Abstract
Enteroviruses (EVs) are recognized as the major etiological agent in meningitis in children and young adults. The use of molecular techniques, such as PCR, has substantially improved the sensitivity of enterovirus detection compared to that of virus culture methods. PCR-based methods also can detect a much wider range of EV variants, including those within species A, as well as human parechoviruses (HPeVs) that often grow poorly in vitro and which previously have been underdiagnosed by traditional methods. To exploit these developments, we developed a real-time one-step reverse transcription-PCR (RT-PCR) for the rapid and sensitive detection of EV and HPeV in clinical specimens. Two commercially available RT-PCR kits were used (method I, Platinum one-step kit; method II, Express qPCR one-step kit) with primers and probes targeting the EV and HPeV 5′-untranslated regions (5′UTR). Amplification dynamics (threshold cycle [CT]values and efficiencies) of absolutely quantified full-length RNA transcripts representative of EV species A to D and HPeV were similar, demonstrating the effectiveness of both assays across the range of currently described human EV and HPeV variants. Probit analysis of multiple endpoint replicates demonstrated comparable sensitivities of the assays for EV and HPeV (method I, approximately 10 copies per reaction for both targets; method II, 20 copies per reaction). CT values were highly reproducible on repeat testing of positive controls within assays and between assay runs. Considering the sample turnaround time of less than 3 h, the multiplexed one-step RT-PCR method provides rapid diagnostic testing for EV and HPeV in cases of suspected central nervous system infections in a clinically relevant time frame.
INTRODUCTION
Human enteroviruses (EV) and parechoviruses (HPeV), within the virus family Picornaviridae, mostly infect children and young adults. Enteroviruses traditionally were divided into polioviruses (PVs; 3 serotypes), coxsackie A viruses (CAVs; 23 serotypes), coxsackie B viruses (CBVs; 6 serotypes), and echoviruses (E; 28 serotypes), mainly on the basis of their pathogenicity in laboratory animals. More recently discovered enteroviruses have been assigned enterovirus type numbers based on the chronological order in which they were identified (34 by the end of 2009). Altogether these 97 human EV types fall into four genetically distinct species, HEV-A to HEV-D, within the Enterovirus genus (16, 23).
The first two HPeV types were isolated more than 50 years ago (25) and were described as echoviruses 22 and 23. Although originally thought to be related to viruses in the Enterovirus genus, sequence analysis has revealed several differences in genome structure and substantially divergent coding sequences that justified their reclassification into a new, separate Parechovirus genus (15). Recently, a further 12 HPeV types known to infect humans have been identified (reviewed in reference 13). Parechovirus infections are enteric and often associated with mild gastrointestinal and respiratory symptoms, although severe neonatal diseases, including sepsis-like illness, meningitis, encephalitis, and hepatitis, have been described. In addition to HPeV type 3, HEV-B variants (including CBVs, CAV9, and echoviruses) are the most commonly identified viral cause of central nervous system (CNS)-associated infection in Europe (10, 11, 18, 26).
The diagnosis of EV and HPeV infections used to rely on often slow, laborious, and insensitive cell culturing, which now has been replaced largely by nucleic acid amplification tests, such as reverse transcription-PCR (RT-PCR) targeting the conserved 5′-untranslated region (5′UTR) (1, 6, 14, 21, 27). HPeVs in particular historically have been problematic to detect by virus culture and cannot be detected by routine enterovirus RT-PCR; thus, HPeV infections long have been underdiagnosed (2). More recently, separate real-time RT-PCR assays for the detection of EV and HPeV have been developed (3, 7, 17, 19, 20); the rapid molecular testing provided by these assays for hospitalized children is important, since it has been shown to reduce antibiotic usage, unnecessary investigations, and duration of hospital stay (3, 17, 26). In the current study, we have multiplexed EV and HPeV into a single, one-step real-time RT-PCR using two assay formats generally used in two diagnostic virology laboratories for the direct testing of clinical specimens. These assays are based on Platinum one-step kit or Express qPCR one-step kit methods. Both showed high sensitivity and would allow the rapid detection of all known EV and HPeV types simultaneously on a variety of clinical specimens.
MATERIALS AND METHODS
In vitro-transcribed RNA for species A to D enteroviruses and parechovirus.
