Abstract
The diverse transcriptional patterns that distinguish metazoan cells are specified by multifactor regulatory complexes containing distinct combinations of factors that assemble at genomic response elements. To investigate combinatorial control, we examined a set of glucocorticoid receptor (GR)-regulated genes bearing nearby regulatory complexes that include both GR and the coregulator Brm, an ATPase subunit of the Swi/Snf chromatin remodeler. We analyzed how GR and Brm affect each other's occupancy and activity by utilizing glucocorticoid treatment and Brm knockdown to modulate GR-mediated transcriptional regulation and Brm-mediated chromatin remodeling, respectively. GR occupancy and activity were altered differentially by Brm knockdown at specific activated and repressed primary GR target genes. Brm knockdown decreased GR occupancy at activated Brm-dependent genes, whereas we identified two classes of repressed genes, at which Brm knockdown either increased or decreased GR occupancy. Glucocorticoid treatment increased both Brm occupancy and chromatin accessibility at Brm-dependent and Brm-independent GR-regulated genes. However, chromatin remodeling activity decreased after Brm knockdown only at genes with Brm-dependent transcription. Our study revealed multiple distinct patterns of GR and Brm interdependence. Thus, monitoring as few as two factors within regulatory complexes is sufficient to reveal functionally distinct assemblies, providing an analytical method for gaining insights into combinatorial regulation.
INTRODUCTION
Transcriptional regulation in multicellular organisms is a sophisticated process governed by combinatorial control (6, 23, 51). According to this view, multifactor regulatory complexes, comprised of DNA-binding regulatory factors and associated coregulators, assemble at specific genomic response elements to control gene transcription (50, 52); occupancy at particular response elements is dependent on cell type and physiological conditions (2, 45). Despite the remarkable specificity of gene expression programs, most regulatory factors are broadly expressed. Thus, by assembling factors in variable combinations at different genomic sites, multifactor regulatory complexes direct diverse transcriptional outputs. This implies that even factors common to multiple regulatory complexes may function in response element-specific ways. However, the rules governing the assembly and function of multifactor regulatory complexes remain unknown. As a first step toward understanding response element-specific combinatorial regulation, we focused on a set of differentially regulated genes that utilize two factors: the glucocorticoid receptor (GR) and the Swi/Snf ATPase Brm.
GR is a ligand-dependent transcriptional regulatory factor that is involved in the precise control of metabolism, development, and the inflammatory response. Upon glucocorticoid binding, GR translocates to the nucleus, where it interacts with primary glucocorticoid response elements (GREs), functional regions of DNA that confer a glucocorticoid response by mediating GR activity through receptor occupancy. GR selectively binds specific genomic regions, which vary depending on cell type (S. B. Cooper and K. R. Yamamoto, unpublished data). Binding to slightly different sequences alters GR conformation and transcriptional activity (27), through differential recruitment of coregulators with distinct activities (26). The composition of these combinatorial GR-assembled regulatory complexes contributes to the nature of the transcriptional response (24).
The Swi/Snf chromatin remodeling complex is a well-characterized GR coregulator. Swi/Snf proteins from budding yeast were shown to interact with rat GR in vitro and to facilitate the transcriptional activation of GR-mediated reporter constructs (53). More recent studies have demonstrated that GR-mediated activation of a chromatin template requires Swi/Snf remodeling activity through the recruitment of its ATPases (Brm or Brg1), which are highly homologous and mutually exclusive subunits (13, 20, 44). Swi/Snf has also been shown to direct GR-mediated transcription in vivo (18, 31). The Brm and Brg1 ATPases regulate transcription by sliding or ejecting nucleosomes, which alters the accessibility of binding sites to regulatory factors such as GR (7, 8, 30, 38).
In this study, we investigated the mechanisms of combinatorial control by which a regulatory factor (GR) functions with a known coregulator (Brm) at specific genes to direct diverse transcriptional responses. Since GR modulates transcription and Brm remodels chromatin structure, these activities can be separately probed through glucocorticoid control of GR and knockdown of Brm. Therefore, we independently examined how GR and Brm affect the activity and occupancy of each other, focusing on transcriptional start sites (TSSs) and GREs.
MATERIALS AND METHODS
Cell culture.
A549 cells were grown in Dulbecco's modified Eagle's medium (DMEM; UCSF Cell Culture Facility) with 5% fetal bovine serum (FBS; Gemini) at 37°C in 8% CO2 in a humidified incubator. One day before hormone treatment, the medium was replaced with DMEM containing 5% charcoal-stripped FBS (Omega). For all experiments, confluent cells were treated with either 0.05% ethanol vehicle or 100 nM dexamethasone (dex; Sigma) dissolved in ethanol. 293T cells were grown in DMEM with 10% FBS and 1× penicillin-streptomycin (UCSF Cell Culture Facility) at 37°C in 5% CO2 in a humidified incubator.
Lentiviral shRNAs.
Lentiviral short hairpin-mediated RNAs (shRNAs) were used to knock down the expression of Brm as described previously (27). The lentiviruses were produced with PCR products containing the U6 promoter and the shRNA of interest, using the following target sequences: Scramble, AAGGGTAGGTTCGACTAGCAGGACTCT; shBrm, AAGCTGACTCAGGTCTTGAACAC; and shBrm 3′ untranslated region (3′UTR), AAGCGCTATTGAATATTGCAATC. The PCR products were subcloned into the pHRCMVPUROWSin18 vector and cotransfected with pMD.G1 and pCMVDR8.91 (generously provided by Didier Trono, University of Geneva) (29) into 293T cells by using Lipofectamine 2000 (Invitrogen). After 48 h and 72 h, the culture medium containing the shRNAs expressing lentivirus was harvested and filtered through 0.45-μm filters. For virus infection, A549 cells were incubated with medium-diluted virus supernatant supplemented with 8 μg/ml Polybrene (Millipore) for 24 h. Cells were then selected with 4 μg/ml puromycin (InvivoGen), and knockdown was confirmed after approximately 1 week.
Immunoblotting.
Protein lysates from equal quantities of cells were separated on 4 to 15% Tris-HCl SDS-PAGE gels and transferred to polyvinylidene difluoride membranes (Millipore) using semidry transfer (Bio-Rad). Membranes were probed with anti-Brm (1:200; BD Transduction), anti-GR (1:4,000; N499 generated by R. M. Nissen, B. Darimont, and K. R. Yamamoto [unpublished data]), and anti-beta-actin (1:2,000; Sigma) primary antibodies, followed by the appropriate secondary antibodies conjugated with horseradish peroxidase (for Brm) or fluorescent dyes (for GR and beta-actin). Proteins were detected by chemiluminescence (ECL Plus; Amersham) or fluorescence (Odyssey; Li-Cor), respectively.
