Abstract
Scrub typhus, caused by antigenically disparate isolates of Orientia tsutsugamushi, is a widely distributed mite-borne human disease in the Asia Pacific region. Information regarding the heterogeneity of the immunodominant 56-kDa type-specific antigen (TSA) gene is crucial for the design and evaluation of scrub typhus-specific diagnostic assays and vaccines. Using indirect immunofluorescence assays (IFA) and PCR assays, O. tsutsugamushi was detected samples from rodents and patients with fever of unknown origin obtained from six provinces of Thailand during 2004 to 2007. Sequences were determined for a fragment of the 56-kDa TSA gene, and the relationship between these sequences and those previously determined were assessed. The phylogenetic analyses of partial 56-kDa TSA gene sequences demonstrated wide diversity and distribution of O. tsutsugamushi genotypes in Thailand. Furthermore, the genetic diversity grouped the scrub typhus agents into two commonly and five infrequently found genotypes within six provinces of Thailand. The two most commonly found genotypes of O. tsutsugamushi described in this study do not associate with the prototype strains that are widely used for the design and evaluation of diagnostic assays and vaccine candidates. Thus, these new genotypes should be considered for future scrub typhus assay and vaccine development.
INTRODUCTION
Orientia tsutsugamushi, formerly known as Rickettsia tsutsugamushi, is the causative agent of scrub typhus, a major cause of acute febrile illnesses in rural southeast Asia (20, 30). This obligate intracellular Gram-negative rod-shaped bacterium (16) is vertically maintained in Leptotrombidium species mite populations and transmitted to humans by the bite of infected larval-stage mites called chiggers (30). The pathogen also may be transmitted horizontally between mites and infected vertebrate hosts (10, 11).
This mite-borne disease is found widely distributed across the Asia Pacific region and causes substantial morbidity in an area including Pakistan, Australia, Japan, South Korea, and Thailand (14). This region in which the disease is endemic often is referred to as the tsutsugamushi triangle, and it hosts approximately 1 billion people (5). Currently, there is no vaccine available against scrub typhus, thus effective management of this disease relies upon rapid diagnosis and antibiotic therapy with doxycycline, tetracycline, or chloramphenicol (30).
Antigenic variation among isolates of O. tsutsugamushi, due to the immunodominant 56-kDa type-specific antigen (TSA) located on the surface of O. tsutsugamushi (29), is commonly and widely used for O. tsutsugamushi serotyping and strain classification (2, 3, 13, 14, 26, 28). Originally, three distinctive antigenic prototypes of O. tsutsugamushi (Karp, Kato, and Gilliam) were described (7). New isolates of O. tsutsugamushi therefore were classified on the basis of reactivity with hyperimmune serum raised against these prototype strains (3). Later, additional antigenic variation of different serotypes was found in many countries (20). Thus, the serotyping of new isolates currently is carried out by indirect immunofluorescence assays (IFA) using strain- or type-specific monoclonal antibodies or hyperimmune sera which recognize the 56-kDa TSA of several unique strains of O. tsutsugamushi. Serotyping is limited by the need for well-characterized strains to produce the required reagents and the breadth of reactivity to detect new strains (9). These antigenic variations were linked to the sequence diversity of the 56-kDa TSA gene (18).
Recently the genotyping of O. tsutsugamushi has been used to characterize unique isolates. This has been accomplished by restriction fragment length polymorphism (RFLP) or sequence analysis of the 56-kDa TSA gene or gene fragment PCR amplicons (17, 29). In contrast to serotyping, molecular methods (8, 18) can be used to determine evolutionary relationships among the different genotypes. This useful information gained regarding genetic diversity and phylogenetic relationships of O. tsutsugamushi based on 56-kDa genotyping is crucial for determining the breadth of antigenic heterogeneity, which is critical for the development of sensitive and specific diagnostic assays as well as effective vaccine candidates.