Plasmids containing the full-length sequence of five different enteroviruses, coxsackievirus A16 (CAV16; species A), echoviruses 7 and 30 (E7 and E30; species B), coxsackievirus A21 (CAV21; species C), enterovirus 70 (EV70; species D), and human parechovirus type 1 (HPeV1) were kindly provided by G. Stanway (CAV16) (22) and D. Evans (E7, E30, CAV21, and EV70). Before transcription each plasmid was purified, linearized at the 3′ end using the appropriate restriction enzyme, and cleaned by phenol-chloroform extraction followed by ethanol precipitation. These linearized plasmids consequently were used as templates for in vitro RNA transcription using T7 RNA polymerase (MEGAscript T7 kit; Ambion, Life Technologies, United Kingdom). After the transcription reaction, the RNA was DNase treated (RQ1 kit; Promega, United Kingdom) and purified using LiCl precipitation. The concentration of the RNAs was determined using a NanoDrop ND-1000 (Thermo Scientific, United Kingdom) photospectrometer. The integrity of RNA transcripts was demonstrated by denaturing agarose gel electrophoresis (data not shown). Tenfold dilution series for assay calibration were made in citrate buffer (pH 6.0; Ambion, Life Technologies, United Kingdom) supplemented with 0.05 μg/ml carrier tRNA and 0.1 U/ml RNAsin inhibitor. Amplification efficiencies were calculated from the gradient of the line of best fit for data points of log10-transformed RNA input copies and CT values using the following formula: 100 × (10−1/gradient − 1). Further dilutions of E7 and HPeV1 RNA were assessed in 24 parallel reactions to determine a 90% detection limit for the assays. Probit analysis was performed using the SAS statistical package.
One-step RT-PCR assays.
For EV amplification, primers and probe (5′-TCC-GGC-CCC-TGA-ATG-CGG-CTA-AT-3′) were as described by Dierssen et al. (7). Primers and probe [5′-AAA-CAC-TAG-TTG-TA(A/T)-GGC-CC-3′] for HPeV corresponded to those described by Benschop et al. (3). In method I, the 5′ and 3′ ends of the EV probes were labeled with cy5 and black hole quencher (BHQ), respectively, whereas 6-carboxyfluorescein (FAM) and minor groove binder were used for the HPeV probe. In method II, 5′ and 3′ labels were FAM and BHQ for the EV probe and Yakima yellow and BHQ for the HPeV probe. All primers and probes were purchased from Applied Biosystems (United Kingdom).
Beside the primers, probes, and oligonucleotides, all other reaction components, including Superscript III, were provided with the Platinum one-step kit (method I; Glasgow) and Express qPCR one-step kit (method II; Edinburgh). For method I, EV primers (1 μM each) and probe (0.025 μM) as well as HPeV primers (1 μM each) and probe (0.05 μM) were added to the master mix (15 μl) as modified from the manufacturer's recommendations (Platinum one-step kit; Invitrogen) (9). Purified RNA (6 μl) was added to the master mix. After reverse transcription at 50°C (15 min) and initial denaturation at 95°C (2 min), amplification was performed in 40 cycles with a standard TaqMan two-step protocol (60°C for 34 s and 95°C for s). For method II, EV primers (0.5 μM each) and probe (0.35 μM) as well as HPeV primers (0.5 μM each) and probe (0.35 μM) were added to the master mix (25 μl) per the manufacturer's recommendations (Express qPCR one-step kit; Invitrogen). Purified RNA (9 μl) was added to the master mix. After reverse transcription at 50°C (15 min) and initial denaturation at 95°C (20 s), amplification was performed in 45 cycles with a standard three-step protocol (55°C for 25 s, 72°C for 10 s, and 95°C for 3 s). Real-time PCR experiments by both methods were run on an ABI Prism 7500 SDS real-time platform (Applied Biosystems, United Kingdom).
Viral strains and quality control panels.
