Abstract
Human papillomavirus type 16 (HPV 16) plays a cardinal role in the pathogenesis of cervical cancer. HPV 16 has intratypic variants which show different geographical distributions and different oncogenic potentials. To analyze the presence of sequence variations of HPV 16 variants in northeast China, 71 cervical carcinomas were identified by HPV typing. HPV 16-positive specimens were analyzed by PCR-directed sequencing in the E6, E7, and L1 genes and the LCR (long control region). The variation data were compared with those of neighboring districts. In this hospital-based study, HPV 16 was the most common type (73.24%). In HPV 16-positive specimens, 67.31% belonged to the European (E) lineage, while 32.69% were Asian (As) variants. The Asian-American (AA), African-1 (Af-1), African-2 (Af-2), and northern American (NA) lineages were not detected. The most frequently observed variation sites were T178G (32.69%) in E6; A647G (34.62%), G666A (38.46%), and T846C (32.69%) in E7; C6826T (36.17%) and G7060A (61.70%) in L1; and G7521A (98.08%) in the LCR. The most prevalent amino acid variations were D25E in E6 and N29S in E7. In addition, 28 novel variations of HPV 16 were reported. Some covariations between different genes were obtained. In this study, HPV 16 variants belonged to the European lineage and the Asian lineage. Compared with neighboring districts, the distribution of HPV 16 variants in northeast China had a typical pattern. As the first report on HPV 16 variants in northeast China, it should be helpful for designing a HPV vaccine and HPV vaccination program in China.
INTRODUCTION
Human papillomavirus type 16 (HPV 16) is the primary etiology of cervical cancer, which is the second most common type of cancer in women worldwide (29). HPV 16 variants, which vary by ≤2% from HPV 16 prototype nucleotide sequences, have been identified as the following six phylogenetic branches: European (E), Asian (As), Asian-American (AA), African-1 (Af-1), African-2 (Af-2), and northern American (NA) variants (18, 52). Several researchers had reported correlations between specific HPV 16 variants and persistent viral infection, followed by the development of malignant lesions (3, 4, 16, 37, 43, 49, 50). Non-European variants were found to be associated with an excess risk of cervical cancer (37). These variants had been found to show different geographic distributions, while some sequence variations had oncogenic potentials. In HPV 16 variants, the L83V mutation in E6 in the Swedish and Italian populations and D25E in E6 in the Japanese population were reported to be associated with the progression of cervical carcinoma (27, 53, 54). The HPV 16 Asian variant was the major causative agent associated with cervical cancer in Japan and northeast Thailand (10).
China has one of the highest incidence rates of cervical cancer, and approximately 13,2300 new cases are reported every year (33). Recent data showed that the mortality of cervical cancer was 2.55/100,000 people in the China mainland. The highest mortality existed in northwest China (10.69/100,000 people in Xinjiang and 9.36/100,000 in Gansu) and central China (4.98/100,000 people in Hunan and 4.90/100,000 in Jiangxi). In southwest China, the mortality of cervical cancer was 1.53/100,000 people in Sichuan. In northeast China, the mortality of cervical cancer was 2.12/100,000 people in Heilongjiang, 1.97/100,000 in Jilin, and 1.35/100,000 in Liaoning (57).
As the most prevalent genotype, the prevalences of HPV 16 among different geographical regions in China were similar (7, 42, 45, 56). However, the distribution of HPV 16 variants in China was studied less. The Asian lineage was reported in southwest China (31.0% in Sichuan) and southern China (50.6% in Hong Kong). The European prototype was reported in southwest China (23.0% in Sichuan), central China (15.52% in Hubei), and southern China (30.0% in Hong Kong) (5, 6, 35). No data on HPV 16 variants and sequence variations were reported in northern China and northeast China.
To characterize the prevalence of HPV 16 variants and nucleotide polymorphisms in northeast China, we have investigated the HPV 16 E6, E7, and L1 genes and the long control region (LCR) in cervical carcinomas. The results were compared with data reported from other neighboring areas.
MATERIALS AND METHODS
Preparation of clinical specimens.
Recruitment of study subjects was conducted from June 2005 through December 2008. The subjects were women with newly diagnosed invasive cervical carcinoma (ICC) that was histological confirmed at Tumor Hospital of Harbin Medical University in Heilongjiang province. The inpatients came from Heilongjiang, Jilin, and the north Inner Mongolia region, which lies in the north district of northeast China. For our database, 71 patients were identified as having squamous carcinomas, except sample 13 (adenocarcinoma) and sample 53 (adenosquamous carcinoma). Cervical samples were obtained from women undergoing surgery. Following the cervical punch biopsy, the biopsy tissue was sent for histological processing and examined by two independent pathologists. Multiple aliquots were cut and stored at −70°C. The study protocol was approved by the institutional ethical committee. Written informed consent was obtained from each study subject.
The quality of extracted DNA was checked by PCR amplification of the β-globin gene (forward primer, 5′-CAACTTCATCCACGTTCACC-3′, and reverse primer, 5′-GAAGAGCCAAGGACAGGTAC-3′). Amplification without a DNA template was used to monitor contamination in both HPV and β-globin reactions. PCRs were performed on a DNA Engine Peltier thermal cycler (Bio-Rad). A 268-bp amplicon was determined with 2% agarose electrophoresis (26).
Detection and typing of HPV.
DNA was extracted by a standard proteinase K digestion and phenol-chloroform extraction method (40). The DNA samples were determined by spectrophotometric quantitation and 0.7% agarose electrophoresis. All specimens were first subjected to PCR amplification with HPV consensus primers GP5+ (5′-TTTGTTACTGTGGTAGATACYAC-3′) and GP6+ (5′-GAAAAATAAACTGTAAATCATATTC-3′). HPV 16- and 18-positive DNA was identified by specific primers (5′-AAGGG[C/A]GTAAC CGAAA[T/A]CGG-3′, 5′-CATATACCTCACGTCGCAG-3′, and 5′-TTCTGCTGGATTCAACGGT-3′). HPV 16 was amplified as a 206-bp fragment, while HPV 18 was identified as a 418-bp fragment. The primers were synthesized by Takara Bioengineering Co. Ltd. in Dalian. After thermal cycling, the amplified DNA fragments were identified by 1.5% agarose electrophoresis.
