Abstract
We report a case of misdiagnosis of Plasmodium falciparum malaria from Brazil with negative PfHRP2-based rapid diagnostic tests (RDTs), leading to inappropriate case management. Genetic tests showed the deletion of both pfhrp2 and pfhrp3 genes. The detection of two distinct P. falciparum target antigens is then advisable.
TEXT
Malaria diagnosis relies on blood smear examination as the reference. Recently, rapid diagnostic tests (RDTs) have been developed to improve the availability of malaria diagnosis (10). RDTs detect malarial antigens such as the Plasmodium falciparum-specific histidine-rich protein 2 (PfHRP2) and lactate dehydrogenase (PfLDH) and the panplasmodial aldolase (pAldolase) and lactate dehydrogenase (pLDH) (8). We report here a case of P. falciparum malaria in which the PfHRP2 test was negative, leading to inappropriate case management. We then provide the molecular basis underlying this false-negative test.
A 38-year-old French man traveled to the Brazilian Amazon region. Three days after returning to Paris, he started experiencing fever and headache. A blood film examination performed in a nonexpert laboratory showed Plasmodium (144,000 parasites/μl; corresponding to 3.2% of infected erythrocytes). In the same laboratory, the Now ICT malaria test (Binax; Inverness Medical, France) was positive for pAldolase but negative for PfHRP2, suggesting non-P. falciparum malaria. The recent travel to Brazil, where P. falciparum chloroquine-resistant malaria is widespread, combined with the RDT results suggested Plasmodium vivax malaria, and the patient was treated with chloroquine. There was no clinical improvement 2 days later, when a second blood film examination in the same laboratory showed a parasitemia of 1,890,000 parasites/μl (corresponding to 42% of infected erythrocytes). The patient was then transferred to our expert malaria center, where a new blood film examination was performed. Old trophozoites with hemozoin and Maurer's clefts were predominant compared to ring-stage parasites. Although unusual, this microscopic-stage pattern of parasites in peripheral blood is specific to P. falciparum. Earlier blood samples were sent to us for verification. We observed parasitized erythrocytes with hemozoin, old trophozoites, and schizonts, specific to P. falciparum. We then performed the Now ICT malaria test, which again failed to detect PfHRP2. The Palutop 4+ test (Alldiag, Strasbourg, France) was negative for PfHRP2 and positive for pLDH, and the OptiMal IT test (Diamed, Cressier, Switzerland) was positive for both pLDH and PfLDH. Quinine was given for 7 days, and the patient achieved full recovery.
During the 7-day quinine course, eight blood samples were collected and analyzed. Parasitemias ranged from 10 to 1,890,000 parasites/μl. RDTs were negative for PfHRP2 at any parasitemias but positive for pLDH and PfLDH until parasitemias dropped below the assay detection limit. The high parasitemias achieved during the infection rule out a false-negative RDT for PfHRP2 because of sensitivity issues. DNA was extracted for all available blood samples. PCR performed according to the method of Snounou (9) showed P. falciparum as the only infecting plasmodial species.
To explain the discordant RDT results, we sought to PCR amplify several fragments of the P. falciparum histidine-rich protein 2 (pfhrp2) and histidine-rich protein 3 (pfhrp3) genes using DNA from three patient isolates collected before quinine initiation (D0) and on day 3 (D3) and day 6 (D6) of quinine therapy, when parasitemias were 1,890,000 parasites/μl, 144,000 parasites/μl, and 1,125 parasites/μl, respectively. Five laboratory-adapted strains were also analyzed as controls: D10 (pfhrp2 deleted), HB3 (pfhrp3 deleted), W2 (no deletion), 7G8 (no deletion), and 3D7 (no deletion).
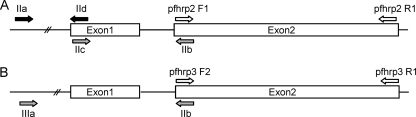
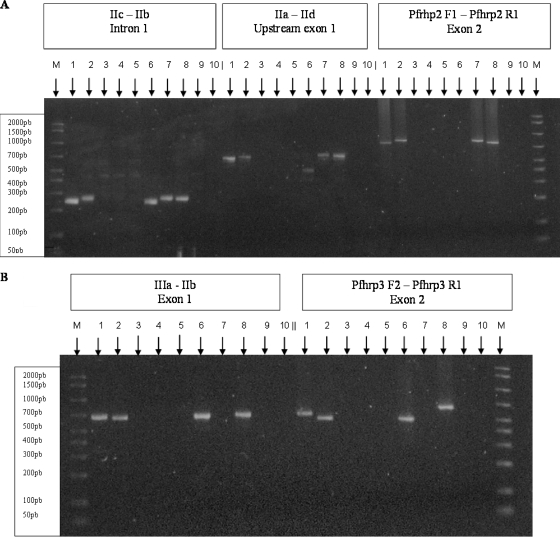
Exon 2 of both pfhrp2 and pfhrp3 genes was amplified in a single round of 40 amplification cycles with the primer pair Pfhrp2-F1 and Pfhrp2-R1 and primer pair Pfhrp3-F2 and Pfhrp3-R1, respectively, and using the PCR cycling conditions described by Baker et al. (2) (Fig. 1). No amplicon was detected for either primer pair with the patient's samples, whereas the expected amplicons (816 bp for pfhrp2 and 665 bp for pfhrp3) were detected with the control strains (Fig. 2). The PCR performed to confirm the malaria species was successful using the same DNA templates, ruling out the presence of PCR inhibitors. These data suggest that both the pfhrp2 and pfhrp3 genes are at least partially deleted in the patient's isolates.
Fig. 1.
Schematic illustration of pfhrp2 and pfhrp3 genomic organizations and primer binding sites. (A) pfhrp2 gene; (B) pfhrp3 gene.
Fig. 2.
Lack of amplification of pfhrp2 and pfhrp3 genes in the patient's isolates. (A) pfhrp2 gene; (B) pfhrp3 gene. Lanes: 1, W2 strain; 2, 7G8 strain; 3, 4, and 5, patient isolates from days 0, 3, and 6, respectively, after quinine initiation; 6, D10 strain; 7, HB3 strain; 8, 3D7 strain; 9, blood negative for malaria; 10, distilled water; M, size ladder.
To confirm these deletion events, additional amplicons were tested with the primer pairs described by Sullivan et al. (11) (Fig. 1): primers IIa-IId (exon 1 of pfhrp2), primers IIc-IIb (intron 1of pfhrp2), and primers IIIa-IIb (exon 1 and intron 1of pfhrp3). PCR cycling conditions were as follows: 94°C denaturation step for 10 min followed by 35 cycles of amplification (94°C for 50 s, 58°C for 50 s, and 60°C for 1 min). Again, no amplicon was detected with the patient's samples, whereas the expected amplicons were detected for the control strains (Fig. 2). Of note, with the D10 strain whose pfhrp2 gene is deleted, 450-bp and 256-bp PCR products were obtained with primer pairs IIa-IId and IIc-IIb, respectively (Fig. 2). Sequencing of these unexpected amplicons showed that they were derived from the pfhrp3 gene, likely because of partial homology between the pfhrp2 and pfhrp3 gene sequences.
The failure to amplify both pfhrp2 and pfhrp3 genes strongly suggests that these two genes were deleted in the patient's isolates. This double gene deletion was associated with the failure to immunodetect the PfHRP2 protein, providing a molecular basis for the false-negative PfHRP2-based RDTs. Recently, a study reported a high frequency of pfhrp2-pfhrp3 double deletion among P. falciparum isolates from the Peruvian Amazonian area (4). These findings are fully consistent with ours, as our patient had traveled to the Brazilian Amazonian region. Importantly, the pfhrp2 single-deletion genotype was not detected among numerous P. falciparum isolates, including isolates originating from African countries (1), suggesting that this double-deletion genotype is much less frequent outside the Amazonian region.
In addition to the gene deletion reported here, false-negative PfHRP2-based tests may be related to some PfHRP2 molecules having various numbers and arrangements of amino acid repeats not recognized by the test antibodies (2, 5). Several studies analyzed the genetic diversity of other malarial antigens used in RDTs (6, 12), and no variability was observed for pAldolase or pLDH. Assessing the molecular variability of parasite antigens used in RDTs should be performed on a large geographic scale to better understand and predict the performance of these extremely useful malaria diagnostic tests.
Importantly, as chloroquine resistance is widespread for P. falciparum and emerging for P. vivax, chloroquine treatment should not be administered in cases where malaria species diagnosis is difficult or undetermined (3).
In summary, although numerous studies found that PfHRP2-based RDTs were highly sensitive (7), extreme caution should be taken when testing a sample originating from an area where the pfhrp2-pfhrp3 double-deletion genotype has been reported. Negative PfHRP2-based RDTs should then be confirmed either by microscopic examination or by another RDT which detects a P. falciparum antigen distinct from PfHRP2.
Footnotes
Published ahead of print on 4 May 2011.
REFERENCES
- 1. Baker J., et al. 2010. Global sequences variation in the histidine-rich proteins 2 and 3 of Plasmodium falciparum; implications for the performance of malaria rapid diagnostic tests. Malar. J. 9:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baker J., et al. 2005. Genetic diversity of Plasmodium falciparum histidine-rich protein 2 (PfHRP2) and its effect on the performance of PfHRP2-based rapid diagnostic tests. J. Infect. Dis. 192:870–877 [DOI] [PubMed] [Google Scholar]
- 3. Douglas N. M., Anstey N. M., Angus B. J., Nosten F., Price R. N. 2010. Artemisinin combination therapy for vivax malaria. Lancet Infect. Dis. 10:405–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gamboa D., et al. 2010. A large proportion of P. falciparum isolates in the Amazon region of Peru lack pfhrp2 and pfhrp3; implications for malaria rapid diagnostic tests. PLoS One 5:e8091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lee N., et al. 2006. Effect of sequence variation in Plasmodium falciparum histidine-rich protein 2 on binding of specific monoclonal antibodies: implications for rapid diagnostic tests for malaria. J. Clin. Microbiol. 44:2773–2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lee N., Baker J., Bell D., McCarthy J., Cheng Q. 2006. Assessing the genetic diversity of the aldolase genes of Plasmodium falciparum and Plasmodium vivax and its potential effect on performance of aldolase-detecting rapid diagnostic tests. J. Clin. Microbiol. 44:4547–4549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Marx A., et al. 2005. Meta-analysis: accuracy of rapid tests for malaria in travellers returning from endemic areas. Ann. Intern. Med. 142:836–846 [DOI] [PubMed] [Google Scholar]
- 8. Moody A. 2002. Rapid diagnostic tests for malaria parasites. Clin. Microbiol. Rev. 15:66–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Snounou G. 1996. Detection and identification of the four malaria parasite species infecting humans by PCR amplification. Methods Mol. Biol. 50:263–291 [DOI] [PubMed] [Google Scholar]
- 10. Stauffer W. M., et al. 2009. Diagnostic performance of rapid diagnostic tests versus blood smears for malaria in US clinical practice. Clin. Infect. Dis. 49:908–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sullivan D. J., Ayala Y. M., Goldberg D. E. 1996. An unexpected 5′ untranslated intron in the P. falciparum genes for histidine-rich proteins II and III. Mol. Biochem. Parasitol. 83:247–251 [DOI] [PubMed] [Google Scholar]
- 12. Talman A. M., et al. 2007. Evaluation of the intra- and inter-specific genetic variability of Plasmodium lactate dehydrogenase. Malar. J. 6:140. [DOI] [PMC free article] [PubMed] [Google Scholar]