RNA isolation, reverse transcription, and real-time qPCR.
The RNA isolation, reverse transcription, and real-time quantitative PCR (qPCR) were performed essentially as previously described (32). After 4 h dex or ethanol treatment, total RNA was isolated from A549 cells using QIAshredder and RNeasy Mini kits (Qiagen). Random-primed cDNA was prepared from 750 ng total RNA using the ProtoScript M-MulV first-strand cDNA synthesis kit (New England BioLabs). Approximately 1/50 of the resultant cDNA was used per 35-μl qPCR mixture containing 0.7 U of AmpliTaq Gold DNA polymerase (Applied Biosystems), 1× PCR buffer II (Applied Biosystems), 2.5 mM MgCl2 solution (Applied Biosystems), 0.15 mM deoxynucleoside triphosphate (dNTP; Invitrogen), 0.2× SYBR green I dye (Invitrogen), 0.4× ROX reference dye (Invitrogen), and 0.5 μM each primer. Primer pairs were described previously (3, 36, 41, 47) or were designed using Primer3 (http://frodo.wi.mit.edu/primer3), and those that efficiently amplified a single product of the expected size were used for qPCR. Primer sequences are provided in Table S1 in the supplemental material. qPCR was performed with a 7300 real-time PCR system (Applied Biosystems) by using standard cycling conditions (10 min at 95°C, followed by 42 cycles of 20 s at 95°C, 30 s at 57°C, and 30 s at 72°C). Data were analyzed using the CT method (Applied Biosystems) and normalized to the expression of the RPL19 gene. Each value was calculated as the median of three technical qPCR replicates and three or four biological replicates.
ChIP.
Chromatin immunoprecipitation (ChIP) assays using the N499 anti-GR antibody were performed as previously described (42). After 30 min or 4 h of dex or ethanol treatment, cells in 15-cm dishes were cross-linked with 1% formaldehyde for 10 min, and then the reaction was stopped with 125 mM glycine for 10 min. Cells were rinsed with ice-cold phosphate-buffered saline (PBS), scraped into conical tubes, collected by centrifugation (600 × g for 5 min at 4°C), and lysed in ice-cold IP lysis buffer (500 mM HEPES-KOH, pH 8, 1 mM EDTA, 0.5 mM EGTA, 140 mM NaCl, 0.5% NP-40, 0.25% Triton X-100, 10% glycerol, supplemented with protease inhibitors) for 10 min at 4°C with nutation. Nuclei were collected by centrifugation (600 × g for 5 min at 4°C), resuspended in 1.5 ml of radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris-HCl pH 8, 1 mM EDTA, 0.5 mM EGTA, 140 mM NaCl, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, supplemented with protease inhibitors), and sonicated with 20-s bursts followed by 1 min cooling on ice for a total sonication time of 3 min per sample. Chromatin samples were immunoprecipitated with 9 μg of N499 anti-GR antibody or normal mouse IgG antibody (Santa Cruz Biotechnology) with protein A/G beads (Santa Cruz Biotechnology). After the beads were washed and cross-links were reversed, DNA fragments were extracted with phenol-chloroform and purified using a QIAquick spin column (Qiagen). GR occupancy was confirmed by qPCR as described above using 2% of the eluted ChIP DNA per reaction. One previously characterized GR binding region (GBR) was investigated per activated gene (41), and multiple repressed GBRs were selected (Cooper and Yamamoto, unpublished data) and tested for this study. Primer pairs were described previously (3, 41) or designed as described above. Primer sequences are presented in Table S2 in the supplemental material. Data were normalized to a region of the HSP70 gene and to an IgG control.
Brm ChIP assays were performed as described above, with the following modifications. After 2 h of dex or ethanol treatment, cells in 15-cm dishes were washed with cold PBS, cross-linked with 10 ml of 1.5 mM ethylene glycolbis (succinimidyl succinate) (EGS) in PBS for 30 min at room temperature, and dual cross-linked with 1% formaldehyde for 15 min at room temperature. Chromatin samples were immunoprecipitated with 9 μg of anti-Brm antibody (Abcam) with protein G-Sepharose 4 fast-flow beads (GE Healthcare). Data were normalized to a region of the HSP70 gene and to an input-only control.
Micrococcal nuclease assay.
Cross-linked mononucleosomes were prepared as previously described (25). After 10 min of dex or ethanol treatment, cells in 15-cm dishes were cross-linked with 1× cross-linking solution (11% formaldehyde, 50 mM HEPES, pH 7.9, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA) for 10 min. Cells were washed twice with PBS, harvested in ice-cold lysis buffer (10 mM Tris-HCl, pH 8, 0.25% Triton X-100, 10 mM EDTA, 0.5 mM EGTA, 100 mM NaCl, supplemented with protease inhibitors), and then left on ice for 10 min. Nuclei were collected by centrifugation (450 × g for 5 min at 4°C) and resuspended in 1 ml of buffer N (10 mM Tris-HCl, pH 8, 0.25 M sucrose, 75 mM NaCl, supplemented with protease inhibitors). Washed nuclei were centrifuged (450 × g for 5 min at 4°C), resuspended in 1 ml of buffer N, and sonicated for 20 s on ice. Nuclear suspensions were adjusted to 3 mM CaCl2, and each sample was split into two 500-μl reaction mixtures, which were digested with either 0 or 125 units of micrococcal nuclease (MNase; Worthington) for 10 min at 37°C. The reaction was stopped with 5 mM EDTA. Samples were rotated for 1 h with 0.5% SDS, and insoluble material was removed by centrifugation at full speed (21,000 × g) in a microcentrifuge for 10 min at 4°C. The chromatin solution was incubated overnight at 65°C, treated with 2 μg/ml DNase-free RNase (Roche) for 1 h at 37°C, and then treated with 200 μg/ml proteinase K (Roche) for 3 h at 37°C. DNA was purified using a QIAquick spin column (Qiagen). Approximately 5 μg of DNA was used per qPCR as described above. The fraction of MNase protection was determined as previously described (33, 39) by calculating a fold difference between MNase-treated and untreated samples. Tiled primers were designed as described above. Primer sequences are provided in Table S3 in the supplemental material.
RESULTS
Brm knockdown affects both activation and repression of specific primary GR target genes.