Sequence analysis of the E6, E7, and L1 genes and the LCR of HPV 16 by PCR-directed sequencing.
HPV 16 L1-E7 fragments and LCR-E7 fragments were amplified by high-fidelity PCR and sequenced. The L1-E7 fragment, with an amplicon size of 3.34 kb, was amplified with primers 5′-TAATATAACTGACCAAGCTCCTTCATTAATTC-3′ and 5′-TACATAAAACCATCCATTACATCCCGTACC-3′, which flank the regions of the L1, E6, and E7 genes and the LCR region (nucleotides [nt] 5502 to 921). The LCR-E7 fragment (nt 7432 to 921) was amplified, with an amplicon size of 1.4 kb, with primers 5′-CTATATTTTGTAGCGCCAG-3′ and 5′-CATCCATTACATCCCGTAC-3′. For HPV 16 L1-E7, PCR was performed in a 50-μl volume containing 1× PCR buffer, 2.5 mM MgCl2, 200 μM each deoxynucleoside triphosphate (dNTP), 0.2 μM sense and antisense primers, 20 ng genomic DNA, and 1.25 U PrimeStar HS DNA polymerase (Takara) with a 35-cycle protocol, as follows: 1 min of denaturation at 94°C, 0.5 min of annealing at 60°C, and 5 min of extension at 68°C, with 5 min of initial denaturation at 94°C and 10 min of final elongation at 68°C. The amplified DNA fragments were sent to Takara Bioengineering Co. Ltd. in Dalian for sequencing after a 2% agarose electrophoresis assay. L1-E7 and LCR-E7 primers were also available for direct sequencing. Sequencing analysis was completed by primer walking. To avoid nucleotide variations resulting from DNA polymerase error, the sequences were confirmed twice by repeat PCR amplification (34). Changes were accepted only when they were reproducible with a second PCR. HPV 16 sequences and base positions were numbered according to the HPV 16 reference (HPV 16R) sequence (30). The sequences were analyzed and determined by DNAMAN version 5.2.2.
Variant identification.
HPV 16 variants were classified into phylogenetic classes and subclasses on the basis of their sequence variation at the E6 open reading frame (ORF), which is the most informative for this purpose. The E6 sequences were then compared with the reference sequences using the HPV 16R and used to classify HPV 16 variant classes. The European (E) lineage has approximately complete homology with the HPV 16R, but with no signature patterns of other classes identified. Changes in the following lineages were noted: Asian (As), T178G; Af-1, G132C, C143G, G145T, T286A, A289G, and C335T; Af-2, T109C, G132T, C143G, G145T, T286A, A289G, C335T, and A403G; NA-1, G145T, T286A, A289G, C335T, and T350G; and AA, G145T, T286A, A289G, C335T, T350G, and A532G (18, 46, 51, 52). Sequence variations in E6, E7, L1, and LCR in this study were compared with other data reported from neighboring geographical districts, which originated from cervical carcinomas in hospital-based studies.
Statistical analysis.
Statistical analysis was performed using the chi-square (χ2) test to determine the differences between northeast China and other geographical regions regarding HPV 16 variants and the European lineage distribution of cancer patients. P values of less than 0.05 were considered statistically significant.
RESULTS
HPV typing.
All 71 cervical carcinomas were β-globin gene positive, indicating the presence of adequate cells in DNA samples. The HPV infection rate of cervical carcinomas was 98.59% (70 of 71). HPV 16 was the most common type (73.24%, 52 of 71), followed by HPV 18 (11.27%, 8 of 71).
HPV 16 variant identification.
In HPV 16-positive specimens, 67.31% (35 of 52) belonged to the European lineage, while 32.69% (17 of 52) belonged to the Asian lineage. The AA, Af-1, Af-2, and NA lineages were not detected. The HPV 16 European lineage contained the European prototype (54.29%, 19 of 35) and European variants (45.71%, 16 of 35) (see Table 3).
Table 3.