E30 isolate cultured in RD cells as well as HPeV1 and HPeV3 isolates cultured on Vero cells were used as assay controls and for the efficiency testing of one-step RT-PCR. The most common enterovirus strains, including 14 HEV-B serotypes (CBV1 to CBV5, CAV9, E3, E6, E7, E11, E13, E18, E21, and E30; obtained from the Health Protection Agency, Colindale, London, United Kingdom), 2 HEV-A serotypes (CAV16 and EV71; obtained from the Health Protection Agency, Colindale, London, United Kingdom), and 24 previously typed clinical parechovirus isolates (13 HPeV1, 4 HPeV3, 2 HPeV4, 4 HPeV5, and 1 HPeV6; kindly provided by Katja Wolthers, Amsterdam Medical Centre, Netherlands) were analyzed. The sensitivity and specificity of one-step EV and HPeV RT-PCR was evaluated using all 12 samples (EVRNA10-1 to EVRNA10-12) from the 2010 proficiency panel obtained from Quality Control on Molecular Diagnostic (QCMD) by both assays.
Clinical samples.
Total RNA from clinical samples was extracted from 200 μl cerebrospinal fluid (CSF) with the Qiagen MDx using the QIAamp viral RNA kit (method II; Qiagen, United Kingdom) or using the bioMérieux easyMag (method I; bioMérieux, France) and was eluted in 50 μl corresponding dilution buffer. Purified RNA (9 μl) was subjected to a study of EV and HPeV RNA detection by the one-step RT-PCR, the established diagnostic real-time RT-PCR for EV (modified from reference 7), and in-house nested RT-PCR assay for HPeV (12). Their diagnostic specificity was confirmed with 120 CSF samples (method II).
RESULTS
Sensitivity of one-step RT-PCR.
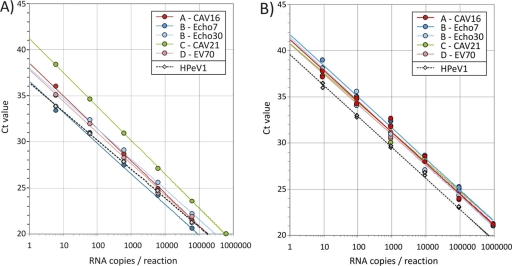
To compare the sensitivities of method I and II one-step RT-PCRs for the four species of human enteroviruses, duplicate dilution series of RNA transcripts derived from the full-length clones of CAV16 (EV-A), E30 E7 (EV-B), CAV21 (EV-C), and EV70 (EV-D) were assayed, and CT values were recorded (Fig. 1). All five enterovirus transcripts showed efficient amplification by both methods; a linear line of the best fit of CT values (y axis) with log-transformed RNA input copies (x axis) showed amplification efficiencies ranging from 92 to 111% (mean, 105%; slopes, −3.08 to −3.54; correlation coefficient [R2], 0.9812 to 0.9998) in method I and 95 to 105.5% (mean, 100%; slopes, −3.21 to −3.44; R2, 0.9949 to 1.000) in method II. Although CT values from method II were approximately 3 cycles greater for a given transcript concentration compared to that of method I, both assays showed highly reproducible detection abilities for transcripts from the four EV species, the only exception being the species C transcript in method I (CT values were 4 cycles higher than those observed with species A, B and D; Fig. 1A, green line). Parechovirus amplification was comparable between the two methods (assay efficiencies were 108 and 97% and R2 was 0.9995 and 0.9977 for methods I and II, respectively) (Fig. 1).
Fig. 1.
Amplification of RNA transcripts of defined concentrations by method I (Platinum RT-PCR) (A) and method II (Express qPCR) (B) multiplex assays. CT values (y axis) were plotted for serial 10-fold dilutions of each transcript (x axis).
By testing a much larger number of replicates around the endpoint of the EV and HPeV assays, the 90% detection frequencies were calculated by probit analysis (Table 1). The 90% detection limit of 7 copies per reaction (95% confidence interval [CI], 4 to 16 copies) for E7 and 9 copies per reaction (95% CI, 5 to 30 copies) for HPeV1 were predicted for method I. For method II, the corresponding 90% detection thresholds were 19 copies (CI, 11 to 50 copies) for E7 and 21 copies (CI, 17 to 32 copies) for HPeV1.
Table 1.