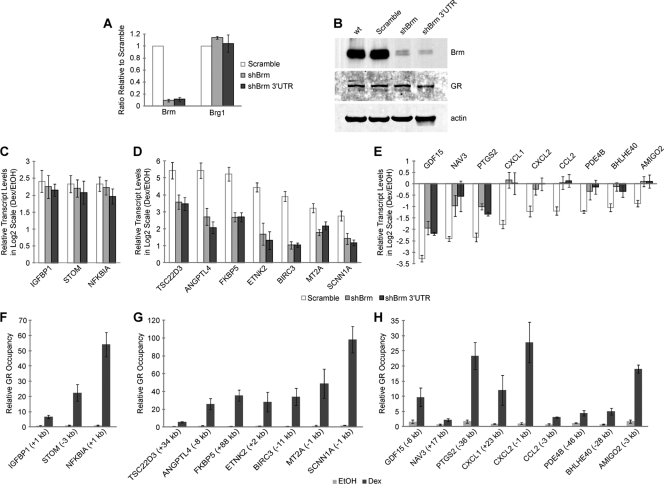
GR binds to specific response elements and recruits multifactor regulatory complexes, composed of various coregulators, to direct combinatorial regulation. To study those complexes containing GR and the coregulator Brm, we knocked down the expression of the Brm Swi/Snf ATPase in A549 cells by using two short hairpin-mediated RNA (shRNA) constructs. One targeted the coding sequence of the gene (shBrm), while the other targeted the 3′ untranslated region (shBrm 3′UTR). Each construct reduced Brm mRNA levels by ∼90%, but no reduction in Brg1 mRNA levels was observed (Fig. 1A), demonstrating that the constructs were specific for the Brm ATPase. Similarly, Brm protein was depleted in shRNA-treated cells, while GR protein levels were not affected (Fig. 1B).
Fig. 1.
Brm knockdown affects both activation and repression of specific primary GR target genes. (A) Short hairpin-mediated RNA (shRNA) knockdown of Brm is specific to that Swi/Snf ATPase, as it does not alter Brg1 mRNA levels. Data are shown relative to scrambled shRNA ± SEM (number of independent experiments [n] = 4). (B) Immunoblots demonstrating that efficient knockdown of Brm does not affect GR protein abundance levels. (C to E) Glucocorticoid-stimulated expression levels after Brm knockdown are Brm independent at some activated genes (C), Brm dependent at other activated genes (D), and Brm dependent at some repressed genes (E). Cells were treated with vehicle (ethanol [EtOH]) or 100 nM dex for 4 h. Data are plotted in a log2 scale relative to EtOH ± SEM (n = 4). (F to H) ChIP showing GR occupancy at activated Brm-independent (F), activated Brm-dependent (G), and repressed Brm-dependent (H) genes. Cells were treated with EtOH or 100 nM dex for 4 h. Data are displayed as enrichment of GBRs near the corresponding gene relative to IgG ± SEM (n = 4). The location of the GBR relative to the transcriptional start site (TSS) is indicated.
In order to investigate the role of Brm in mediating the transcriptional activity of GR, we looked at the effects of Brm knockdown on the expression levels of candidate genes selected from previous studies profiling GR expression and binding (36, 41, 47; S. B. Cooper and K. R. Yamamoto, unpub-lished data). Treatment of cells with the synthetic glucocorticoid dexamethasone (dex) led to transcriptional activation (Fig. 1C and D) or repression (Fig. 1E). Knockdown of Brm did not affect the regulation of some activated genes (Fig. 1C), while it reduced the activation of others (Fig. 1D). These observations indicate that GR requires Brm for proper regulation of only a subset of activated genes. Unlike the activated genes, all of the repressed genes we investigated showed decreased repression upon Brm knockdown (Fig. 1E). The two shRNA constructs showed agreement at all tested genes. Our results suggest that the Brm Swi/Snf ATPase is an important coregulator of GR-mediated repression. In contrast, the Brg1 ATPase is thought to only play a minor role in the repression of GR target genes (18).
Primary GR targets are glucocorticoid-regulated genes with a nearby GR binding region (GBR). We assume that one or more of the proximal GBRs are functional GREs. Chromatin immunoprecipitation (ChIP) confirmed GR occupancy near the activated Brm-independent (Fig. 1F), activated Brm-dependent (Fig. 1G), and repressed Brm-dependent (Fig. 1H) genes in a dex-dependent manner, suggesting that all these genes are primary GR targets. Patterns of GR occupancy were confirmed to be similar at two different times following dex treatment (see Fig. S1 in the supplemental material). Multiple GBRs have been verified for some activated (41) and repressed (see Fig. S2 in the supplemental material) genes. These results indicate that Brm modulates both the transcriptional activation and repression activities of GR at specific GR primary targets.
Brm knockdown decreases GR occupancy at GR-activated Brm-dependent genes and has disparate effects on GR occupancy at GR-repressed Brm-dependent genes.
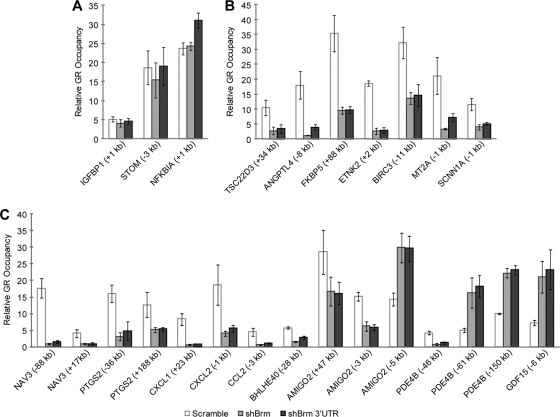
To understand the role of Brm on GR occupancy, we performed GR ChIP after Brm knockdown. As expected, ChIP of activated Brm-independent genes showed no change in GR occupancy upon Brm knockdown (Fig. 2A). In contrast, all the activated Brm-dependent genes showed a decrease in GR occupancy upon Brm knockdown (Fig. 2B), indicating that Brm facilitates GR recruitment at these genes. This decreased GR occupancy may be insufficient to produce the level of transcriptional response seen in control cells. Since the Brm ATPase subunit of Swi/Snf has chromatin remodeling activity, we hypothesized that the observed deficiency in GR occupancy could be due to altered chromatin structure. In particular, GR may bind chromatinized GBRs with varying affinity, and some of this initial binding may require GR-independent Brm-mediated chromatin remodeling. GR occupancy may be accompanied by recruitment of Brm, which would produce increased local Brm activity and result in more accessible DNA around the GBR. This would in turn allow additional GR to bind.
Fig. 2.