Comparison of HPV 16 variant distributions and sequence variations with neighboring districts and countries
| Variant (sample no.) | Reference | No. of cases sequenced | No. (%) of cervical carcinoma casesa |
|||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HPV 16 variant |
E lineage |
Variation site |
||||||||||||||||||||||
| E | As | Af-1 | Af-2 | AA | NA | E prototype | E variant | D25E in E6 | T350G (L83V) in E6 | G94A in E6 | T178A in E6 | A276G (N58S) in E6 | C335T (H78Y) in E6 | A442C (E113D) in E6 | T183G (I27R) in E6 | A647G (N29S) in E7 | C749T (S63F) in E7 | G666A in E7 | C6826T (silence) in L1 | G7521A in LCR | G7842A in LCR | |||
| Northeast China (1) | 52 | 35 (67.31) | 17 (32.69) | 0 | 0 | 0 | 0 | 19 (54.29) | 16 (45.71) | 22 (42.31) | 3 (5.77) | 6 (11.54) | 5 (9.62) | 2 (3.85) | 0 | 1 (1.92) | 0 | 18 (34.62) | 0 | 20 (38.46) | 17 (36.17) | 51 (98.08) | 16 (30.77) | |
| Southwest China—Sichuan (2) | 35 | 113 | 78 (69.03) | 35 (30.97) | 0 | 0 | 0 | 0 | 26 (33.33) | 52 (66.67) | 35 (30.97) | 35 (30.97) | 7 (6.19) | 0 | 4 (3.54) | 14 (12.39) | 7 (6.19) | 0 | — | — | — | — | — | — |
| Southwest China—Sichuan (3) | 41 | 21 | 16 (76.19) | 5 (23.81) | 0 | 0 | 0 | 0 | 4 (25.00) | 12 (75.00) | 5 (23.81) | 2 (9.52) | 0 | 0 | 0 | 2 (9.52) | 1 (4.76) | 0 | 3 (14.29) | 0 | 3 (14.29) | — | 6 (60.00) | 3 (14.29) |
| Central China—Hubei (4) | 5 | 58 | 23 (39.66) | 35 (60.34) | 0 | 0 | 0 | 0 | 9 (39.13) | 14 (60.87) | 36 (62.07) | 3 (5.17) | — | 1 (1.72) | 0 | 2 (3.45) | 5 (8.62) | 3 (5.17) | — | — | — | — | — | — |
| Central and southern China—Jiangxi, Guangdong (5) | 47 | 55 | 19 (34.55) | 36 (65.45) | 0 | 0 | 0 | 0 | 5 (26.32) | 14 (73.68) | 37 (67.27) | 2 (3.64) | 0 | 1 (1.82) | 0 | 3 (5.45) | 8 (14.55) | 0 | 33 (70.21) | 24 (51.06) | 2 (4.26) | — | — | — |
| Japan (6) | 27 | 43 | 24 (55.81) | 19 (44.19) | 0 | 0 | 0 | 0 | 5 (20.83) | 19 (79.17) | 19 (44.19) | 14 (32.56) | 0 | 0 | 1 (2.33) | 3 (6.98) | 11 (25.58) | 1 (2.33) | — | — | — | — | — | — |
| Northeast Thailand (7) | 10 | 23 | 6 (26.09) | 17 (73.91) | 0 | 0 | 0 | 0 | 1 (16.67) | 5 (83.33) | 17 (73.9) | — | 0 | 0 | — | — | — | — | — | — | — | — | — | — |
| Russia (8) | 19 | 32 | 30 (93.75) | 0 | 0 | 0 | 0 | 2 (6.25) | 5 (16.67) | 25 (83.33) | 0 | 27 (84.38) | 0 | 0 | 0 | 2 (6.25) | 0 | 0 | — | — | — | — | — | — |
| Indonesia (9) | 11 | 22 | 19 (86.36) | 2 (9.09) | 0 | 1 (4.55) | 0 | 0 | 7 (36.84) | 12 (63.16) | 2 (9.09) | 1 (4.55) | 0 | 0 | 9 (40.91) | 1 (4.55) | 0 | 0 | 5 (22.73) | 0 | 16 (72.73) | 17 (77.27) | — | — |
| North India (10) | 32 | 60 | 52 (86.67) | 0 | 0 | 0 | 2 (3.33) | 6 (10.00) | 18 (34.62) | 34 (65.38) | 0 | 42 (70.00) | 0 | 0 | 0 | 6 (10.00) | 0 | 0 | 0 | 0 | 1 (1.67) | 0 | 41 (68.33) | 0 |
A dash indicates that no data were provided. Chi-square (χ2) tests were applied in this study. Regarding HPV 16 variant distribution, the differences had no significance between samples 1 and 2, 1 and 3, 2 and 3, 1 and (2 + 3), 4 and 5, and 1 and 6 (P value > 0.05), while statistical significance existed between samples 1 and 4/5/7/8/9/10, 1 and (4 + 5), (2 + 3) and 6/7/8/9/10, and (4 + 5) and 6/7/8/9/10 (P value < 0.05). Regarding the distribution of the HPV 16 European prototype and European variant, statistical significance existed between 1 and 2/3/4/5/6/7/8/9/10.
E6 gene sequence variations.
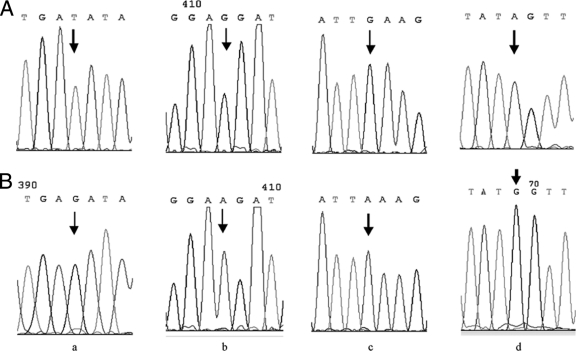
In HPV 16-positive samples, the nucleotide variation rate of HPV 16 E6 was 63.46% (33 of 52) (Table 1), whereas the amino acid variation rate was 57.69% (30 of 52). Nucleotide variation positions of E6 included three silent mutations (G94A, A131C, and T241G) and six missense mutation positions (G176A, T178G/T178A, A276G, T341G, T350G, and A442C), which led to amino acid variations (D25N, D25E, N58S, C80G, L83V, and E113D). The most frequent sequence variation site was nt 178 (42.31%, 22 of 52), which contained T178G (17 of 52) and T178A (5 of 52) (Fig. 1 a). Other common mutations were G94A (11.54%, 6 of 52) and T350G (5.77%, 3 of 52). The rank orders of incidence of the HPV 16 prototype and E6 variants were as follows: D25E (42.31%), the prototype (36.54%), L83V (5.77%), D25N (5.77%), and N58S (3.85%). Only one novel mutation (T341G) in E6 was found for the first time, which resulted in amino acid substitution C80G (Table 2).
Table 1.
Cases exhibiting the HPV 16 prototype or variant in E6, E7, the LCR, and L1
| Gene/region | No. (%) of cases |
|
|---|---|---|
| Prototype | Variant | |
| E6 | 19 (36.54) | 33 (63.46) |
| E7 | 8 (15.38) | 44 (84.62) |
| LCRa | 0 (0.00) | 52 (100.00) |
| L1b | 2 (4.26) | 45 (95.74) |
Variant analysis for the partial LCR sequence was done in samples 48, 49, 50, 51, and 52 (Fig. 2).