Utilizing sequences recently obtained from the 56-kDa TSA gene of human- and rodent-derived O. tsutsugamushi, we discovered genetic diversity that grouped the scrub typhus agent into two commonly and five infrequently found genotypes located in six provinces of Thailand. The detection of identical genotypes in different hosts, including humans, implies that the potential transmission of scrub typhus occurs in any area where there is an appearance of one of the infected hosts. The commonly identified genotypes described herein do not phylogenetically associate with the prototypes (Karp, Kato, and Gilliam); therefore, the design and evaluation of scrub typhus diagnostic assays as well as vaccines candidates which relies on original prototypes may not be effective in Thailand.
MATERIALS AND METHODS
Sample collection.
A total of 604 blood samples from patients with fever of unknown origin (FUO) were obtained from clinical centers in central (Bangkok, 203 cases), northeastern (Nakhonratchasima, Sisaket, and Sakonnakhon, 183 cases), and northern (Chiangmai and Tak, 218 cases) regions of Thailand between February 2004 and April 2007. Patients provided informed consent before their blood samples were collected and used in this study. Blood samples subsequently were shipped at ≤−70°C to the Armed Forces Research Institute of Medical Sciences, Bangkok (AFRIMS), for further studies. This study was approved by the Institutional Review Board, Royal Thai Army Medical Department, Thailand. Culture isolates of O. tsutsugamushi were obtained from 3 of 69 rodents collected in Phangna (southern region of Thailand) in 2006 using the methods described previously (25). All captured rodents were processed by following appropriate laboratory animal procedures (6). This study was approved by the Animal Research Committee of the Royal Thai Army Component, Armed Forces Research Institute of Medical Sciences.
IFA.
All human blood samples initially were tested for O. tsutsugamushi infection using IFA (4, 24). Briefly, pooled antigens from the prototype strains (Karp, Kato, and Gilliam) cultured and passaged in the mouse fibroblast cell line (L929) were used to screen and detect O. tsutsugamushi-specific antibodies. Fluorescein isothiocyanate (FITC) conjugated to polyclonal rabbit anti-human immunoglobulins IgM and IgG (Dako, Glostrup, Denmark) were used to detect O. tsutsugamushi-specific antibodies in humans. Serum samples initially were screened against the pooled antigens at a 1:50 dilution. If the serum samples showed positive reactivity with O. tsutsugamushi antigen, the titer of antibodies in those samples was determined using a series of 2-fold dilutions of 1:100, 1:200, 1:400, 1:800, 1:1,600, 1:3,200, 1:6,400, and 1:12,800 of the sera in phosphate-buffered saline (PBS). Positive and negative controls were used in all tests, and a cutoff titer of 400 was used to identify seropositivity. Paired serum samples were not available from the FUO patients.
PCR and DNA sequencing.
Genomic DNA of FUO patients' EDTA blood and culture isolates from rodents were extracted using a DNeasy blood kit and a DNeasy tissue kit (Qiagen, Hilden, Germany), respectively. The nested PCR corresponding to the 56-kDa TSA gene spanning three of the four major variable regions was performed by utilizing two sets of primers. The outer primers were JG-OtF584 (5′-CAA TGT CTG CGT TGT CGT TGC) and RTS9 (5′-ACA GAT GCA CTA TTA GGC AA), and the inner primers were F (5′-AGC GCT AGG TTT ATT AGC AT) and RTS8 (5′-AGG ATT AGA GTG TGG TCC TT). The PCR profile was determined with the following steps: denaturation at 94°C for 3 min, follow by 40 cycles of 94°C for 30 s, 57°C for 40 s, and 72°C for 1 min, and a final incubation at 72°C for 7 min. PCR products of the expected amplicon size were purified using a High Pure PCR template preparation kit (Roche, Indianapolis, IN). The purified PCR products were directly sequenced using the dye terminator method (BigDye Terminator sequencing kit; Applied Biosystems, Foster City, CA) with an ABI Prism 377 DNA sequencer (Applied Biosystems).
Phylogenetic analysis.