Brm knockdown decreases GR occupancy at GR-activated Brm-dependent genes and has disparate effects on GR occupancy at GR-repressed Brm-dependent genes. (A to C) ChIP of GR after Brm knockdown at activated Brm-independent (A), activated Brm-dependent (B), and repressed Brm-dependent (C) genes. Cells were treated with 100 nM dex for 4 h. Data are displayed as enrichment of GBRs near the corresponding gene relative to IgG ± SEM (n = 4). The location of the GBR relative to the TSS is indicated.
Interestingly, we observed two distinct classes of GR occupancy at repressed Brm-dependent genes upon Brm knockdown (Fig. 2C). Several GBRs showed decreased GR occupancy, as was seen at the activated genes. However, some GBRs showed increased GR occupancy. This suggests that two different mechanisms exist for Brm-mediated repression of primary GR target genes. It is possible that these disparate effects on GR occupancy are due to distinct modes of Brm-mediated chromatin remodeling, such that the DNA becomes more or less accessible at different GBRs. Alternatively, chromatin remodeling may increase the occupancy of factors that compete with GR binding. At two of the repressed genes (AMIGO2 and PDE4B), we witnessed both classes of GR occupancy upon Brm knockdown at different GBRs near the same gene. Consistent results were observed with both of the shRNA constructs at all tested GBRs. In summary, we found that Brm alters GR occupancy at both activated and repressed Brm-dependent genes, and Brm-mediated repression may occur through two different mechanisms. Thus, the role of Brm in modulating GR occupancy appears to be important for regulating the expression of specific primary GR target genes.
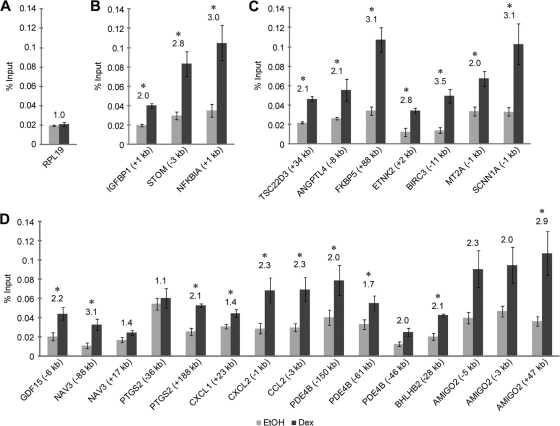
Brm occupies GR binding regions of GR-regulated genes in a dex-stimulated manner.
To examine how GR modulates Brm occupancy, we performed ChIP assays in the presence and absence of dex stimulation to look for Brm-containing Swi/Snf complexes at the GBRs near our genes of interest. As a negative control, a region of the housekeeping gene RPL19 was analyzed by ChIP and showed only basal levels of Brm occupancy in the presence of ethanol or dex (Fig. 3A). However, ChIP showed increased Brm occupancy at the GBRs of activated Brm-independent genes after dex treatment (Fig. 3B), even though GR activity and occupancy were both unaffected by Brm knockdown. At these genes, Brm-containing Swi/Snf complexes may be inactive or performing functions that do not affect the transcriptional response. Alternatively, redundancy may exist such that another remodeler (possibly a Brg1-containing Swi/Snf complex) can compensate for Brm knockdown.
Fig. 3.
Brm occupies GR binding regions of GR-regulated genes in a dex-stimulated manner. (A) RPL19 serves as a negative control for Brm ChIP. (B to D) ChIP showing Brm occupancy at activated Brm-independent (B), activated Brm-dependent (C), and repressed Brm-dependent (D) genes. This antibody does not cross-react with Brg1 (9, 11). Cells were treated with EtOH or 100 nM dex for 2 h. Data are displayed as enrichment of GBRs near the corresponding gene normalized to input samples ± SEM (n ≥ 3). The location of the GBR relative to the TSS is indicated. Fold change is displayed above the bar graphs. *, P ≤ 0.05 by Welch's t test.
ChIP confirmed dex-stimulated Brm occupancy at all tested GBRs near activated Brm-dependent genes (Fig. 3C). Dex-stimulated Brm occupancy was also observed at one or more GBRs for each repressed Brm-dependent gene (Fig. 3D). This result suggests that GR can localize Brm-containing Swi/Snf complexes to GBRs of both activated and repressed primary GR target genes. Two GBRs (NAV3 [+17 kb] and PDE4B [−46 kb]) showed a dex-stimulated increase in Brm occupancy, but this occupancy did not significantly exceed the basal levels observed in RPL19. Conversely, one GBR (PTGS2 [−36 kb]) experienced no increase in Brm occupancy upon dex treatment but was well above RPL19 levels in the presence of ethanol or dex. Brm-containing Swi/Snf complexes may be present at basal levels throughout the genome, and GR appears to increase the local concentration of these complexes at specific regions. These results show that Brm occupancy of at least one GBR of the studied activated and repressed GR-regulated genes is dex dependent, suggesting that GR may be involved in recruiting Brm to these regions. Since Brm is an ATPase subunit of the Swi/Snf chromatin remodeling complex, we propose that the Brm-dependent effects are due to chromatin remodeling.
Chromatin becomes more accessible upon dex treatment at GR-regulated genes and undergoes decreased remodeling at Brm-dependent genes following Brm knockdown.
To monitor changes in chromatin accessibility (the readout for remodeling activity), we employed an assay using micrococcal nuclease (MNase), which preferentially cleaves linker DNA between nucleosomes, as well as nucleosome-free DNA. The MNase assay measures the relative protection at a given region of DNA by comparing the amount of MNase-digested DNA to an undigested control (33, 39). We titrated the concentration of MNase to yield predominantly mononucleosome-sized fragments (see Fig. S3 in the supplemental material) and tiled primers amplifying roughly 100-bp regions that were 50 bp apart along the regions of interest. Analysis of the housekeeping gene RPL19 did not show dex-induced changes in chromatin structure (see Fig. S4 in the supplemental material).