Variant analysis for L1 was done using only 47 cases.
Fig. 1.
A sequencing electropherogram showed frequently detected nucleotide variations in different genomic segments of HPV 16. (a) E6 T178A; (b) E7 G666A; (c) L1 G7060A; (d) LCR G7521A. (Top) Prototype sequences indicated by arrows; (bottom) variant sequences indicated by arrows.
Table 2.
Novel variants of HPV 16 E6 and L1 and the LCR according to nucleotide position, variant, altered amino acid, and mutation
| Gene/region | Nt position | Nt |
Amino acid |
Codon no. | Type of mutationa | No. of mutations | ||
|---|---|---|---|---|---|---|---|---|
| Prototype | Variant | Original | Altered | |||||
| E6 | 341 | T | G | Cysteine | Glycine | 80 | M, Tv | 1 |
| L1 | 5569 | G | A | Valine | Valine | 3 | S, Ts | 1 |
| 5596 | A | T | Threonine | Threonine | 12 | S, Tv | 1 | |
| 5836 | A | G | Serine | Serine | 92 | S, Ts | 1 | |
| 5883 | A | C | Lysine | Threonine | 108 | M, Tv | 1 | |
| 5902 | C | A | Threonine | Threonine | 114 | S, Tv | 1 | |
| 6085 | G | A | Methionine | Isoleucine | 175 | M, Ts | 1 | |
| 6164 | A | G | Threonine | Alanine | 202 | M, Ts | 1 | |
| 6166 | C | A | Threonine | Threonine | 202 | S, Tv | 1 | |
| 6427 | G | A | Arginine | Arginine | 289 | S, Ts | 1 | |
| 6505 | A | G | Serine | Serine | 315 | S, Ts | 2 | |
| 6658 | A | T | Threonine | Threonine | 366 | S, Tv | 3 | |
| 6667 | A | T | Serine | Serine | 369 | S, Tv | 9 | |
| 6938 | C | G | Proline | Alanine | 460 | M, Tv | 1 | |
| 7142 | A | C | Lysine | Glutamine | 528 | M, Tv | 1 | |
| LCR | 7168 | A | G | Ts | 17 | |||
| 7174 | A | C | Tv | 9 | ||||
| 7233 | A | G | Ts | 1 | ||||
| 7419 | A | G | Ts | 2 | ||||
| 7429 | G | A | Ts | 1 | ||||
| 7660 | A | G | Ts | 2 | ||||
| 7667 | A | C | Tv | 1 | ||||
| 7700 | T | G | Tv | 1 | ||||
| 7781 | T | A | Tv | 1 | ||||
| 7855 | G | A | Ts | 1 | ||||
| 7859 | A | C | Tv | 1 | ||||
| 16 | G | A | Ts | 1 | ||||
| 48 | G | A | Ts | 2 | ||||
M, missense; S, silence; Ts, transition; Tv, transversion.
E7 gene sequence variations.
Forty-four (84.62%) patients showed six mutation sites for the E7 gene, of which two, A645C and A647G, were missense mutations: asparagine→serine (N29S) and leucine→phenylalanine (L28F). The remaining four, G666A (Fig. 1b), T760C, T843C, and T846C, led to silent mutations. The amino acid variation rate was 36.54% (19 in 52). The most frequently observed mutations were G666A (38.46%, 20 of 52), A647G (34.62%, 18 of 52), and T846C (32.69%, 17 of 52), while the most frequent amino acid mutation was N29S (34.62%).
L1 gene sequence variations.
The HPV 16 L1 genes (nt 5559 to 7154) from 47 HPV 16-positive cervical cancers patients were determined. Of all 19 nucleotide variation positions, six were missense mutations (A5883C, G6085A, A6164G, A6180C, C6938G, and A7142C) which led to K108T, M175I, T202A, N207T, P460A, and K528Q, respectively. The remaining 13 were silent mutations. There were three common nucleotide variations at nt 6667, nt 6826, and nt 7060, which were silent variations (Fig. 1c). The most frequent amino acid mutation was N207T (6.38%, 3 out of 47). In 47 HPV 16-positive cervical carcinomas, the nucleotide variation rate in the HPV 16 L1 region was 95.74% (45 of 47), whereas the amino acid variation rate was 14.89% (7 of 47). There were 14 novel nucleotide mutations in the L1 gene (Table 2).
LCR sequence variations.
Forty-seven whole LCR fragments and five partial LCR fragments were sequenced. A total of 32 nucleotide variation positions were detected (Fig. 2). The most commonly observed LCR variation was the transition replacement G7521A (98.08%), which is shown in Fig. 1d. We reported a total of 13 novel nucleotide variation sites in LCR, which were observed here for the first time (Table 2). The nucleotide variation rate in the LCR was 100% in 47 HPV 16-positive cervical carcinomas.
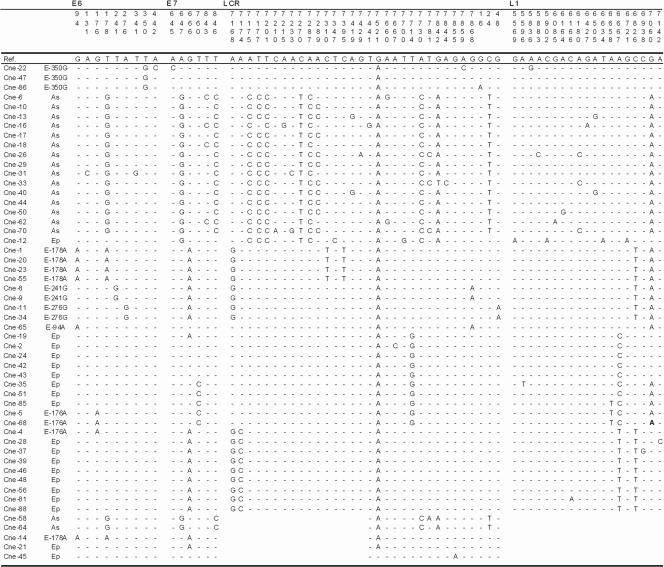
Fig. 2.
Nucleotide sequence variations among the HPV 16 isolates. The E6, E7, L1, and LCR variants were identified in 52 samples from northeast China. The nucleotide positions of detected mutations are written vertically across the top and are indicated by the corresponding nucleotide letter. The absence of variations relative to the prototype is represented by dashes. The identification codes of the samples are indicated on the left. 16R, HPV 16 reference; Ep, European prototype; As, Asian.
Covariations.
In the HPV 16 Asian branch, covariations of T178G, A647G, T846C, A7175C, T7177C, T7201C, C7270T, A7287C, A7289C, A7730C, G7842A, and C24T were detected, resulting in covariations of predicted amino acids D25E and N29S (32.69%, 17/52) (Fig. 2). Covariations of G666A (E7), C6826T (L1), and A7168G (LCR) were detected (36.17%, 17 of 47) in the E branch in this study.
Variant comparison with neighboring districts.
HPV 16 variant distribution in this study was different from that in central and southern China, Russia, Indonesia, and north India, whereas it was not different from that in Sichuan (southwest China) and Japan. The difference had no significance among northeast China and Sichuan; however, Japan was different from Sichuan in terms of statistics. No differences existed between Hubei (central China), Jiangxi, and Guangdong (central and southern China). Furthermore, the distribution of the European prototype and European variants in northeast China was different from that in any other area by statistical analysis (P < 0.05). The frequencies of mutation for D25E and N29S in northeast China were higher than those in Sichuan but lower than those in central and southern China. Other mutation frequencies varied among different districts (Table 3).
DISCUSSION
Located in the center of northeast Asia, northeast China is close to north China and neighbors Korea, Russia, Mongolia, and Japan. In history, more communication lies between China, Mongolia, Korea, Russia, and Japan. Northeast China has a population of around 110 million people, who immigrated mainly from north China (Shandong and Hebei). However, no HPV 16 variant data from north China were provided, where most of the population originated. Since HPV 16 variants are associated with geographic diversity and oncogenic risk, we analyzed the data with China origin (southwest China, central China, and southern China) and those from neighboring countries (Russia and Japan) (52). Hong Kong, Mongolia, and Korea were excluded from the analysis since no individual data on cervical carcinoma samples were reported (6, 8, 9).
In this hospital-based study, the samples contained two HPV 16 lineages, the European lineage (67.31%) and the Asian lineage (32.69%), which suggested that the European and Asian lineages were the main lineages in northeast China. In Table 3, the European and Asian lineages in these studies were circulated, while the AA, Af-1, Af-2, and NA lineages were reported less often. The European lineage was the main lineage in Russia, north India, and Indonesia. By statistical analysis, the variant distribution in northeast China was the same as those in Sichuan (southwest China) and Japan and different from those in central and southern China and neighboring countries, except Japan.
Southwest China (Sichuan) and northeast China were areas with a moderate mortality of invasive cervical cancer, while central China (Hubei and Jiangxi) had a high mortality (57). Northeast China and southwest China have the same HPV 16 variant distribution by statistical analysis, while Hubei, Jiangxi, and Guangdong have higher Asian variant frequencies. Interestingly, it was reported that HPV 16 Asian variants might be linked to a high incidence of cervical cancer in China (48). We wondered if the distribution character of HPV 16 variants, especially Asian variants, had a relationship with the mortality of invasive cervical cancer in these geographical areas. But this consideration needs further investigation by population-based study in more districts in China.
The European and Asian lineages were the main patterns in the Chinese population, as shown in Table 3. There were two specific groups with HPV 16 variant distribution. The first group contained southwest China (Sichuan) and northeast China, and the second group contained central China and southern China (Hubei, Jiangxi, and Guangdong). The difference had no significance within each group. The two groups were different from neighboring countries. In this analysis, the Japan sample was different from that of the first group, although it had no differences with the northeast China sample. Data from east China (Zhejiang) belonged to the second group (20). Data from Hong Kong (southern China) also conformed to the second group, which belonged to a population-based study (6). However, the distributions of the European prototype and European variants in northeast China were specific, because the data were different from data of any other area by statistical analysis. Identification of HPV 16 variants may be helpful to evaluate the potential impact of HPV vaccine and design diagnostics and therapeutic methods on cervical cancer (15, 44). Although further confirmation of this study is still needed, the results may be helpful for the study of the HPV vaccination program in China.
Covariation of nucleotide sequences among these genes was analyzed, since the HPV 16 E6, E7, and L1 genes and the LCR were sequenced at one DNA molecule by primer walking. Among the Asian lineage, covariations of predicted amino acids D25E and N29S might be a typical pattern. Covariations of G666A (E7) and C6826T (L1), reported as Javanese variants, were found in 72.73% of Indonesian samples (Fig. 2) (11). Validation of Javanese variants needed more confirmation, because G176A, T178A, and T241G were detected in this variant apart from A276G.