Consensus sequences were constructed using Sequencher 3.1 (GeneCodes, Ann Arbor, MI). The 56-kDa TSA gene identification was done by BLAST based on pairwise similarity search (http://ncbi.nlm.nih.gov/blastn) (1). Nucleotide identity was calculated to sort and group closely related O. tsutsugamushi strains (>99%) using the MegAlign tool of Lasergene (DNASTAR Inc., Madison, WI). Table 1 shows four 56-kDa TSA partial gene sequences of culture isolates from rodents from Chonburi, in the eastern region of Thailand, collected in our previous study (25), that were included in the analysis of this study. Nucleotide sequences of human and rodent samples were aligned with reference O. tsutsugamushi sequences (3, 8, 23) using the MegAlign tool of Lasergene by a cluster W algorithm based on amino acid translation. Neighbor-joining (NJ), maximum-parsimony (MP), and maximum-likelihood (ML) methods were used to generate phylogenetic relationships using PAUP 4.0b10 (27). The overall transition-transversion ratio was calculated using the general time-reversible method (31) chosen by MODELTEST 3.06 (21, 22).
Table 1.
Representatives of each Orientia tsutsugamushi strain used for phylogenetic characterization
| Strain | Source | Location (province) | Region | No. of collections per strain | Genotype | Passage no. | GenBank accession no. |
|---|---|---|---|---|---|---|---|
| CM445h-NR | Human | Chiangmai | Northern | 1 | SEA1 | HM777456 | |
| PG6r-SR | Tupaia glis | Phangnga | Southern | 1 | SEA1 | 4 | HM777469 |
| CM438h-NR | Human | Chiangmai | Northern | 19 | SEA2 | HM777455 | |
| CM464h-NR | Human | Chiangmai | Northern | 1 | SEA2 | HM777457 | |
| MH440h-NR | Human | Maehongson | Northern | 2 | SEA2 | HM777462 | |
| MH650h-NR | Human | Maehongson | Northern | 1 | SEA2 | HM777465 | |
| CB52r-ER (19) | Menetes berdmorei | Chonburi | Eastern | 1 | SEA2 | 3 | GU068054 |
| CB62r-ER (19) | Bandicota indica | Chonburi | Eastern | 1 | SEA2 | 3 | GU068055 |
| PG9r-SR | Rattus rattus | Phangnga | Southern | 1 | SEA2 | 4 | HM777470 |
| BK131h-CR | Human | Bangkok | Central | 1 | LA | HM777451 | |
| MH465h-NR | Human | Maehongson | Northern | 2 | LA | HM777464 | |
| NR284h-NER | Human | Nakhonratchasima | Northeastern | 1 | LA | HM777466 | |
| CM414h-NR | Human | Chiangmai | Northern | 2 | SEA3 | HM777452 | |
| CM415h-NR | Human | Chiangmai | Northern | 11 | SEA3 | HM777453 | |
| CM492h-NR | Human | Chiangmai | Northern | 1 | SEA3 | HM777459 | |
| MH437h-NR | Human | Maehongson | Northern | 1 | SEA3 | HM777461 | |
| NR338h-NER | Human | Nakhonratchasima | Northeastern | 1 | SEA3 | HM777467 | |
| PG3r-SR | Rattus rattus | Phangnga | Southern | 1 | SEA3 | 4 | HM777468 |
| CM432h-NR | Human | Chiangmai | Northern | 1 | KATO | HM777454 | |
| CM606h-NR | Human | Chiangmai | Northern | 1 | TH1 | HM777460 | |
| CM471h-NR | Human | Chiangmai | Northern | 1 | TH2 | HM777458 | |
| MH446h-NR | Human | Maehongson | Northern | 1 | TH2 | HM777463 | |
| CB19r-ER (19) | Rattus rattus | Chonburi | Eastern | 1 | TH2 | 5 | GU068058 |
| CB35r-ER (19) | Bandicota indica | Chonburi | Eastern | 1 | TH2 | 3 | GU068053 |
Nucleotide sequence accession numbers.
Selected sequences determined in the course of this work have been deposited in GenBank under the accession numbers listed in Table 1.
