Abstract
Highly sensitive techniques, such as PCR, have greatly improved the detection of respiratory viruses. However, the sensitivity of PCR tests also complicates clinical interpretation, as the presence of small amounts of viral targets may not necessarily have clinical relevance. We performed a prospective case-control study in asymptomatic and symptomatic young children. PCR detection of 14 respiratory viruses was performed in nasal washes, and results were quantified in copies per milliliter. A total of 141 cases and 157 controls were included. In 72% of the cases and 28% of the controls, at least one virus was identified. When stratified for age, at least one virus was identified in 47% of the controls younger than 1 year old. Rhinovirus (RV) was frequently detected in both symptomatic and asymptomatic individuals. Receiver operating characteristic analysis for quantitative rhinovirus detection showed that cutoff values for clinical relevance are feasible for RV. In contrast to rhinovirus, respiratory syncytial virus (RSV) was rarely detected in controls, suggesting that a positive RSV test result is almost always of clinical relevance, independent of viral quantity. In conclusion, our study shows that asymptomatic carriage of a respiratory virus occurs frequently in young children. However, significant differences in the amount of virus present were observed between cases and controls. This suggests that defining cutoff levels should be feasible and represents the next necessary step for diagnosing viral respiratory infections using molecular tests.
INTRODUCTION
Molecular techniques, such as PCR, have revolutionized the diagnosis of respiratory viruses. PCR has enabled rapid and highly sensitive detection of a large number of respiratory viruses, including those that are difficult to culture, such as human bocavirus (hBoV) and human metapneumovirus (hMPV) (7, 22, 24). The high sensitivity of PCR allows the detection of minimal amounts of viral nucleic acids, but there are questions concerning the clinical relevance of positive test results. For example, small amounts of a respiratory virus could represent asymptomatic colonization or postinfectious shedding. Appraisal of asymptomatic persons, in comparison to patients with respiratory illnesses, is essential for better understanding the significance of detected viral nucleic acids and for improved interpretation of diagnostic results.
Until now, only a few studies employing PCR specifically looked at the presence of respiratory viruses in asymptomatic persons. In most of these studies only one or two viral species or specific populations, such as children with chronic asthma or adults, were addressed (5, 8, 13, 14, 17, 21). We performed a prospective case-control study to (i) determine the prevalence of respiratory viruses in asymptomatic and symptomatic young children and (ii) to evaluate whether a quantitative approach could aid in the interpretation of diagnostic results.
MATERIALS AND METHODS
This prospective case-control study was performed at the Academic Medical Centre (AMC) in Amsterdam, which serves the local area and acts as a tertiary referral center. A total of 141 cases and 157 controls were included during the winter seasons (November to April) of 2007-2008 and 2008-2009 (Table 1). Cases were children up to 6 years old who were admitted with a clinical suspicion of an acute respiratory infection. Controls were children of the same age range without respiratory symptoms or fever for at least 7 days prior to inclusion who visited an outpatient clinic of the AMC for nonrespiratory disease in the same period as the cases. Asymptomatic children accompanying a sibling who visited the outpatient clinic were also asked to participate as controls. After written informed consent was obtained from one of the parents, clinical data (except for influenza vaccination status) and a nasal wash (NW) were collected. Influenza vaccination status was not recorded for patients or controls. Moreover, subjects did not receive live attenuated influenza vaccine (FluMist), since this is not available in Netherlands and none of the participants received palavizumab.
Table 1.
Clinical characteristics of cases and controls
| Characteristica | No. (%) in group with characteristic |
P value | |
|---|---|---|---|
| Cases (n = 141) | Controls (n = 157) | ||
| Median age in yrs (IQR) | 0.9 (0.3–2.4) | 2.9 (1.2–4.6) | <0.01 |
| Girls | 49 (35) | 66 (42) | 0.2 |
| Reason for clinic visit | |||
| Cases | |||
| URTI and imminent dehydration | 16 (11) | ||
| Wheezing illness | 32 (23) | ||
| Pneumonia | 12 (9) | ||
| Impending respiratory failure | 18 (13) | ||
| Dyspnea not further classified | 15 (11) | ||
| Fever of unknown origin | 25 (17) | ||
| Other (not classifiable) | 23 (16) | ||
| Controls | |||
| Orthopedics or surgical disease | 42 (27) | ||
| Gastrointestinal disease | 12 (8) | ||
| Neurological disease | 7 (4) | ||
| Cardiac disease | 16 (10) | ||
| Respiratory disease (noninfectious) | 7 (4) | ||
| Uro-genital disease | 5 (3) | ||
| Accompanying sibling | 43 (27) | ||
| