). Using a tagged version of the full-length protein enabled us to test for growth on raffinose as well as aggregation, two important [SWI+] phenotypes. When imaged for mCherry fluorescence, approximately 95% of swi1Δ [SWI+]/p415TEF-SWI11-37GFP/p416TEF-SWI1mCherry cells exhibited a punctate pattern of fluorescence, a percentage comparable to that of the [SWI+] control strain (Fig. 5B and C). This indicates that Swi11-37 alone can induce aggregation of soluble, full-length Swi1 protein. These cells were then spotted to medium containing raffinose as the sole carbon source. The swi1Δ [SWI+]/p415TEF-SWI11-37GFP/p416TEF-SWI1mCherry strain showed a Raf+/− phenotype, indicating that Swi11-37 can in fact transmit the [SWI+] phenotype to full-length Swi1 (Fig. 5D). Finally, the swi1Δ [SWI+]/p415TEF-SWI11-37GFP/p416TEF-SWI1mCherry strain was passaged twice on medium containing 5 mM GdnHCl, which cures the [SWI+] prion (15). After GdnHCl treatment, growth of these cells on raffinose was restored to levels comparable to that of [swi−] cells (Fig. 5D), indicating that the Raf+/− phenotype was based on a prion-like phenomenon.
Fig. 5.
Swi11-37 can transmit [SWI+] to full-length Swi1. (A) Schematic of the experimental design. A swi1Δ [SWI+]/p415TEF-SWI11-37GFP strain was transformed with p416TEF-SWI1-mCherry. (B) Microscopy images (×100) of cells expressing Swi1mCherry. [swi−] and [SWI+] control cells are included. Ten transformants each from 3 individual transformations were imaged for mCherry aggregation (swi1Δ [SWI+]/pSWI11-37GFP); five transformants from three individual transformations were imaged for [swi−] and [SWI+] control strains. Representative images are shown. (C) Quantification of transformants with aggregates. One hundred cells from three individual transformants were counted for each group. The error bars indicate standard deviations. (D) One transformant from each of three individual transformations was tested for reduced growth on raffinose-containing medium (-ura+raff). These same cells were also passaged twice on selective medium containing 5 mM GdnHCl and then restreaked to raffinose-containing (-ura+raff) and dropout (-ura) media. The images shown are representative of all three colonies tested.
Swi11-37 can transmit [SWI+] to endogenous Swi1.
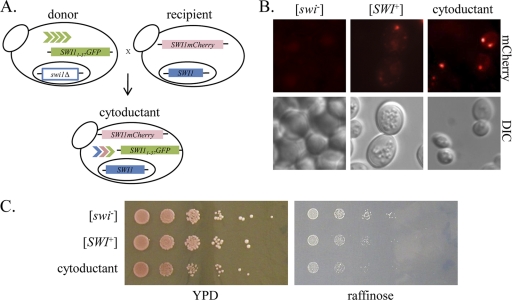
The results described above demonstrate that aggregated Swi11-37 can propagate the [SWI+] prion conformation within a carefully designed system. We next asked whether Swi11-37 can also transmit [SWI+] to soluble Swi1 in a naïve strain with a wild-type genetic background. To address this question, we cytoduced the swi1Δ [SWI+]/p415TEF-SWI11-37GFP strain generated as described above (donor) to a [swi−]/p416TEF-SWI1mCherry reporter strain (recipient) (Fig. 6 A). The recipient strain carries the kar1-1 mutation, which prevents nuclear fusion; the resulting “cytoductants” therefore contain either the donor or recipient nucleus and the cytoplasmic contents of both donor and recipient cells. Seven cytoductants containing only the recipient nucleus were tested for Swi1mCherry aggregation and reduced growth on raffinose-containing media. When these cells were imaged for mCherry fluorescence, five out of seven cytoductants showed Swi1mCherry aggregation (one representative cytoductant with aggregation is shown in Fig. 6B). The expression of the Swi1mCherry fusion protein is low, which may account for the lack of visible aggregates in two of the seven cytoductants. We previously showed that the expression of exogenous Swi1 is tightly controlled by the cell, resulting in very low expression levels, even from strong constitutive promoters (14). All seven cytoductants showed reduced growth on raffinose (Raf+/−), indicating transmission of the [SWI+] conformation to Swi1mCherry (one representative cytoductant is shown in Fig. 6C). Thus, we conclude that Swi11-37 aggregates can transmit [SWI+] conformation to wild-type cells expressing endogenous Swi1 by cytoplasmic mixing.
Fig. 6.