RESULTS AND DISCUSSION
Initial IFA demonstrated that antibodies specific for O. tsutsugamushi were found in 4.8% (IgM) and 4.3% (IgG) of the FUO patients' blood samples (n = 604), whereas O. tsutsugamushi DNA was found in 7.95% of these samples (Table 2). The highest infection prevalence was found in samples from the northern provinces (n = 218; IFA detection rates for IgM and IgG, 11.98 and 10.55%, respectively; PCR detection rate, 20.64%), whereas evidence of infection was rarely found in samples from the central (n = 203; IFA rate for IgM and IgG, 0.49 and 0.49%, respectively; PCR rate, 0.49%) and northeastern provinces (n = 183; IFA rate for IgM and IgG, 1.09 and 1.09%, respectively; PCR rate, 1.09%) (Table 2). These data corresponded with incidence rates reported for scrub typhus during 2004 to 2007 (data from the Center of Epidemiological Information, Bureau of Epidemiology, Ministry of Public Health, Thailand). The concordance of incidence cases reported and this study indicate that scrub typhus is common and found mostly in the northern region of Thailand. According to the nonnormal distribution of the samples (Table 2) (a, b, and c by Kolmogorov-Smirnov test, P < 0.001, respectively), a Mann-Whitney U test (U) was employed for the statistical analysis of different methods of northern samples. The detection efficiency using IFA (IgMa, 11.93%; IgGb, 10.55%) was significantly lower than that with PCRc (20.64%) (a and b versus c; U, P = 0.031, 0.01, respectively), but there was no significant difference between IgMa and IgGb (a versus b; U, P = 0.666). The comparison demonstrates that PCR can detect O. tsutsugamushi infection better than IFA when the size of the infected population is increasing, such as in the northern provinces. Thus, the PCR assay is a good choice for scrub typhus detection, especially when conventional IFA cannot prove current infection.
Table 2.
Evidence of Orientia tsutsugamushi infection in blood samples collected from FUO patients using IFA and PCR
| Region | Febrile illness patient samples tested for evidence of O. tsutsugamushi infection |
|||
|---|---|---|---|---|
| Total no. of study cases | % (no.) IFA positive |
% (no.) PCRc positive | ||
| IgMa | IgGb | |||
| Central | 203 | 0.49 (1) | 0.49 (1) | 0.49 (1) |
| Northeastern | 183 | 1.09 (2) | 1.09 (2) | 1.09 (2) |
| Northern | 218 | 11.93 (26) | 10.55 (23) | 20.64 (45) |
| Total | 604 | 4.80 (29) | 4.30 (26) | 7.95 (48) |
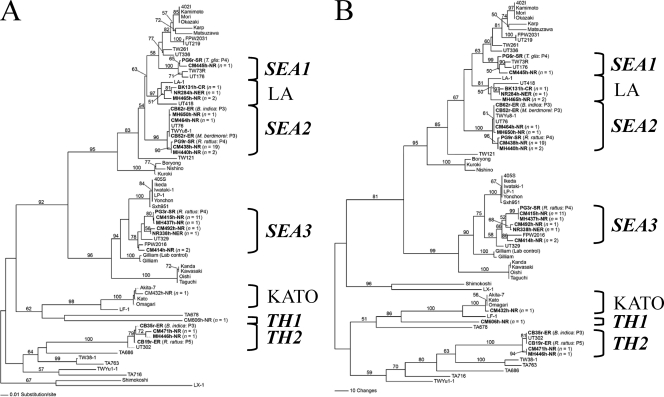
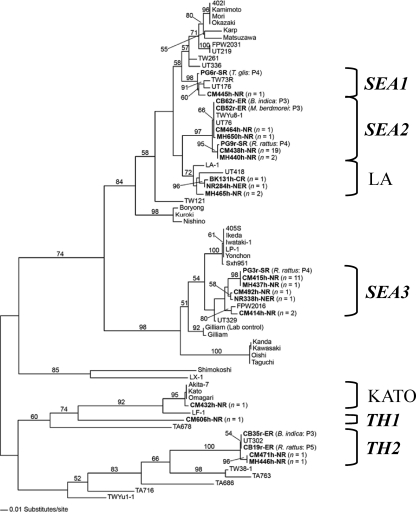
The genetic characterization of 51 consensus sequences of the partial 56-kDa TSA gene showed that the human (n = 48) and rodent (n = 3) samples were highly similar to the 56-kDa TSA gene of O. tsutsugamushi sequences listed in GenBank. The selected sequences (Table 1) (n = 24) were aligned with 46 reference O. tsutsugamushi sequences retrieved from GenBank. Figures 1 and 2 illustrate phylogenetic relationships generated by NJ, MP, and ML methods. Tree topologies generated by these three methods show the concordance of node divergence, except that there is an exchange between Shimokoshi and Kato clades (Fig. 1A and B) in the NJ tree but not in MP and ML trees (Fig. 1B and 2). A low bootstrap value (<50%) is responsible for the occurrence of uncertainty divergence of these two nodes. The phylogenetic trees generated by the three methods have similar tree topologies and demonstrate that, in six provinces of Thailand, the 56-kDa TSA genes of O. tsutsugamushi are diverse and indicate polyphyletic evolution. The limitation of the use of hypervariable regions based on three of the four major variable regions (VD I, II, and III) of the 56-kDa TSA gene might represent an even higher level of hypervariation than that of the whole gene itself. Nevertheless, topology based on these three regions does not significantly differ from whole-gene topologies employing the same method (Fig. 1A) (8, 15). Slight differences in topology show that there may have been an exchange between Gilliam and Kato assemblages, which was observed in some works with low bootstrap values (<50%) (3). Thus, this may be responsible for uncertainty divergence. We have chosen the ML tree for describing gene evolutionary relationships among O. tsutsugamushi strains, because the ML tree is generated based on evolutionary and tree searching methods which best meet optimality criterion by evaluating individual trees (12).
Fig. 1.
Phylogenetic trees of Orientia tsutsugamushi based on partial 56-kDa TSA gene sequences demonstrating the relationships among human and rodent strains found in Thailand and others listed in GenBank. The tree was midpoint rooted. Bootstrap values of >50% are labeled above branches (1,000 replicates). Human and rodent strains described in this report are shown in boldface. (A) Neighbor-joining tree. The tree was generated by the Kimura two-parameter method with gamma correction (gamma shape parameter, 0.9561). (B) Maximum-parsimony tree. The tree was generated using a heuristic search with random stepwise addition (10 replicates).
Fig. 2.
Maximum-likelihood tree based on partial 56-kDa TSA gene sequences of Orientia tsutsugamushi demonstrating the relationships among human and rodent strains found in Thailand and others listed in GenBank. The tree was generated by the stepwise addition method using a general time-reversible method with consideration of gamma-distributed rate heterogeneity across sites (−lnL = 6904.15; gamma shape parameter, 0.9561). The tree was midpoint rooted. Bootstrap values of >50% are labeled above branches (1,000 replicates). Human and rodent strains described in this report are shown in boldface. Thai genotypes are shown by boldface italics. SEA, southeastern Asia; TH, Thailand; SR, southern region; ER, eastern region; NR, northern region; NER, northeastern region.
Figure 2 illustrates an enlargement of the ML tree in which human- and rodent-infected O. tsutsugamushi strains in six provinces are labeled with assigned genotypes. The newly characterized genotypes of closely related O. tsutsugamushi (including human and rodent strains) obtained from different Thai provinces are listed in Table 1 and are shown to cluster into seven assemblages (Fig. 2). Two assemblages are located within the well-known LA and KATO genotypes (8), whereas five assemblages cluster in distinctive genotypes that we have named SEA1, SEA2, SEA3, TH1, and TH2 according to their original locations and distribution. Four of these assemblages cluster near the genotypes Karp (SEA1 and SEA2), Ikeda (SEA3), and UT302 (TH2). The genotypes SEA1, SEA2, Karp, and LA show monophyletic divergence, indicating a common ancestor and close relationships among these O. tsutsugamushi strains. Therefore, SEA1 should be related to Karp. The trees in this study have demonstrated that SEA2 clustered closer to LA, which was reported previously as a different genotype (8) from the Karp prototype (Fig. 1 and 2). Thus, it might be more appropriate that the SEA2 genotype is designated LA related rather than Karp related. SEA3 shows monophyletic evolution with the Ikeda group, which previously has been denoted the JG type (14), so that the JG-related classification should be applied to SEA3. In contrast to the other genotypes, TH1 formed a separately deep-node divergence distant from but linked to the Kato and LF-1 genotypes.
Interestingly, the previously reported strains, e.g., UT76, UT176, and UT418 (3), which were classified as Karp like, are more closely associated with SEA1, SEA2, and LA. These results suggest that these strains should belong to SEA1, SEA2, and LA rather than the Karp genotype. The nucleotide identity of these UT genotypes to Karp was less than 95% (3), which corresponded to our analysis. The genotype classification of O. tsutsugamushi isolates in a Karp-like grouping is unclear and complicated by the numerous disparate isolates recently identified, and therefore the definition of Karp-like strains continues to evolve (9, 14).
The prevalence of O. tsutsugamushi genotypes described herein appear to be either common (SEA2 and SEA3) or infrequent (KATO, LA, SEA1, TH1, and TH2). The most commonly found genotypes, SEA2 and SEA3 (n = 23 and 16, respectively), collectively encompassed 81.25% of O. tsutsugamushi isolates causing scrub typhus illness (39/48) described in this study, whereas the other five genotypes collectively were responsible for only 18.75% of the scrub typhus cases. The use of specific reagents to different genotypes will increase the specificity and sensitivity of O. tsutsugamushi detection in scrub typhus patients (9). Therefore, the development of vaccines and diagnostic kits, especially those to be used in Thailand, should take into account these O. tsutsugamushi genotypes.
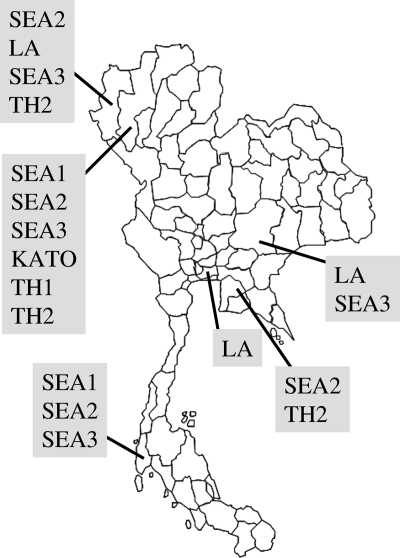
There appears to be no occurrence of geographic restriction among O. tsutsugamushi strains within six provinces of Thailand. The O. tsutsugamushi strains collected from the same location did not necessarily cluster together in the same genotype (Fig. 2). Moreover, different genotypes were variously distributed in six provinces (Fig. 3). In addition, there was a lack of correlation between location and genotype, and there was no correlation between host and genotype. For example, the O. tsutsugamushi isolates from rodents (CB62r-ER and CB52r-ER) (25), humans (UT76, CM464h-NR, and MH650h-NR) (3), and a mite (TWYu8-1) (23) were obtained from distinct locations but grouped together in SEA2 and share the same divergence assemblage with high nucleotide and amino acid similarity (100% identity). The similarity of a genotype in different hosts, including humans, implies that scrub typhus transmission occurs in any area where there is an appearance of one of the infected hosts.
Fig. 3.
Genotype distribution of O. tsutsugamushi in six provinces of Thailand.
Parola et al. (19) have demonstrated that specific antigens, i.e., Gilliam, Kato, and Kawasaki, were cross-reactive against antibodies derived against different strains identified in Laos patients' sera. The different strains were closely related based upon the 56-kDa TSA gene sequences determined in previous work (3, 23) and clustered within new genotypes. The occurrence of antibodies cross-reactive to different serotypes has been observed in many studies (9, 19); however, this may not always be the case when infection occurs with diverse strains that are not included in the antigen panel (9). Therefore, specific antigens, including the three prototypes, i.e., Karp, Kato, and Gilliam, and new genotypes that were observed in this study should be considered for testing against any Thai patient sera to ensure the detection of antibodies produced against different O. tsutsugamushi serotypes in the future.
In this study, we have demonstrated 56-kDa TSA genotype diversity in six provinces of Thailand. Two genotypes (SEA2 and SEA3) were commonly distributed among these provinces. The understanding of the diversity of O. tsutsugamushi, especially of the two common genotypes, is important especially in view of the future design and evaluation of sensitive and specific diagnostic assays and vaccine candidates. Previous human trials with scrub typhus vaccine candidates in the field have failed due to the inability of the vaccines to protect against exposure to disparate O. tsutsugamushi strains. Thus, according to their dominance and distribution, the genotypes described in this report should be considered for incorporation into future scrub typhus vaccines along with other commonly found isolates causing human disease.