Detection limit of one-step RT-PCR as determined by serial endpoint dilutions of in vitro-transcribed E7 and HPeV1 RNAa
| Method and no. of RNA copies/reaction | No. (%) of positive results for virus: |
|
|---|---|---|
| E7 | HPeV1 | |
| I | ||
| 600 | 24/24 (100) | 8/8 (100) |
| 60 | 4/4 (100) | 8/8 (100) |
| 6 | 21/24 (88) | 20/24 (83) |
| 0.6 | 2/24 (8) | 5/24 (21) |
| II | ||
| 900 | 4/4 (100) | 4/4 (100) |
| 90 | 20/20 (100) | 4/4 (100) |
| 20 | 22/24 (92) | 21/24 (88) |
| 6 | 15/24 (63) | 1/24 (4) |
| 0.9 | 5/24 (21) | 0/24 (0) |
The 90% detection limit with a 95% confidence interval (CI) was calculated by probit analysis. Method I and II 90% detection limits (95% CI) for E7 were 7 (4 to 16) and 19 (11 to 50) RNA copies/reaction, respectively, and for HPeV1 they were 9 (5 to 30) and 21 (17 to 31) RNA copies/reaction, respectively.
A panel consisting of 16 EV serotypes (CBV1 to CBV5, CAV9, CAV16, E3, E6, E7, E11, E13, E18, E21, E30, and EV71) and 5 HPeV types (HPeV1, HPeV3, HPeV4, HPeV5, and HPeV6) was tested. All (sero)types were efficiently amplified and sensitively detected, as evidenced by low CT values (data not shown). The one-step RT-PCR method was used to screen the QCMD panels for the detection of enteroviruses and parechoviruses. In the duplicate testing of the 2010 QCMD panel sample, EVRNA10-12 (10−7 dilution of E11) was detected only once by method II, whereas all other EV (10−7 dilution of E30, CAV9, and EV71; 10−6 dilution of CBV3; 10−5 dilution of E11, E30, CAV9, and EV71)- and HPeV (10−6 and 10−4 dilutions of HPeV3)-positive samples, and a negative sample, were scored correctly in both assays.
Comparison with singleplex RT-PCR for EV and HPeV.
Multiplexed RT-PCR for EV and HPeV was compared to the single RT-PCR assays to investigate whether multiplexing the PCR influenced assay sensitivity. Using the published one-step RT-PCR for EV detection (7), all five RNA transcripts tested in duplicate were positive for 80 copies (10/10 combined), 7/10 were positive for 8 copies, and 0/10 were positive for 0.8 copies of RNA used per reaction. These are comparable to detection frequencies in both multiplexed assays (Table 1). Further sensitivity comparisons between assays (multiplexed RT-PCR for EV and HPeV detection and singleplex RT-PCR for EV and HPeV detection) were made by assaying RNA extracted from a 10-fold dilution series of culture supernatant from an in vitro isolate of EV species B, E30, and two HPeVs (types 1 and 3) by methods I and II. Both assays and methods showed equal endpoint dilutions (10−5 for E30 and 10−7 for HPeV1 and HPeV3), providing further evidence that multiplexing did not affect the sensitivity of RT-PCR.
Specificity and reproducibility of one-step RT-PCR for EV and HPeV detection.
Cross-reactivity with viruses commonly present in clinical specimens was analyzed by both methods. The common respiratory pathogens (including influenza viruses [A, A/H1N1, and B], parainfluenza viruses [types 1 to 4], respiratory syncytial virus, human metapneumovirus, coronaviruses [229E, OC43, NL63, and HKU1], adenovirus, Pneumocystis jirovecii, and Mycoplasma pneumoniae), viruses causing gastroenteritis (norovirus and rotavirus) and rash (measles virus), various herpesviruses (herpes simplex virus types 1 and 2, varicella-zoster virus, Epstein-Barr virus, and cytomegalovirus), and mumps virus all tested negative by methods I and II. Similarly, the interassay reproducibility was evaluated by monitoring CT values from the positive controls (E30 and HPeV3) across 20 RT-PCR runs and intra-assay reproducibility by testing the controls in 20 wells on a single RT-PCR run by both methods (Table 2).
Table 2.