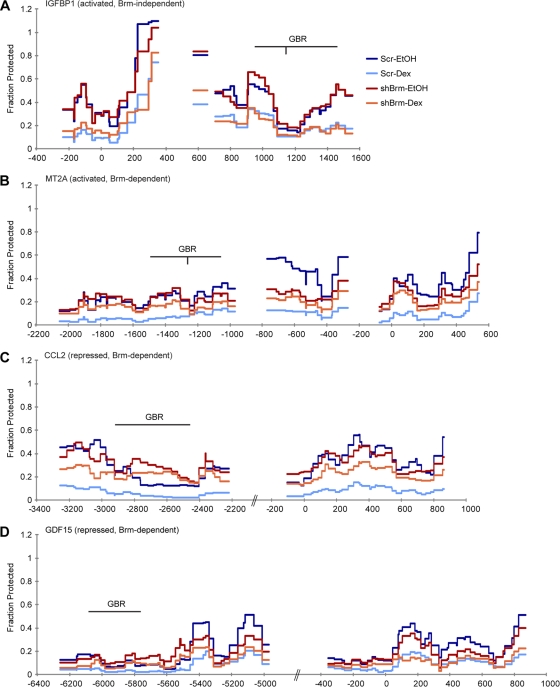
IGFBP1, an activated Brm-independent gene, displayed a dex-dependent decrease in protection around both the TSS and GBR (Fig. 4A, dark blue versus light blue line). (These graphs are shown with error bars in Fig. S5 in the supplemental material.) This corresponding increase in accessibility is consistent with activation, as it would allow GR and the transcriptional machinery increased access to the gene. We performed the MNase assay after Brm knockdown to investigate the role of Brm on these dex-dependent changes in chromatin structure. IGFBP1 showed no Brm-dependent remodeling (Fig. 4A, red and orange versus dark blue and light blue lines; see Fig. S6A in the supplemental material), which concurs with the Brm-independent GR transcriptional activity (Fig. 1C) and occupancy (Fig. 2A) observed at this gene. Interestingly, Brm is recruited to IGFBP1 (Fig. 3B) even though it does not appear to function as a chromatin remodeler at the regions we observed with the MNase assay.
Fig. 4.
Chromatin becomes more accessible upon dex treatment at GR-regulated genes and undergoes decreased remodeling at Brm-dependent genes following Brm knockdown. (A to D) MNase assay showing fraction protected at IGFBP1 (A), MT2A (B), CCL2 (C), and GDF15 (D). Cells were treated with EtOH or 100 nM dex for 10 min. Data are displayed as fraction of MNase protection by calculating a fold difference between MNase-treated and untreated samples (y axis), plotted against the location relative to the TSS (x axis) (n = 3). Error bars are omitted for clarity, but they are shown in Fig. S5 in the supplemental material. The horizontal line with GBR written above it represents the region of GR binding, and the vertical line shows the location of the canonical GR binding motif. The motif is not present at CCL2 or GDF15.
At MT2A, an activated Brm-dependent gene, we also witnessed a decrease in protection around the TSS and GBR upon dex treatment (Fig. 4B, dark blue versus light blue line), consistent with GR-mediated transcriptional activation. In addition, we observed strongly diminished dex-dependent remodeling upon Brm knockdown at both the TSS and GBR (Fig. 4B, red versus orange line; see Fig. S6B in the supplemental material), suggesting that Brm-mediated chromatin remodeling accounts for the Brm-dependent GR transcriptional activity (Fig. 1D) and occupancy (Fig. 2B) at MT2A. Brm knockdown in ethanol-treated samples had no significant effect on some regions (particularly around the GBR), whereas it decreased protection at other regions (particularly upstream of the TSS) (Fig. 4B, dark blue versus red line). These results imply that Brm is protecting chromatin in the MT2A promoter from MNase digestion in the absence of dex. Presumably, this decrease in MNase accessibility is due to Brm-mediated repositioning of nucleosomes into the promoter (17). Although Brm occupancy increased at the GBR of MT2A in a dex-stimulated manner (Fig. 3C), a basal activity of Brm may account for this GR-independent remodeling. Upon dex treatment, all regions were more protected in shBrm cells than in control cells (Fig. 4B, orange versus light blue line), suggesting that Brm is deployed in a GR-dependent manner to make DNA more accessible. Thus, at MT2A, the chromatin-remodeling activity of Brm appears to protect the DNA from MNase digestion in the absence of dex and to decrease this protection in the presence of dex.
CCL2, a repressed Brm-dependent gene with reduced GR occupancy upon Brm knockdown, displayed a dex-dependent decrease in protection around the TSS and GBR, similar to what was observed at the activated genes IGFBP1 and MT2A (Fig. 4C, dark blue versus light blue line). However, at CCL2, this Brm-mediated increase in accessibility leads to repression, rather than activation. Brm knockdown again resulted in decreased remodeling following dex treatment (Fig. 4C, red versus orange line; see Fig. S6C in the supplemental material), indicating that Brm contributes both to the regulation of GR-mediated transcription (Fig. 1E) and to GR occupancy (Fig. 2C) at CCL2 by altering chromatin structure. Although Brm knockdown in ethanol-treated samples had no effect on most regions, it did cause increased protection at a region around the GBR (Fig. 4C) (bp −2800 to −2400, dark blue versus red line), suggesting that basal levels of Brm can act in a GR-independent manner to remove or reposition nucleosomes (38). In both ethanol- and dex-treated cells, protection from MNase increased upon Brm knockdown relative to control cells at the GBR (Fig. 4C) (bp −2800 to −2400, red and orange versus dark blue and light blue lines). Thus, decreased access of GR to its GBR upon Brm depletion may account for the decreased GR occupancy at this site after Brm knockdown (Fig. 2C).
As with the other GR-regulated genes, the DNA around the TSS and GBR of GDF15, a repressed Brm-dependent gene with increased GR occupancy upon Brm knockdown, showed a dex-dependent decrease in protection (Fig. 4D, dark blue versus light blue line). In contrast to the other Brm-dependent genes, only a mild decrease in chromatin remodeling was observed at GDF15 upon Brm knockdown (Fig. 4D, red versus orange line; see Fig. S6D in the supplemental material). While some regions experienced a mild degree of chromatin remodeling, the GBR and TSS were largely unprotected regardless of Brm knockdown or dex treatment (Fig. 4D, bp −6300 to −5600 and −400 to 0, all lines). Thus, GR occupancy at this GBR appears relatively independent of chromatin remodeling. Since the DNA in this region appears to be particularly accessible under all observed conditions, the increase in GR occupancy following Brm knockdown (Fig. 2C) may be due to binding site competition between GR and some other factor, such as Brm.
In summary, dex treatment decreased DNA protection from MNase digestion in every GR-regulated region we examined (also see Fig. S7 in the supplemental material). Brm knockdown decreased the dynamic range of chromatin accessibility for both activated and repressed genes with Brm-dependent transcriptional regulation. This diminished chromatin remodeling correlated with decreased transcriptional activation or repression (Fig. 1D and E). The two classes of GR occupancy upon Brm knockdown at repressed genes (Fig. 2C) may reflect different native chromatin states at the GBRs of these genes (Table 1). At the CCL2 GBR, Brm remodeled chromatin structure in such a way to decrease protection, thus exposing the GBR to facilitate GR binding (Fig. 4C; see Fig. S6C in the supplemental material). Conversely, at GDF15, the GBR was packaged in relatively accessible chromatin, so Brm performed only mild remodeling activity (Fig. 4D; see Fig. S6D in the supplemental material).