ACKNOWLEDGMENTS
We thank Atthaya Ruangpung and Yuwadee Yusuk for technical assistance. We also thank Juntima Chanprasert for her useful comments and technical support.
This work was supported by a Thanphuying Viraya Chavakul Foundation for Medical Armed Forces Research grant (2008 to 2009) and the Thailand Tropical Diseases Research Funding Program (T-2). Part of this work (by A.L.R.) was supported by the Global Emerging Infections Surveillance and Response System, a Division of the Armed Forces Health Surveillance Center.
None of the authors or organization members has a relationship(s) which could be perceived as constituting a conflict of interest.
A.L.R. is an employee of the U.S. government, and this work was prepared as part of his official duties.
Footnotes
Published ahead of print on 18 May 2011.
REFERENCES
- 1. Altschul S. F., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Blacksell S. D., et al. 2007. Scrub typhus serologic testing with the indirect immunofluorescence method as a diagnostic gold standard: a lack of consensus leads to a lot of confusion. Clin. Infect. Dis. 44:391–401 [DOI] [PubMed] [Google Scholar]
- 3. Blacksell S. D., et al. 2008. Genetic typing of the 56-kDa type-specific antigen gene of contemporary Orientia tsutsugamushi isolates causing human scrub typhus at two sites in north-eastern and western Thailand. FEMS Immunol. Med. Microbiol. 52:335–342 [DOI] [PubMed] [Google Scholar]
- 4. Bozeman F. M., Elisberg B. L. 1963. Serological diagnosis of scrub typhus by indirect immunofluorescence. Proc. Soc. Exp. Biol. Med. 112:568–573 [DOI] [PubMed] [Google Scholar]
- 5. Chattopadhyay S., Richards A. L. 2007. Scrub typhus vaccines: past history and recent developments. Hum. Vaccines 3:73–80 [DOI] [PubMed] [Google Scholar]
- 6. Clark D. C., et al. 1996. Guide for the care and use of laboratory animals. National Academy Press, Washington, DC [Google Scholar]
- 7. Eisemann C. S., Osterman J. V. 1985. Identification of strain-specific and group-reactive antigenic determinants on the Karp, Gilliam and Kato strains of Rickettsia tsutsugamushi. Am. J. Trop. Med. Hyg. 34:1173–1178 [DOI] [PubMed] [Google Scholar]
- 8. Enatsu T., Urakami H., Tamura A. 1999. Phylogenetic analysis of Orientia tsutsugamushi strains based on the sequence homologies of 56-kDa type-specific antigen genes. FEMS Microbiol. Lett. 180:163–169 [DOI] [PubMed] [Google Scholar]
- 9. Fournier P. E., et al. 2008. Detection of new genotypes of Orientia tsutsugamushi infecting humans in Thailand. Clin. Microbiol. Infect. 14:168–173 [DOI] [PubMed] [Google Scholar]
- 10. Frances S. P. 2005. Potential for horizontal transmission of Orientia tsutsugamushi by chigger mites (Acari: Trombiculidae). Int. J. Acarol. 31:75–82 [Google Scholar]
- 11. Frances S. P., Watcharapichat P., Phulsuksombati D. 2001. Vertical transmission of Orientia tsutsugamushi in two lines of naturally infected Leptotrombidium deliense (Acari: Trombiculidae). J. Med. Entomol. 38:17–21 [DOI] [PubMed] [Google Scholar]
- 12. Hall B. G. 2001. Phylogenetic trees made easy: a how to manual for molecular biologists. Sinauer Associates, Inc., Sunderland, MA [Google Scholar]
- 13. Horinouchi H., et al. 1996. Genotypic identification of Rickettsia tsutsugamushi by restriction fragment length polymorphism analysis of DNA amplified by the polymerase chain reaction. Am. J. Trop. Med. Hyg. 54:647–651 [DOI] [PubMed] [Google Scholar]