Inter- and intra-assay reproducibility of multiplexed EV/HPeV RT-PCR
| Method and reproducibility type | Virus |
CTa |
CVb | |||
|---|---|---|---|---|---|---|
| Mean | SD | Max | Min | |||
| Intertest | ||||||
| I | EV | 26.23 | 0.57 | 27.94 | 24.51 | 0.0217 |
| HPeV | 27.02 | 0.48 | 28.45 | 25.6 | 0.0178 | |
| II | EV | 31.96 | 0.36 | 32.66 | 31.27 | 0.0113 |
| HPeV | 26.86 | 0.32 | 27.31 | 26.23 | 0.0119 | |
| Intratest | ||||||
| I | EV | 24.84 | 0.34 | 25.86 | 23.83 | 0.0137 |
| HPeV | 27.34 | 0.28 | 28.81 | 26.51 | 0.0102 | |
| II | EV | 29.01 | 0.48 | 29.74 | 28.13 | 0.0165 |
| HPeV | 27.54 | 0.35 | 28.03 | 27.02 | 0.0127 | |
Maximum (max) and minimum (min) values are within 3 SD from the mean CT.
CV, coefficient of variation.
Evaluation of one-step RT-PCR for diagnostic EV and HPeV detection in CSF samples.
A total of 120 anonymous CSF samples obtained from patients with suspected meningitis or neonatal sepsis-like disease were evaluated. After RNA extraction from CSF, results from EV and HPeV RNA detection by the one-step multiplexed RT-PCR (method II) were compared to the established diagnostic real-time RT-PCR for EV (modified from reference 7) and in-house nested RT-PCR assay for HPeV (12). Both RT-PCR methods gave concordant results (8 EV positive, 7 HPeV positive, and 105 negative), indicating the 100% sensitivity and specificity of one-step RT-PCR compared to those of single real-time RT-PCR for EVs and nested RT-PCR for HPeVs. Molecular typing by VP1 sequencing for EVs and VP3/VP1 for HPeVs were achieved in all samples: CAV9 (5), CAV6 (1), E6 (1), E30 (1), and HPeV3 (7).
DISCUSSION
The newly developed one-step real-time RT-PCR protocol(s) enables the rapid and simultaneous diagnosis of enterovirus and human parechovirus RNA in clinical specimens. The one-step RT-PCR protocol combined reverse transcription and DNA amplification in a single closed tube and allowed convenient real-time detection of specific amplicons for assay specificity. The sensitivity and specificity of the assay implemented in the widely used Platinum one-step and Express qPCR one-step kits were equivalent despite considerable differences in thermal cycling, annealing temperatures, and reaction buffer components. These results indicate that the multiplexed format as well as primer and probe combinations are relatively robust and can be incorporated into a wider range of assay formats.
Previous evaluations of the EV primers and probe in a nonmultiplexed one-step RT-PCR format demonstrated a sensitivity of 100 copies of EV RNA per reaction, corresponding to approximately 3,800 copies/ml of CSF (7). Nonmultiplexed detection for HPeV by one-step real-time RT-PCR showed a 100% detection limit of 30 RNA copies per reaction, derived from testing 30 replicates (19). Although an optimized two-step real-time RT-PCR assay for EVs was shown to be more sensitive than the one-step method, reaching a detection limit of about 10 to 50 genomes per reaction (17), the recent studies using SuperScript III confirmed that the one-step RT-PCR method can have sensitivity equal to (if not better than) that of two-step reactions (19, 24). This was indeed seen in our study, with 90% detection limits of approximately 10 copies for both analytes in method I and 20 copies in method II. This corresponds to approximately 420 copies/ml of CSF for method I with currently used test volumes (RNA extracted from 200 μl CSF into an elution volume of 50 μl; 6 μl was used in the PCR; the effective test volume was 24 μl) and 550 copies/ml in method II (200 μl CSF, with an elution volume of 50 μl; 9 μl was used in the PCR; the effective test volume was 36 μl). A 5- to 10-fold increase in diagnostic sensitivity could be achieved with large sample volume extractions and increasing the amount of RNA added to the PCR (150- to 300-μl effective volumes; 30 to 70 RNA copies/ml in CSF using method I, 70 to 130 copies/ml using method II).
The equivalent amplification efficiency and sensitivity of the assay for target sequences of all four EV species provides reassurance that a wide range of EV variants can be detected by the multiplexed assay. In particular, it directly addresses the concern arising from the previous use of cell culture-based diagnostic methods that species A variants often were underdiagnosed because of their poor in vitro replication (2, 4). Similarly, the absence of species C or D nonpoliovirus EV types in diagnostic specimens, screened to provide direct evidence for their lack of CNS-associated presentations (or sepsis-like illness) in the target population, does not reflect a lack of assay sensitivity (10). This is despite the wide circulation of species C enteroviruses in the community (4), as also determined by recent environmental surveillance (E. C. M. Leitch, J. S. Calvert, H. Harvala, and P. Simmonds, unpublished data). Furthermore, for species C this is particularly important in view of the previous transportation of wild poliovirus serotype 1 into Europe, the first report since the European regions were declared polio-free in 2002 (5, 8), and validates the assay for current enhanced enterovirus surveillance.