Amino acid mutations in HPV 16 variants should be considered in the vaccine design and evasion of the natural immune response (12, 17, 47). As the target of humoral and cellular immune responses, the amino acid changes of HPV 16 E6 may have effects (2, 13, 39). D25E and L83V are associated with the elevated risk of cervical carcinomas and vary geographically due to genetic differences between populations (1, 3, 18, 22, 25, 27, 54, 55). The D25E variant distributes mainly in Asian populations rather than in Russia (0%) (6, 28, 31, 52). Based on our results, it was supposed that genetic movement of D25E mutations might exist in these neighboring areas, for northeast China lies geographically between high-frequency areas (central and southern China and southeast Asia) and low-distribution areas (Russia and Mongolia). T178A, which led to D25E amino acid changes, was found in the Chinese and Japanese populations (5, 6, 47). The L83V distribution was lower in the Asian population and higher in the American and European populations. In Table 3, L83V mutations in northeast China accounted for 5.77%, close to those in central China and southern China, in contrast to that in southwest China (27.61% in Sichuan), 32.56% in Japan, and 84.38% in Russia (Moscow) (5, 6, 8, 9, 19, 47, 52). G94A was also reported in southwest China and Mongolia (8, 35). Sequence variation at E7 displays geographic dependence (14, 36). The E7 N29S replacement may be associated with a higher oncogenic risk (12, 23, 38). In this study, N29S accounted for 34.62% of mutations, which varied from 14.29% (southwest China) to 70.21% (central and southern China). G666A varied between northeast China, southwest China, central and southern China, and Indonesia (11, 41, 47).
Compared with E6 gene variations, the HPV 16 L1 gene product is more conservative in terms of its amino acid sequence. HPV 16 L1 nucleotide variations were found in 95.74% of specimens, while amino acid mutations were located only in 14.89% of specimens. The amino acid changes of K108T, M175I, T202A, N207T, P460A, and K528Q were reported previously. The silent variation C6826T has been reported from southeast Asia (52). To our knowledge, 14 novel mutations in this study were not reported previously.
The LCR is the most variable segment of the HPV 16 genome in different populations (21, 22, 24, 43, 52). The LCR is the binding site of various cellular and viral transcription factors. In this study, the nucleotide variation rate in the LCR was 100%. A total of 5 of 32 mutation positions (15.63%) impacted cellular transcription factor binding sites: C7310T lies in AP1, C7395T lies in GRE, T7441G lies in YY11, T7714G lies in NF1, and G7842A and G7842T lie in Oct-1. Over half of the isolates (32/52) showed the above-mentioned mutation sites. The exact oncogenic potentials of these nucleotide mutations still need further investigation.
In this study, we investigated comprehensively sequence variations of the HPV 16 E6, E7, and L1 genes and the LCR based on long fragment sequencing. Five HPV 16-positive samples which lost the L1 ORF were sequenced on LCR-E7 fragments. Age was not considered in this study, for age was reported to have no relationship with the distribution of HPV 16 variants among women in the same geographic region (34). As a hospital-based sample study, our study was still limited by sample size. The conclusions in this study need further investigation by a large population-based study.
In the present study, it was demonstrated that HPV 16 variants in northeast China have typical distribution. HPV 16 variants belonged to the European lineage and the Asian lineage. D25E variation in E6 and N29S variation in E7 were the most prevalent variations. Twenty-eight novel variations of HPV 16 were reported. This is the first report on HPV 16 variant distribution and sequence variations in northeast China. Based on this study, we supposed that northeast China may be an area with moderate HPV 16 Asian variant prevalence. Further investigation should be emphasized on Asian variants in east Asia. We suggested that an HPV-preventive vaccine should be applied to control the HPV 16 infection in northeast China. These data laid the foundation for the HPV vaccination program in northeast China and are important for diagnostic technique development and vaccine design for eradication of cervical cancers.
ACKNOWLEDGMENTS
This research was supported by grant 11551236 from the Heilongjiang Higher Education Institutions, funding (grants LC2009C26 and D201065) from the Natural Science Foundation of Heilongjiang Province, and grants 81000726 and 30901706 from the National Natural Science Foundation of China, People's Republic of China. We have no conflicts of interest to declare.
Footnotes
Published ahead of print on 18 May 2011.
REFERENCES
- 1. Andersson S., et al. 2000. Uneven distribution of HPV 16 E6 prototype and variant (L83V) oncoprotein in cervical neoplastic lesions. Br. J. Cancer 83:307–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bartholomew J. S., et al. 1994. Identification of a naturally processed HLA A0201-restricted viral peptide from cells expressing human papillomavirus type 16 E6 oncoprotein. Eur. J. Immunol. 24:3175–3179 [DOI] [PubMed] [Google Scholar]
- 3. Berumen J., et al. 2001. Asian-American variants of human papillomavirus 16 and risk for cervical cancer: a case-control study. J. Natl. Cancer Inst. 93:1325–1330 [DOI] [PubMed] [Google Scholar]
- 4. Bontkes H. J., et al. 1998. HPV 16 infection and progression of cervical intra-epithelial neoplasia: analysis of HLA polymorphism and HPV 16 E6 sequence variants. Int. J. Cancer 78:166–171 [DOI] [PubMed] [Google Scholar]
- 5. Cai H. B., Chen C. C., Ding X. H. 2010. Human papillomavirus type 16 E6 gene variations in Chinese population. Eur. J. Surg. Oncol. 36:160–163 [DOI] [PubMed] [Google Scholar]
- 6. Chan P. K., et al. 2002. Human papillomavirus type 16 intratypic variant infection and risk for cervical neoplasia in southern China. J. Infect. Dis. 186:696–700 [DOI] [PubMed] [Google Scholar]
- 7. Chen W., et al. 2009. Human papillomavirus type-distribution in cervical cancer in China: the importance of HPV 16 and 18. Cancer Causes Control 20:1705–1713 [DOI] [PubMed] [Google Scholar]
- 8. Chimeddorj B., Pak C. Y., Damdin A., Okamoto N., Miyagi Y. 2008. Distribution of HPV-16 intratypic variants among women with cervical intraepithelial neoplasia and invasive cervical cancer in Mongolia. Asian Pac. J. Cancer Prev. 9:563–568 [PubMed] [Google Scholar]
- 9. Choi B. S., Kim S. S., Yun H., Jang D. H., Lee J. S. 2007. Distinctive distribution of HPV 16 E6 D25E and E7 N29S intratypic Asian variants in Korean commercial sex workers. J. Med. Virol. 79:426–430 [DOI] [PubMed] [Google Scholar]
- 10. Chopjitt P., et al. 2009. Prevalence of human papillomavirus type 16 and its variants in abnormal squamous cervical cells in Northeast Thailand. Int. J. Infect. Dis. 13:212–219 [DOI] [PubMed] [Google Scholar]
- 11. de Boer M. A., et al. 2004. Human papillomavirus type 16 E6, E7, and L1 variants in cervical cancer in Indonesia, Suriname, and The Netherlands. Gynecol. Oncol. 94:488–494 [DOI] [PubMed] [Google Scholar]
- 12. Duensing S., Münger K. 2002. The human papillomavirus type 16 E6 and E7 oncoproteins independently induce numerical and structural chromosome instability. Cancer Res. 62:7075–7082 [PubMed] [Google Scholar]
- 13. Ellis J. R., et al. 1995. The association of an HPV16 oncogene variant with HLA-B7 has implications for vaccine design in cervical cancer. Nat. Med. 1:464–470 [DOI] [PubMed] [Google Scholar]
- 14. Eschle D., et al. 1992. Geographical dependence of sequence variation in the E7 gene of human papillomavirus type 16. J. Gen. Virol. 73:1829–1832 [DOI] [PubMed] [Google Scholar]
- 15. Harper D. M., et al. 2006. Sustained efficacy up to 4.5 years of a bivalent L1 virus-like particle vaccine against human papillomavirus types 16 and 18: follow-up from a randomised control trial. Lancet 367:1247–1255 [DOI] [PubMed] [Google Scholar]
- 16. Hildesheim A., et al. 2001. Human papillomavirus type 16 variants and risk of cervical cancer. J. Natl. Cancer Inst. 93:315–318 [DOI] [PubMed] [Google Scholar]
- 17. Hildesheim A., Wang S. S. 2002. Host and viral genetics and risk of cervical cancer: a review. Virus Res. 89:229–240 [DOI] [PubMed] [Google Scholar]
- 18. Ho L., et al. 1991. Sequence variants of human papillomavirus type 16 in clinical samples permit verification and extension of epidemiological studies and construction of a phylogenetic tree. J. Clin. Microbiol. 29:1765–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hu X., et al. 2001. HPV 16 E6 gene variations in invasive cervical squamous cell carcinoma and cancer in situ from Russian patients. Br. J. Cancer 84:791–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hu Y., Zhu Y. Y., Zhang S. H., Zhu H., Shuai C. X. 2011. Human papillomavirus type 16 E6 gene variations in young Chinese women with cervical squamous cell carcinoma. Reprod. Sci. 18:406–412 [DOI] [PubMed] [Google Scholar]
- 21. Hubert W. G. 2005. Variant upstream regulatory region sequences differentially regulate human papillomavirus type 16 DNA replication throughout the viral life cycle. J. Virol. 79:5914–5922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kämmer C., Warthorst U., Torrez-Martinez N., Wheeler C. M., Pfister H. 2000. Sequence analysis of the long control region of human papillomavirus type 16 variants and functional consequences for P97 promoter activity. J. Gen. Virol. 81:1975–1981 [DOI] [PubMed] [Google Scholar]
- 23. Kast W. M., et al. 1994. Role of HLA-A motifs in identification of potential CTL epitopes in human papillomavirus type 16 E6 and E7 proteins. J. Immunol. 152:3904–3912 [PubMed] [Google Scholar]
- 24. Kurvinen K., Yliskoski M., Saarikoski S., Syrjänen K., Syrjänen S. 2000. Variants of the long control region of human papillomavirus type 16. Eur. J. Cancer 36:1402–1410 [DOI] [PubMed] [Google Scholar]
- 25. Lee K., et al. 2008. Human papillomavirus 16 E6, L1, L2 and E2 gene variants in cervical lesion progression. Virus Res. 131:106–110 [DOI] [PubMed] [Google Scholar]
- 26. Lefevre J., Hankins C., Pourreaux K., Voyer H., Coutlée F., for the Canadian Women's HIV Study Group 2003. Real-time PCR assays using internal controls for quantitation of HPV-16 and beta-globin DNA in cervicovaginal lavages. J. Virol. Methods 114:135–144 [DOI] [PubMed] [Google Scholar]
- 27. Matsumoto K., et al. 2000. Enhanced oncogenicity of human papillomavirus type 16 (HPV 16) variants in Japanese population. Cancer Lett. 156:159–165 [DOI] [PubMed] [Google Scholar]
- 28. Matsumoto K., et al. 2003. Human papillomavirus type 16 E6 variants and HLA class II alleles among Japanese women with cervical cancer. Int. J. Cancer 106:919–922 [DOI] [PubMed] [Google Scholar]
- 29. Muñoz N., et al. 2003. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med. 348:518–527 [DOI] [PubMed] [Google Scholar]
- 30. Myers G., et al. 1997. Human papillomaviruses 1997: a compilation and analysis of nucleic acid and amino acid sequences. Theoretical Biology and Biophysics, Los Alamos National Laboratory, Los Alamos, NM [Google Scholar]