Swi11-37 can transmit [SWI+] to endogenous Swi1. (A) Schematic of experimental design. A swi1Δ [SWI+]/p415TEF-SWI11-37GFP strain (donor) was cytoduced to a [swi−] kar1-1/p416TEF-SWI1-mCherry strain (recipient). Cytoductants were selected for on media lacking uracil and leucine. Authentic cytoductants were selected based on the presence of the recipient cell nuclear marker (Met+) and the absence of the donor cell nuclear marker (Lys+). (B) Microscopy images (×100) of cytoductant, as well as [swi−] and [SWI+] control strains containing p416TEF-SWI1mCherry. Seven cytoductants were imaged; a representative image of a cytoductant exhibiting Swi1-mCherry aggregation is shown. (C) Log-phase cells were serially diluted 1:10 and spotted to YPD and raffinose media. Seven cytoductants were tested; representative results are shown.
DISCUSSION
The PrD of a yeast prion protein is the minimal region required for propagation and transmission of the prion. In this study, we have demonstrated that the first 37 amino acids of Swi1 represent the minimal PrD of the protein. Swi11-37 forms prion-like aggregates in the cytoplasm of [SWI+] cells, but not [swi−] cells, indicating that it can decorate preexisting [SWI+] aggregates (Fig. 3A). The peptide remains aggregated in the absence of full-length Swi1, indicating that by itself it is sufficient for propagation of the prion fold (Fig. 4). Aggregated Swi11-37 is also able to induce full-length Swi1 to adopt the prion conformation, thus demonstrating that it is capable of transmitting [SWI+] (Fig. 5). Finally, Swi11-37 can transmit the [SWI+] conformation to endogenous Swi1 by cytoplasmic mixing (Fig. 6). Due to the lack of a positive selection system for [SWI+] cells, we were unable to test whether Swi11-37 is sufficient for de novo induction of [SWI+] in naïve cells. While Swi11-31 did not form visible aggregates in [SWI+] cells, we cannot exclude the possibility that smaller oligomeric forms could propagate the prion.
The minimal Swi1 PrD defined here is unique among known PrDs in that it contains no glutamine residues (Fig. 1B). The yeast PrDs defined to date are highly enriched in both glutamine and asparagine, and screens used to detect new prion candidates have been based on parameters considering both amino acids to be present in large quantities (1, 26). Our results show that an asparagine-rich, glutamine-free PrD is capable of propagating and transmitting a yeast prion. Interestingly, Swi11-37 also lacks glycine residues (which are biased for in mammalian PrP and the PrDs of Sup35 and Rnq1 [18]), as well as tyrosine and serine residues (which are mildly biased for in some yeast prion PrDs [18]). In a recent genome-wide screen for yeast prion protein candidates, the authors noted that the asparagine-rich candidates they identified were more likely to be amyloidogenic than glutamine-rich candidates (1). Our results correlate well with this observation. While one study did show that a polyasparagine peptide [poly(N104)] could form SDS-insoluble, Hsp104-dependent aggregates in yeast, this peptide does not demonstrate the heritability that is characteristic of prions and, at 104 amino acids, was much larger than the asparagine-rich PrD described here (35).
Another notable feature of the Swi1 PrD is its highly homogeneous amino acid composition: 60% of the residues comprising this domain are asparagine (N) and 30% are threonine (T), with non-N or -T residues representing only 10% of the total composition, making the Swi1 PrD unique in its pronounced enrichment for a single residue. While a role for threonine in prion formation has not been previously described, one study did note enrichment for T residues in the Ure2 PrD (50). This observation, and the high prevalence of T residues in the Swi1 PrD, suggests that T may also play a role in prion formation and propagation. Interestingly, the Ure2 PrD has a greater content of N versus Q than the well-defined Sup35 and Rnq1 PrDs (10). This raises the intriguing possibility that Swi1 and Ure2 may represent a subclass of prion proteins with specialized requirements for PrD composition. We anticipate that the identification and characterization of novel prion proteins in the future will shed light on this hypothesis.
One of the most surprising findings from this study is the strikingly small size of the Swi1 PrD. At a mere 37 amino acids, this domain is smaller than any prion domain defined to date: Sup351-61 can propagate [PSI+] in vitro (7, 21), although up to 83 amino acids are required for propagation of weaker [PSI+] variants (46); the Ure2 PrD was defined as amino acids 1 to 65 (25), and the smallest defined region of Rnq1 required for [PIN+] decoration and transmission is even larger (55). However, to our knowledge, no genetic evidence has been reported showing that any of the above-mentioned PrDs are able to propagate their corresponding prions in vivo. This means that Swi11-37 is the smallest domain defined to date that is capable of propagating and transmitting a yeast prion in vivo. A recent study by Fiumara et al. identified a marked enrichment for coiled-coil motifs in prion proteins and their interactors (16). The authors suggested that a coiled-coil structure may explain the pathogenic threshold for poly(Q) length in the Huntington protein (Htt): a pathogenic expansion region of 36 or more Q residues contains five heptads (a repeat typical of coiled-coil domains), and greater expansion results in an increased tendency to polymerize. Since the Swi1 PrD aggregation threshold is virtually the same length, it is tempting to speculate that coiled-coil structure may also play a role in [SWI+] formation. If this is true, then further truncation to 31 amino acids would eliminate the fifth heptad, which could account for its inability to propagate the prion conformation. However, prediction software (Coils and Paricoil2) indicates a very low probability of coiled-coil structure within this region (E. T. Crow, unpublished data), and the amino acid sequence does not exhibit the heptad repeat motif characteristic of coiled-coil regions (Fig. 1B). Indeed, all the regions of Swi1 that have a high probability of forming coiled coils are downstream of Swi11-327, a domain that we have already shown to be crucial for [SWI+] propagation (14). If coiled-coil structure does not in fact play a role in [SWI+] aggregation, then the fact that removal of six additional residues from the C terminus of Swi11-37 renders the peptide unable to decorate [SWI+] aggregates suggests that these residues might be critical for interactions between monomeric and aggregated Swi1. It is possible that these residues are localized within the core region of [SWI+] amyloids. Further structural studies will shed light on the importance of individual residues within Swi11-37 in prionogenesis.
The location of the Swi1 PrD is also surprising: since up to 536 amino acids can be deleted from the amino terminus of the protein without significantly affecting readily assayable Swi1 functions (15), this suggests that the intervening region between the prion domain and the carboxyl terminus may be largely without function. One study demonstrated that the second “quarter” of Swi1 (amino acids 329 to 657) facilitates SWI/SNF interaction with transcriptional activators but also showed that this activity is largely redundant with that of a region of Snf5, another member of the SWI/SNF complex (37), indicating that the middle region of Swi1 is nonessential. A recent paper by Kadnar et al. suggests that multiple prion determinants may exist within a yeast prion domain (20). Our deletion analysis clearly demonstrates that the extreme N terminus of Swi1 is essential for aggregation (since deletion of the first 55 amino acids completely abolishes decoration of [SWI+] aggregates); however, we cannot rule out the possibility that downstream regions may have a modifying effect on [SWI+] formation and propagation.
In conclusion, we have shown here that the minimal prion domain of Swi1 consists of the 37 amino acids at the extreme amino terminus of the protein. This domain is unusual in both its size and composition and as such has significant implications for understanding the nature of prion aggregates and for the identification of novel prion proteins.
ACKNOWLEDGMENTS
We thank T. Volpe (Northwestern University) and D. Chakravarti (Northwestern University) for plasmids.
This work was supported by a grant from the U.S. National Institutes of Health (R01NS056086) to L.L.
Footnotes
Published ahead of print on 13 June 2011.
REFERENCES
- 1. Alberti S., Halfmann R., King O., Kapila A., Lindquist S. 2009. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 137:146–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bahler J., et al. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14:943–951 [DOI] [PubMed] [Google Scholar]
- 3. Benkemoun L., et al. 2006. Methods for the in vivo and in vitro analysis of [Het-s] prion infectivity. Methods 39:61–67 [DOI] [PubMed] [Google Scholar]
- 4. Boeke J. D., Trueheart J., Natsoulis G., Fink G. R. 1987. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154:164–175 [DOI] [PubMed] [Google Scholar]
- 5. Brown J. C., Lindquist S. 2009. A heritable switch in carbon source utilization driven by an unusual yeast prion. Genes Dev. 23:2320–2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Burke D., Dawson D., Stearns T. 2000. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 7. Chang H. Y., Lin J. Y., Lee H. C., Wang H. L., King C. Y. 2008. Strain-specific sequences required for yeast [PSI+] prion propagation. Proc. Natl. Acad. Sci. U. S. A. 105:13345–13350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chernoff Y. O. 2001. Mutation processes at the protein level: is Lamarck back? Mutat. Res. 488:39–64 [DOI] [PubMed] [Google Scholar]
- 9. Cox B. 1965. [PSI], a cytoplasmic suppressor of super-suppression in yeast. Heredity 20:505–521 [Google Scholar]
- 10. Crow E., Du Z., Li L. 2008. New insights into prion biology from the novel [SWI+] system. Prion 2:141–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Derdowski A., Sindi S. S., Klaips C. L., DiSalvo S., Serio T. R. 2010. A size threshold limits prion transmission and establishes phenotypic diversity. Science 330:680–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Derkatch I. L., Bradley M. E., Hong J. Y., Liebman S. W. 2001. Prions affect the appearance of other prions: the story of [PIN+]. Cell 106:171–182 [DOI] [PubMed] [Google Scholar]
- 13. Derkatch I. L., Bradley M. E., Zhou P., Chernoff Y. O., Liebman S. W. 1997. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics 147:507–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Du Z., Crow E. T., Kang H. S., Li L. 2010. Distinct subregions of Swi1 manifest striking differences in prion transmission and SWI/SNF function. Mol. Cell. Biol. 30:4644–4655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Du Z., Park K. W., Yu H., Fan Q., Li L. 2008. Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae. Nat. Genet. 40:460–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fiumara F., Fioriti L., Kandel E. R., Hendrickson W. A. 2010. Essential role of coiled coils for aggregation and activity of Q/N-rich prions and PolyQ proteins. Cell 143:1121–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Glover J. R., Lum R. 2009. Remodeling of protein aggregates by Hsp104. Protein Pept. Lett. 16:587–597 [DOI] [PubMed] [Google Scholar]
- 18. Harrison P. M., Gerstein M. 2003. A method to assess compositional bias in biological sequences and its application to prion-like glutamine/asparagine-rich domains in eukaryotic proteomes. Genome Biol. 4:R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Higurashi T., Hines J. K., Sahi C., Aron R., Craig E. A. 2008. Specificity of the J-protein Sis1 in the propagation of 3 yeast prions. Proc. Natl. Acad. Sci. U. S. A. 105:16596–16601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kadnar M. L., Articov G., Derkatch I. L. 2010. Distinct type of transmission barrier revealed by study of multiple prion determinants of Rnq1. PLoS Genet. 6:e1000824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. King C. Y., Diaz-Avalos R. 2004. Protein-only transmission of three yeast prion strains. Nature 428:319–323 [DOI] [PubMed] [Google Scholar]
- 22. Kryndushkin D. S., Alexandrov I. M., Ter-Avanesyan M. D., Kushnirov V. V. 2003. Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J. Biol. Chem. 278:49636–49643 [DOI] [PubMed] [Google Scholar]
- 23. Lacroute F. 1971. Non-Mendelian mutation allowing ureidosuccinic acid uptake in yeast. J. Bacteriol. 206:519–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li L., Lindquist S. 2000. Creating a protein-based element of inheritance. Science 287:661–664 [DOI] [PubMed] [Google Scholar]
- 25. Masison D. C., Maddelein M. L., Wickner R. B. 1997. The prion model for [URE3] of yeast: spontaneous generation and requirements for propagation. Proc. Natl. Acad. Sci. U. S. A. 94:12503–12508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Michelitsch M. D., Weissman J. S. 2000. A census of glutamine/asparagine-rich regions: implications for their conserved function and the prediction of novel prions. Proc. Natl. Acad. Sci. U. S. A. 97:11910–11915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mumberg D., Muller R., Funk M. 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156:119–122 [DOI] [PubMed] [Google Scholar]
- 28. Neigeborn L., Carlson M. 1984. Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics 108:845–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Neigeborn L., Carlson M. 1987. Mutations causing constitutive invertase synthesis in yeast: genetic interactions with snf mutations. Genetics 115:247–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nelson R., et al. 2005. Structure of the cross-beta spine of amyloid-like fibrils. Nature 435:773–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Reference deleted. [Google Scholar]