- 14. Kelly D. J., Fuerst P. A., Ching W. M., Richards A. L. 2009. Scrub typhus: the geographic distribution of phenotypic and genotypic variants of Orientia tsutsugamushi. Clin. Infect. Dis. 48(Suppl. 3):S203–S230 [DOI] [PubMed] [Google Scholar]
- 15. Nakayama K., et al. 2010. Genome comparison and phylogenetic analysis of Orientia tsutsugamushi strains. DNA Res. 17:281–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ohashi N., Fukuhara M., Shimada M., Tamura A. 1995. Phylogenetic position of Rickettsia tsutsugamushi and the relationship among its antigenic variants by analyses of 16S rRNA gene sequences. FEMS Microbiol. Lett. 125:299–304 [DOI] [PubMed] [Google Scholar]
- 17. Ohashi N., et al. 1996. Demonstration of antigenic and genotypic variation in Orientia tsutsugamushi which were isolated in Japan, and their classification into type and subtype. Microbiol. Immunol. 40:627–638 [DOI] [PubMed] [Google Scholar]
- 18. Ohashi N., Nashimoto H., Ikeda H., Tamura A. 1992. Diversity of immunodominant 56-kDa type-specific antigen (TSA) of Rickettsia tsutsugamushi. Sequence and comparative analyses of the genes encoding TSA homologues from four antigenic variants. J. Biol. Chem. 267:12728–12735 [PubMed] [Google Scholar]
- 19. Parola P., et al. 2008. Genotyping of Orientia tsutsugamushi from humans with scrub typhus, Laos. Emerg. Infect. Dis. 14:1483–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Phongmany S., et al. 2006. Rickettsial infections and fever, Vientiane, Laos. Emerg. Infect. Dis. 12:256–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Posada D. 2003. Using MODELTEST and PAUP* to select a model of nucleotide substitution. Curr. Protoc. Bioinformatics 6:6.5. [DOI] [PubMed] [Google Scholar]
- 22. Posada D., Crandall K. A. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817–818 [DOI] [PubMed] [Google Scholar]
- 23. Qiang Y., et al. 2003. Phylogenetic characterization of Orientia tsutsugamushi isolated in Taiwan according to the sequence homologies of 56-kDa type-specific antigen genes. Microbiol. Immunol. 47:577–583 [DOI] [PubMed] [Google Scholar]
- 24. Robinson D. M., Brown G., Gan E., Huxsoll D. L. 1976. Adaptation of a microimmunofluorescence test to the study of human Rickettsia tsutsugamushi antibody. Am. J. Trop. Med. Hyg. 25:900–905 [DOI] [PubMed] [Google Scholar]
- 25. Rodkvamtook W., et al. 2011. Isolation and characterization of Orientia tsutsugamushi from rodents obtained at the central Thailand military training base, Bothong district, Chonburi. Am. J. Trop. Med. Hyg. 84:599–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shishido A. 1962. Identification and serological classification of the causative agent of scrub typhus in Japan. Jpn. J. Sci. Biol. 15:308–321 [Google Scholar]
- 27. Swofford D. L. 2002. PAUP*. Phylogenetic analysis using pasimony (* and other methods), version 4.0, beta 10. Sinauer Associates, Sunderland, MA [Google Scholar]
- 28. Tamura A. 1988. Invasion and intracellular growth of Rickettsia tsutsugamushi. Microbiol. Sci. 5:228–232 [PubMed] [Google Scholar]
- 29. Tamura A., et al. 1997. Characterization of Orientia tsutsugamushi isolated in Taiwan by immunofluorescence and restriction fragment length polymorphism analyses. FEMS Microbiol. Lett. 150:225–231 [DOI] [PubMed] [Google Scholar]
- 30. Watt G., Parola P. 2003. Scrub typhus and tropical rickettsioses. Curr. Opin. Infect. Dis. 16:429–436 [DOI] [PubMed] [Google Scholar]
- 31. Yang Z. 1994. Estimating the pattern of nucleotide substitution. J. Mol. Evol. 39:105–111 [DOI] [PubMed] [Google Scholar]