Table 1.
Summary of GR and Brm interdependence
| Gene | GR response | Change in responsea |
|||
|---|---|---|---|---|---|
| Brm dependent |
GR dependent |
||||
| GR occupancy | GR regulation | Brm occupancy | Brm remodeling | ||
| IGFBP1 | Activation | No change | No change | + | No change |
| MT2A | Activation | + | + | + | ++ |
| CCL2 | Repression | + | + | + | ++ |
| GDF15 | Repression | − | + | + | + |
+ indicates an increase, and − a decrease, relative to what was seen in the absence of Brm or GR.
DISCUSSION
Different metazoan cell types are distinguished by extensive gene- and cell-specific transcription. This remarkable complexity of expression is not accomplished by a corresponding expansion in gene number (1) or by cell-specific regulatory factors (51). Instead, these selective and sophisticated patterns of transcription are generated by multifactor regulatory complexes comprised of different combinations of broadly expressed regulatory factors (4, 23). Although this concept of combinatorial regulation is long established (6), we understand relatively little about how common factors assemble into regulatory complexes that differ in composition, geometry, and function. Indeed, even the total number of proteins associated in a given complex, and the dynamics of their interactions, have not been determined. However, it is apparent that mammalian transcriptional regulatory complexes, for example, may contain 50 to 100 or more different proteins that associate on demand at genomic response elements (15).
In this study, we sought to determine whether as few as two proteins, GR and Brm, both residing in a select set of regulatory complexes, could serve as probes to functionally distinguish different roles of those complexes. The activities of GR and Brm can be monitored separately, since GR activates or represses transcription, whereas Brm remodels chromatin. Within a set of GR-activated and GR-repressed genes, we independently controlled the actions of the two proteins using glucocorticoid dependence of GR and knockdown of Brm, and we monitored the effects of GR and Brm on each other's occupancy and activity. At genes with GR-dependent Brm occupancy, we observed striking differences in the effects of Brm on GR occupancy and regulation (Table 1). Investigation of chromatin architecture at four such differentially controlled genes revealed distinct and unanticipated classes of chromatin remodeling. For example, in the absence of dex, Brm maintained either increased protection or increased accessibility, dex-dependent chromatin remodeling was Brm independent or Brm dependent, and the magnitude of Brm-dependent remodeling spanned a broad range. Overall, our analysis of four genes in A549 cells revealed four distinct patterns of transcriptional response, factor occupancy, and activity (Table 1). Thus, monitoring only two proteins within regulatory complexes is sufficient to identify functionally discrete assemblies.
At IGFBP1, an activated gene, Brm occupancy increased at the GBR in a GR-dependent manner, yet we cannot identify any activity associated with the recruitment of Brm to this regulatory region. Brm knockdown had no effect on GR occupancy, transcription, or chromatin remodeling, although Brm may remodel chromatin outside of the GBR and TSS regions that we analyzed. Previous studies have shown that increased GR and coregulator occupancy do not always correlate with their known activities (24, 48). In these cases, GR and coregulators may bind unproductively, or they may serve as scaffolds to recruit additional factors. Brm may be inactive at this site due to altered Swi/Snf subunit composition (34, 37, 49) or posttranslational modifications of the Swi/Snf complex (5, 40). Since the two Swi/Snf ATPases possess distinct functions in certain settings (12, 21, 28, 35), future studies could determine whether Brg1-containing Swi/Snf complexes are responsible for the dex-dependent remodeling at IGFBP1. Alternatively, both Brm and Brg1 may participate in chromatin remodeling at this gene, but functional redundancy between these ATPases may allow Brg1-containing Swi/Snf complexes to compensate for Brm knockdown (10, 43). The choice between two ATPases with both redundant and distinct functions may increase the specificity and versatility of Swi/Snf-containing multifactor regulatory complexes. Preliminary ChIP assays indicate that Brg1 occupancy increases upon dex treatment at a subset of GBRs in tandem with Brm (see Fig. S8 in the supplemental material).
Interestingly, two genes with opposite transcriptional responses (MT2A and CCL2) displayed similar patterns of GR and Brm interdependence. Upon dex treatment, Brm occupancy and Brm-mediated chromatin remodeling increased at both genes. In the presence of Brm, GR occupancy increased, leading to the transcriptional activation or repression of MT2A or CCL2, respectively. Therefore, it appears that the regulatory complexes acting at these two genes similarly regulate GR and Brm occupancy as well as chromatin remodeling despite mediating opposite transcriptional responses. Perhaps differential recruitment of DNA-binding activators or repressors, due to Brm-mediated changes in the accessibility of their target sequences, dictates transcriptional activation or repression by stabilizing the associated chromatin conformation (22). Alternatively, the overall composition of factors within the regulatory complex at the response element may specify the transcriptional output. For instance, differential recruitment of coregulators may produce an activating or repressing complex (24), or differential recruitment of chromatin-modifying proteins may lead to activating or repressing histone marks (16, 46). Future experiments could identify differences within the MT2A and CCL2 regulatory complexes that might account for their opposite transcriptional outputs.
The reciprocal roles of GR in Brm occupancy and Brm in GR occupancy suggest positive feedback in which GR binds to chromatinized GBRs with low affinity, recruiting Brm-containing Swi/Snf complexes to these regulatory regions, causing local chromatin remodeling, which allows more GR, and in turn more Swi/Snf, to bind accessible GBRs. Even before dex treatment, Brm appears to associate and function at a low level throughout the genome. This basal level of Brm remodeling activity may be important for initial “pioneer” GR binding events to relatively inaccessible chromatin, as we observed GR-independent Brm-mediated chromatin remodeling at the GBR of CCL2. Consistent with previous studies, the GBRs of the genes we investigated appear to be in regions that are relatively accessible (18, 19). Although we examined Brm occupancy only at GBRs, others have suggested that Brm is also recruited to TSSs of GR-regulated genes in a dex-dependent manner (31), possibly through looping interactions between regulatory complexes and the promoter (14). These long-range interactions could produce the Brm-dependent remodeling that we observe around the TSSs.