For more than for any other sample type, high sensitivity and specificity is needed for CSF screening. Viral loads frequently are extremely low, close to or below assay sensitivity levels due to the limited penetration of virus into the CNS and because sampling often is conducted in the resolution phase of the acute infection, when symptoms may be most pronounced but systemic viral loads are rapidly falling. In further contrast to other specimen types, because of its ability to cross the blood-brain barrier, the detection of EV or HPeV in CSF specimens usually should be regarded as significant and potentially causative of the disease presentations. The introduction of the highly sensitive, rapid screening of patients presenting with meningitis or other neurological symptoms thus has the ability to considerably improve their clinical management. Although EV and HPeV infections cannot be directly treated, rapid test results (within hours of obtaining a sample) will, for example, reduce hospital attendance and shorten the duration of antibiotic treatment (17, 26). The screening assay described in the current study provides the means to rapidly detect the two viruses most closely linked with CNS-associated infections, including viral meningitis, and its format represents a useful template for multiplexing further diagnostic targets.
ACKNOWLEDGMENTS
Full-length clones were kindly provided by Glyn Stanway, University of Essex, United Kingdom, and David Evans, University of Warwick, United Kingdom. The probit analysis was kindly performed by Margo Chase-Topping, University of Edinburgh, United Kingdom. We thank Peter McCullough, Nadine Wilkinson, Alison Hardie, and Naomi Gadsby for technical assistance with the assay. The transcripts were created in a diagnostic development program funded by the United Kingdom clinical virology network.
The work was funded in part by a grant to H.H. from the Society for General Microbiology Specialist Trainee award.
Footnotes
Published ahead of print on 18 May 2011.
REFERENCES
- 1. Arola A., Santti J., Ruuskanen O., Halonen P., Hyypia T. 1996. Identification of enteroviruses in clinical specimens by competitive PCR followed by genetic typing using sequence analysis. J. Clin. Microbiol. 34:313–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Benschop K., et al. 2010. Detection of enterovirus and human parechovirus genotypes from clinical stiool samples; PCR and direct molecular typing, culture characteristics and serotyping. Diagn. Microbiol. Infect. Dis. 68:166–173 [DOI] [PubMed] [Google Scholar]
- 3. Benschop K., Molenkamp R., van der Ham A., Wolthers K., Beld M. 2008. Rapid detection of human parechoviruses in clinical samples by real-time PCR. J. Clin. Virol. 41:69–74 [DOI] [PubMed] [Google Scholar]
- 4. Blomqvist S., Paananen A., Savolainen-Kopra C., Hovi T., Roivainen M. 2008. Eight years of experience with molecular identification of human enteroviruses. J. Clin. Microbiol. 46:2410–2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. CDC 2010. Outbreaks following wild poliovirus importations-Europe, Africa, and Asia, January 2009-September 2010. MMWR Morb. Mortal. Wkly. Rep. 59:1393–1399 [PubMed] [Google Scholar]
- 6. Chapman N. M., Tracy S., Gauntt C. J., Fortmueller U. 1990. Molecular detection and identification of enteroviruses using enzymatic amplification and nucleic acid hybridization. J. Clin. Microbiol. 28:843–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dierssen U., Rehren F., Henke-Gendo C., Harste G., Heim A. 2008. Rapid routine detection of enterovirus RNA in cerebrospinal fluid by a one-step real-time RT-PCR assay. J. Clin. Virol. 42:58–64 [DOI] [PubMed] [Google Scholar]
- 8. European Centre for Disease Prevention and Control 2010. Outbreak of poliomyelitis in Tajikistan in 2010: risk for importation and impact on polio surveillance in Europe? Eurosurveillance 15:19558. [DOI] [PubMed] [Google Scholar]