- 32. Patel B. K., Gavin-Smyth J., Liebman S. W. 2009. The yeast global transcriptional co-repressor protein Cyc8 can propagate as a prion. Nat. Cell Biol. 11:344–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Perutz M. F., Finch J. T., Berriman J., Lesk A. 2002. Amyloid fibers are water-filled nanotubes. Proc. Natl. Acad. Sci. U. S. A. 99:5591–5595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Perutz M. F., Pope B. J., Owen D., Wanker E. E., Scherzinger E. 2002. Aggregation of proteins with expanded glutamine and alanine repeats of the glutamine-rich and asparagine-rich domains of Sup35 and of the amyloid beta-peptide of amyloid plaques. Proc. Natl. Acad. Sci. U. S. A. 99:5596–5600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Peters T. W., Huang M. 2007. Protein aggregation and polyasparagine-mediated cellular toxicity in Saccharomyces cerevisiae. Prion 1:144–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Peterson C. L., Dingwall A., Scott M. P. 1994. Five SWI/SNF gene products are components of a large multisubunit complex required for transcriptional enhancement. Proc. Natl. Acad. Sci. U. S. A. 91:2905–2908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Prochasson P., Neely K. E., Hassan A. H., Li B., Workman J. L. 2003. Targeting activity is required for SWI/SNF function in vivo and is accomplished through two partially redundant activator-interaction domains. Mol. Cell 12:983–990 [DOI] [PubMed] [Google Scholar]
- 38. Prusiner S. B. 1982. Novel proteinaceous infectious particles cause scrapie. Science 216:136–144 [DOI] [PubMed] [Google Scholar]
- 39. Prusiner S. B. 1998. Prions. Proc. Natl. Acad. Sci. U. S. A. 95:13363–13383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Roberts B. T., Wickner R. B. 2003. Heritable activity: a prion that propagates by covalent autoactivation. Genes Dev. 17:2083–2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rogoza T., et al. 2010. Non-Mendelian determinant [ISP+] in yeast is a nuclear-residing prion form of the global transcriptional regulator Sfp1. Proc. Natl. Acad. Sci. U. S. A. 107:10573–10577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ross E. D., Minton A., Wickner R. B. 2005. Prion domains: sequences, structures and interactions. Nat. Cell Biol. 7:1039–1044 [DOI] [PubMed] [Google Scholar]
- 43. Ross E. D., Toombs J. A. 2010. The effects of amino acid composition on yeast prion formation and prion domain interactions. Prion 4:60–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Saifitdinova A. F., et al. 2010. [NSI (+)]: a novel non-Mendelian nonsense suppressor determinant in Saccharomyces cerevisiae. Curr. Genet. 56:467–478 [DOI] [PubMed] [Google Scholar]
- 45. Shewmaker F., Mull L., Nakayashiki T., Masison D. C., Wickner R. B. 2007. Ure2p function is enhanced by its prion domain in Saccharomyces cerevisiae. Genetics 176:1557–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shkundina I. S., Kushnirov V. V., Tuite M. F., Ter-Avanesyan M. D. 2006. The role of the N-terminal oligopeptide repeats of the yeast Sup35 prion protein in propagation and transmission of prion variants. Genetics 172:827–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shorter J., Lindquist S. 2005. Prions as adaptive conduits of memory and inheritance. Nat. Rev. Genet. 6:435–450 [DOI] [PubMed] [Google Scholar]
- 48. Sondheimer N., Lindquist S. 2000. Rnq1: an epigenetic modifier of protein function in yeast. Mol. Cell 5:163–172 [DOI] [PubMed] [Google Scholar]
- 49. Sudarsanam P., Iyer V. R., Brown P. O., Winston F. 2000. Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 97:3364–3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Thual C., et al. 2001. Stability, folding, dimerization, and assembly properties of the yeast prion Ure2p. Biochemistry 40:1764–1773 [DOI] [PubMed] [Google Scholar]
- 51. Toombs J. A., McCarty B. R., Ross E. D. 2010. Compositional determinants of prion formation in yeast. Mol. Cell. Biol. 30:319–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tuite M. F. 2004. Cell biology: the strain of being a prion. Nature 428:265–267 [DOI] [PubMed] [Google Scholar]
- 53. Uptain S. M., Lindquist S. 2002. Prions as protein-based genetic elements. Annu. Rev. Microbiol. 56:703–741 [DOI] [PubMed] [Google Scholar]
- 54. Urakov V. N., et al. 2006. N-terminal region of Saccharomyces cerevisiae eRF3 is essential for the functioning of the eRF1/eRF3 complex beyond translation termination. BMC Mol. Biol. 7:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Vitrenko Y. A., Pavon M. E., Stone S. I., Liebman S. W. 2007. Propagation of the [PIN+] prion by fragments of Rnq1 fused to GFP. Curr. Genet. 51:309–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wickner R. B. 1994. [URE3] as an altered Ure2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science 264:566–569 [DOI] [PubMed] [Google Scholar]
- 57. Wickner R. B., Dyda F., Tycko R. 2008. Amyloid of Rnq1p, the basis of the [PIN+] prion, has a parallel in-register beta-sheet structure. Proc. Natl. Acad. Sci. U. S. A. 105:2403–2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wickner R. B., et al. 1999. Prions in Saccharomyces and Podospora spp.: protein-based inheritance. Microbiol. Mol. Biol. Rev. 63:844–861 [DOI] [PMC free article] [PubMed] [Google Scholar]