In contrast to the similarities between repressed CCL2 and activated MT2A, CCL2 and GDF15 are both repressed genes, yet they have very different patterns of GR and Brm interdependence. Both genes showed a dex-dependent increase in Brm occupancy, leading to greater GR-mediated transcriptional repression. While robust Brm-mediated chromatin remodeling at CCL2 allowed increased GR occupancy, the mild remodeling observed at GDF15 was associated with decreased GR occupancy. The multifactor regulatory complexes acting at these two genes apparently dictate transcriptional repression through different mechanisms. At CCL2, changes in DNA accessibility regulate GR occupancy. However, GR occupancy at GDF15 appears fairly independent of chromatin remodeling, as the GBR is relatively accessible under all observed conditions. Instead, the Brm-dependent decrease in GR occupancy may reflect competition for binding with Brm or an unknown factor that is recruited by Brm. If Brm can indeed exclude a factor such as GR without remodeling chromatin, this would represent a fundamentally new mechanism of Brm action. Alternatively, local chromatin rearrangements not detectable by MNase digestion might account for the Brm-dependent decrease in GR occupancy.
Our investigation of two proteins present in functionally different regulatory complexes, each bearing perhaps 50 times as many components, revealed their distinctive roles and interdependencies in those complexes. In doing so, we laid the groundwork for an experimental system to monitor combinatorial assembly and function, in which we can independently modify and measure factor occupancy and activity. Since four genes showed four different patterns of transcriptional response and GR and Brm interdependence, we conclude that these two factors interact through multiple distinct mechanisms. Thus, our results underscore the power and specificity of combinatorial regulation.
By what mechanisms might factors engage different physical interactions that create distinctive regulatory complex geometries? One way to think about these results comes from the view of proteins as “mosaics” of potentially functional surfaces, each of which may be more “active” in some contexts than in others. For example, Rogatsky et al. (36) found that GR activates the transcription of different genes in human U2OS cells by utilizing gene-specific patterns of functional surfaces. These differential patterns likely reflect, in part, allosteric effects on GR conformation driven by the precise DNA sequence with which GR interacts at each target gene (27). However, there exist many contextual influences, including ligand chemistry, posttranslational modifications, and occupancy by other DNA-binding regulatory factors and non-DNA-binding coregulators. In turn, the functional surfaces so created or stabilized may confer enzymatic activities, serve as sites for posttranslational modification or interact with other factors, thereby greatly expanding combinatorial diversity. We demonstrate that profiling the interactions and functions of two factors is sufficient to separate regulatory complexes into distinct classes, implicating additional context-specific interactions. Overall, this strategy provides a general route toward the discovery of functional surfaces and components within regulatory complexes and showcases how combinatorial control produces remarkable regulatory specificity. Understanding the functional relationships between regulatory complex components will eventually enable prediction, rather than mere observation, of the diverse outputs of combinatorial regulation.
Supplementary Material
ACKNOWLEDGMENTS
We thank Sheng-hong Chen, Ben Engel, Linet Mera, Geeta Narlikar, Ben Schiller, Kevan Shokat, and Alex So for critical comments on the manuscript. We are grateful to Danielle Bittencourt for assistance with Brm ChIP and Alex So for inspiration during the initiation of this project.
K.B.E. was supported by an NIH NIGMS Chemistry-Biology Training Grant and was a predoctoral fellow of the Graduate Dean's Health Science Fellowship and the Tobacco-Related Disease Research Program (17DT-0177). Research support was from NIH (CA020535) to K.R.Y.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
Published ahead of print on 6 June 2011.
REFERENCES
- 1. Baltimore D. 2001. Our genome unveiled. Nature 409:814–816 [DOI] [PubMed] [Google Scholar]
- 2. Biddie S. C., John S., Hager G. L. 2010. Genome-wide mechanisms of nuclear receptor action. Trends Endocrinol. Metab. 21:3–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bolton E. C., et al. 2007. Cell- and gene-specific regulation of primary target genes by the androgen receptor. Genes Dev. 21:2005–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boube M., Joulian L., Cribbs D. L., Bourbon H. M. 2002. Evidence for a mediator of RNA polymerase II transcriptional regulation conserved from yeast to man. Cell 110:143–151 [DOI] [PubMed] [Google Scholar]
- 5. Bourachot B., Yaniv M., Muchardt C. 2003. Growth inhibition by the mammalian SWI-SNF subunit Brm is regulated by acetylation. EMBO J. 22:6505–6515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Britten R. J., Davidson E. H. 1969. Gene regulation for higher cells: a theory. Science 165:349–357 [DOI] [PubMed] [Google Scholar]
- 7. Cairns B. R. 2009. The logic of chromatin architecture and remodeling at promoters. Nature 461:193–198 [DOI] [PubMed] [Google Scholar]
- 8. Clapier C. R., Cairns B. R. 2009. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78:273–304 [DOI] [PubMed] [Google Scholar]
- 9. Cohet N., et al. 2010. SWI/SNF chromatin remodeling enzyme ATPases promote cell proliferation in normal mammary epithelial cells. J. Cell. Physiol. 223:667–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. de la Serna I. L., Carlson K. A., Imbalzano A. N. 2001. Mammalian SWI/SNF complexes promote MyoD-mediated muscle differentiation. Nat. Genet. 27:187–190 [DOI] [PubMed] [Google Scholar]
- 11. Fish J. E., et al. 2010. Hypoxic repression of endothelial nitric-oxide synthase transcription is coupled with eviction of promoter histones. J. Biol. Chem. 285:81–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Flowers S., Nagl N. G., Jr., Beck G. R., Jr., Moran E. 2009. Antagonistic roles for BRM and BRG1 SWI/SNF complexes in differentiation. J. Biol. Chem. 284:10067–10075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fryer C. J., Archer T. K. 1998. Chromatin remodeling by the glucocorticoid receptor requires the BRG1 complex. Nature 393:88–91 [DOI] [PubMed] [Google Scholar]
- 14. Hakim O., et al. 2009. Glucocortoid receptor activation of the Ciz1-Lcn2 locus by long range interactions. J. Biol. Chem. 284:6048–6052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Holstege F. C., et al. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717–728 [DOI] [PubMed] [Google Scholar]