- 31. Nindl I., Rindfleisch K., Lotz B., Schneider A., Dürst M. 1999. Uniform distribution of HPV 16 E6 and E7 variants in patients with normal histology, cervical intra-epithelial neoplasia and cervical cancer. Int. J. Cancer 82:203–207 [DOI] [PubMed] [Google Scholar]
- 32. Pande S., et al. 2008. Human papillomavirus type 16 variant analysis of E6, E7, and L1 genes and long control region in biopsy samples from cervical cancer patients in north India. J. Clin. Microbiol. 46:1060–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Parkin D. M., Pisani P., Ferlay J. 1999. Estimates of the worldwide incidence of 25 major cancers in 1990. Int. J. Cancer 80:827–841 [DOI] [PubMed] [Google Scholar]
- 34. Pista A., et al. 2007. Molecular variants of human papillomavirus type 16 and 18 and risk for cervical neoplasia in Portugal. J. Med. Virol. 79:1889–1897 [DOI] [PubMed] [Google Scholar]
- 35. Qiu A. D., et al. 2007. HPV prevalence, E6 sequence variation and physical state of HPV 16 isolates from patients with cervical cancer in Sichuan, China. Gynecol. Oncol. 104:77–85 [DOI] [PubMed] [Google Scholar]
- 36. Radhakrishna Pillai M., Sreevidya S., Pollock B. H., Jayaprakash P. G., Herman B. 2002. Human papillomavirus type 16 E6 and E7 gene variations in Indian cervical cancer. Gynecol. Oncol. 87:268–273 [DOI] [PubMed] [Google Scholar]
- 37. Sichero L., et al. 2007. High grade cervical lesions are caused preferentially by non-European variants of HPVs 16 and 18. Int. J. Cancer 120:1763–1768 [DOI] [PubMed] [Google Scholar]
- 38. Song Y. S., et al. 1997. Major sequence variants in E7 gene of human papillomavirus type 16 from cervical cancerous and noncancerous lesions of Korean women. Gynecol. Oncol. 66:275–281 [DOI] [PubMed] [Google Scholar]
- 39. Stacey S. N., et al. 1994. Scanning the structure and antigenicity of HPV-16 E6 and E7 oncoproteins using antipeptide antibodies. Oncogene 9:635–645 [PubMed] [Google Scholar]
- 40. Steinke M., Geissler U., Gehrisch S., Jross W. 1995. Nachweis und differenzierung von humanen papillomviren der typen 6, 11, 16, 18 und 33 in cervix-abstrichen mittels PCR. Lab. Med. 19:128–133 [Google Scholar]
- 41. Stephen A. L., Thompson C. H., Tattersall M. H., Cossart Y. E., Rose B. R. 2000. Analysis of mutations in the URR and E6/E7 oncogenes of HPV 16 cervical cancer isolates from central China. Int. J. Cancer 86:695–701 [DOI] [PubMed] [Google Scholar]
- 42. Sun Z. R., et al. 2010. Characteristics of HPV prevalence among women in Liaoning province, China. Int. J. Gynaecol. Obstet. 109:105–109 [DOI] [PubMed] [Google Scholar]
- 43. Villa L. L., et al. 2000. Molecular variants of human papillomavirus types 16 and 18 preferentially associated with cervical neoplasia. J. Gen. Virol. 81:2959–2968 [DOI] [PubMed] [Google Scholar]
- 44. Villa L. L., et al. 2006. Immunologic responses following administration of a vaccine targeting human papillomavirus types 6, 11, 16, and 18. Vaccine 24:5571–5583 [DOI] [PubMed] [Google Scholar]
- 45. Wang S. M., Li J., Qiao Y. L. 2010. HPV prevalence and genotyping in the cervix of Chinese women. Front. Med. China. 4:259–263 [DOI] [PubMed] [Google Scholar]
- 46. Wheeler C. M., Yamada T., Hildesheim A., Jenison S. A. 1997. Human papillomavirus type 16 sequence variants: identification by E6 and L1 lineage-specific hybridization. J. Clin. Microbiol. 35:11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wu Y., et al. 2006. Analysis of mutations in the E6/E7 oncogenes and L1 gene of human papillomavirus 16 cervical cancer isolates from China. J. Gen. Virol. 87:1181–1188 [DOI] [PubMed] [Google Scholar]
- 48. Wu Y., et al. 2007. HPV 16 E6 variants and HLA class II polymorphism among Chinese women with cervical cancer. J. Med. Virol. 79:439–446 [DOI] [PubMed] [Google Scholar]
- 49. Xi L. F., et al. 1997. Genomic variation of human papillomavirus type 16 and risk for high grade cervical intraepithelial neoplasia. J. Natl. Cancer Inst. 89:796–802 [DOI] [PubMed] [Google Scholar]
- 50. Xi L. F., et al. 2007. Risk for high-grade cervical intraepithelial neoplasia associated with variants of human papillomavirus types 16 and 18. Cancer Epidemiol. Biomarkers Prev. 16:4–10 [DOI] [PubMed] [Google Scholar]
- 51. Yamada T., et al. 1995. Human papillomavirus type 16 variant lineages in United States populations characterized by nucleotide sequence analysis of the E6, L2, and L1 coding segments. J. Virol. 69:7743–7753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yamada T., et al. 1997. Human papillomavirus type 16 sequence variation in cervical cancers: a worldwide perspective. J. Virol. 71:2463–2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zehbe I., Voglino G., Delius H., Wilander E., Tommasino M. 1998. Risk of cervical cancer and geographical variations of human papillomavirus 16 E6 polymorphisms. Lancet 352:1441–1442 [DOI] [PubMed] [Google Scholar]
- 54. Zehbe I., Wilander E., Delius H., Tommasino M. 1998. Human papillomavirus 16 E6 variants are more prevalent in invasive cervical carcinoma than the prototype. Cancer Res. 58:829–833 [PubMed] [Google Scholar]
- 55. Zehbe I., et al. 2003. Association between human papillomavirus 16 E6 variants and human leukocyte antigen class I polymorphism in cervical cancer of Swedish women. Hum. Immunol. 64:538–542 [DOI] [PubMed] [Google Scholar]
- 56. Zhao Y., Lin H., Shen D., Xuan Y., Lin Z. 2008. Distribution of HPV genotypes in uterine cervical lesions in Yanbian Northern China. Pathol. Int. 58:643–647 [DOI] [PubMed] [Google Scholar]
- 57. Zhou M. G., et al. 2010. Geographical distribution of cancer mortality in China, 2004-2005. China J. Prev. Med. 44:303–308 (In Chinese.) [PubMed] [Google Scholar]