- 9. Gunson R. N., Collins T. C., Carman W. F. 2006. Practical experience of high throughput real time PCR in the routine diagnostic virology setting. J. Clin. Virol. 35:355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Harvala H., et al. 2011. Comparison of human parechovirus and enterovirus detection frequencies in cerebrospinal fluid samples collected over a 5-year period in Edinburgh-HPeV type 3 identified as the most common picornavirus type. J. Med. Virol. 83:889–896 [DOI] [PubMed] [Google Scholar]
- 11. Harvala H., et al. 2009. Aetiological role of human parechovirus type 3 in neonatal sepsis identified by direct typing assay on cerebrospinal fluid. J. Infect. Dis. 199:1753–1760 [DOI] [PubMed] [Google Scholar]
- 12. Harvala H., et al. 2008. Epidemiology and clinical associations of human parechovirus respiratory infections. J. Clin. Microbiol. 46:3446–3453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Harvala H., Wolthers K., Simmonds P. 2010. Parechoviruses in children: understanding a new infection. Curr. Opin. Infect. Dis. 23:224–230 [DOI] [PubMed] [Google Scholar]
- 14. Hyypiä T., Auvinen P., Maaronen M. 1989. Polymerase chain reaction for human picornaviruses. J. Gen. Virol. 70:3261–3268 [DOI] [PubMed] [Google Scholar]
- 15. Hyypiä T., et al. 1992. A distinct picornavirus group identified by sequence analysis. Proc. Natl. Acad. Sci. U. S. A. 89:8847–8851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hyypiä T., Hovi T., Knowles N. J., Stanway G. 1997. Classification of enteroviruses based on molecular and biological properties. J. Gen. Virol. 78:1–11 [DOI] [PubMed] [Google Scholar]
- 17. Lai K. K., Cook L., Wendt S., Corey L., Jerome K. R. 2003. Evaluation of real-time PCR versus PCR with liquid-phase hybridization for detection of enterovirus RNA in cerebrospinal fluid. J. Clin. Microbiol. 41:3133–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Leitch E. C., et al. 2009. Direct identification of human enterovirus serotypes in cerebrospinal fluid by amplification and sequencing of the VP1 region. J. Clin. Virol. 44:119–124 [DOI] [PubMed] [Google Scholar]
- 19. Nix W. A., et al. 2008. Detection of all known parechoviruses by real time-PCR. J. Clin. Microbiol. 46:2519–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Noordhoek G. T., Weel J. F., Poelstra E., Hooghiemstra M., Brandenburg A. H. 2008. Clinical validation of a new real-time PCR assay for detection of enteroviruses and parechoviruses, and implications for diagnostic procedures. J. Clin. Virol. 41:75–80 [DOI] [PubMed] [Google Scholar]
- 21. Olive D. M., et al. 1990. Detection and differentiation of picornaviruses in clinical samples following genomic amplification. J. Gen. Virol. 71:2141–2147 [DOI] [PubMed] [Google Scholar]
- 22. Pöyry T., et al. 1994. Molecular analysis of coxsackievirus A16 reveals a new genetic group of enteroviruses. Virology. 202:982–987 [DOI] [PubMed] [Google Scholar]
- 23. Stanway G., et al. 2005. Family Picornaviridae, p. 757–778 In Fauquet C. M., Mayo M. A., Maniloff J., Desselberger U., Ball L. A. (ed.), Virus taxonomy. Eighth Report of the International Committee on Taxonomy of Viruses. Elsevier/Academic Press, London, United Kingdom [Google Scholar]
- 24. Wacker M. J., Godard M. P. 2005. Analysis of one-step and two-step real-time RT-PCR using SuperScript III. J. Biomol. Tech. 16:266–271 [PMC free article] [PubMed] [Google Scholar]
- 25. Wigand R., Sabin A. B. 1961. Properties of ECHO types 22, 23 and 24 viruses. Arch. Gesamte Virusforsch. 11:224–247 [DOI] [PubMed] [Google Scholar]
- 26. Wolthers K. C., et al. 2008. Human parechoviruses as an important viral cause of sepsislike illness and meningitis in young children. Clin. Infect. Dis. 47:358–363 [DOI] [PubMed] [Google Scholar]
- 27. Zoll G. J., et al. 1992. General primer-mediated polymerase chain reaction for detection of enteroviruses: application for diagnostic routine and persistent infections. J. Clin. Microbiol. 30:160–165 [DOI] [PMC free article] [PubMed] [Google Scholar]