- 16. Huang Z. Q., Li J., Sachs L. M., Cole P. A., Wong J. 2003. A role for cofactor-cofactor and cofactor-histone interactions in targeting p300, SWI/SNF and Mediator for transcription. EMBO J. 22:2146–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jaskelioff M., Gavin I. M., Peterson C. L., Logie C. 2000. SWI-SNF-mediated nucleosome remodeling: role of histone octamer mobility in the persistence of the remodeled state. Mol. Cell. Biol. 20:3058–3068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. John S., et al. 2008. Interaction of the glucocorticoid receptor with the chromatin landscape. Mol. Cell 29:611–624 [DOI] [PubMed] [Google Scholar]
- 19. John S., et al. 2011. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat. Genet. 43:264–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Johnson T. A., Elbi C., Parekh B. S., Hager G. L., John S. 2008. Chromatin remodeling complexes interact dynamically with a glucocorticoid receptor-regulated promoter. Mol. Biol. Cell 19:3308–3322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kadam S., Emerson B. M. 2003. Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol. Cell 11:377–389 [DOI] [PubMed] [Google Scholar]
- 22. Kingston R. E., Narlikar G. J. 1999. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 13:2339–2352 [DOI] [PubMed] [Google Scholar]
- 23. Levine M., Tjian R. 2003. Transcription regulation and animal diversity. Nature 424:147–151 [DOI] [PubMed] [Google Scholar]
- 24. Luecke H. F., Yamamoto K. R. 2005. The glucocorticoid receptor blocks P-TEFb recruitment by NFkB to effect promoter-specific transcriptional repression. Genes Dev. 19:1116–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. MacDonald N., et al. 2005. Molecular basis for the recognition of phosphorylated and phosphoacetylated histone h3 by 14-3-3. Mol. Cell 20:199–211 [DOI] [PubMed] [Google Scholar]
- 26. McKenna N. J., O'Malley B. W. 2002. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell 108:465–474 [DOI] [PubMed] [Google Scholar]
- 27. Meijsing S. H., et al. 2009. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science 324:407–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Miao J., et al. 2009. Functional specificities of Brm and Brg-1 Swi/Snf ATPases in the feedback regulation of hepatic bile acid biosynthesis. Mol. Cell. Biol. 29:6170–6181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Naldini L., et al. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263–267 [DOI] [PubMed] [Google Scholar]
- 30. Narlikar G. J., Fan H. Y., Kingston R. E. 2002. Cooperation between complexes that regulate chromatin structure and transcription. Cell 108:475–487 [DOI] [PubMed] [Google Scholar]
- 31. Paakinaho V., Makkonen H., Jaaskelainen T., Palvimo J. J. 2010. Glucocorticoid receptor activates poised FKBP51 locus through long-distance interactions. Mol. Endocrinol. 24:511–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pantoja C., Huff J. T., Yamamoto K. R. 2008. Glucocorticoid signaling defines a novel commitment state during adipogenesis in vitro. Mol. Biol. Cell 19:4032–4041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Petesch S. J., Lis J. T. 2008. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell 134:74–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Phelan M. L., Sif S., Narlikar G. J., Kingston R. E. 1999. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol. Cell 3:247–253 [DOI] [PubMed] [Google Scholar]
- 35. Reyes J. C., et al. 1998. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2α). EMBO J. 17:6878–6991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rogatsky I., et al. 2003. Target-specific utilization of transcriptional regulatory surfaces by the glucocorticoid receptor. Proc. Natl. Acad. Sci. U. S. A. 100:13845–13850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ryme J., Asp P., Bohm S., Cavellan E., Farrants A. K. 2009. Variations in the composition of mammalian SWI/SNF chromatin remodeling complexes. J. Cell. Biochem. 108:565–576 [DOI] [PubMed] [Google Scholar]
- 38. Schwabish M. A., Struhl K. 2007. The Swi/Snf complex is important for histone eviction during transcriptional activation and RNA polymerase II elongation in vivo. Mol. Cell. Biol. 27:6987–6995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sekinger E. A., Moqtader Z., Struhl K. 2005. Intrinsic histone-DNA interactions and low nucleosome density are important for preferential accessibility of promoter regions in yeast. Mol. Cell 18:735–748 [DOI] [PubMed] [Google Scholar]
- 40. Sif S. P. T., Stukenberg, Kirschner M. W., Kingston R. E. 1998. Mitotic inactivation of a human SWI/SNF chromatin remodeling complex. Genes Dev. 12:2842–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. So A. Y., Chaivorapol C., Bolton E. C., Li H., Yamamoto K. R. 2007. Determinants of cell- and gene-specific transcriptional regulation by the glucocorticoid receptor. PLoS Genet. 3:e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. So A. Y., Bernal T. U., Pillsbury M. L., Yamamoto K. R., Feldman B. J. 2009. Glucocorticoid regulation of the circadian clock modulates glucose homeostasis. Proc. Natl. Acad. Sci. U. S. A. 106:17582–17587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Strobeck M. W., et al. 2002. Compensation of BRG-1 function by Brm: insight into the role of the core SWI-SNF subunits in retinoblastoma tumor suppressor signaling. J. Biol. Chem. 277:4782–4789 [DOI] [PubMed] [Google Scholar]
- 44. Trotter K. W., Archer T. K. 2004. Reconstitution of glucocorticoid receptor-dependent transcription in vivo. Mol. Cell. Biol. 24:3347–3358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tuch B. B., Galgoczy D. J., Hernday A. D., Li H., Johnson A. D. 2008. The evolution of combinatorial gene regulation in fungi. PLoS Biol. 6:e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Underhill C., Qutob M. S., Yee S. P., Torchia J. 2000. A novel nuclear receptor corepressor complex, N-CoR, contains components of the mammalian SWI/SNF complex and the corepressor KAP-1. J. Biol. Chem. 275:40463–40470 [DOI] [PubMed] [Google Scholar]
- 47. Wang J. C., et al. 2004. Chromatin immunoprecipitation (ChIP) scanning identifies primary glucocorticoid receptor target genes. Proc. Natl. Acad. Sci. U. S. A. 101:15603–15608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang J. C., et al. 2006. Novel arylpyrazole compounds selectively modulate glucocorticoid receptor regulatory activity. Genes Dev. 20:689–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang W., et al. 1996. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 10:2117–2130 [DOI] [PubMed] [Google Scholar]
- 50. Wolberger C. 1998. Combinatorial transcription factors. Curr. Opin. Genet. Dev. 8:552–559 [DOI] [PubMed] [Google Scholar]
- 51. Yamamoto K. R. 1989. A conceptual view of transcriptional regulation. Amer. Zool. 29:537–547 [Google Scholar]
- 52. Yamamoto K. R., Pearce D., Thomas J., Miner J. N. 1992. Combinatorial regulation at a mammalian composite response element, p. 1169–1192 In McKnight S. L., Yamamoto K. R. (ed.), Transcriptional regulation. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 53. Yoshinaga S. K., Peterson C. L., Herskowitz I., Yamamoto K. R. 1992. Roles of SWI1, SWI2, and SWI3 proteins for transcriptional enhancement by steroid receptors. Science 258:1598–1604 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.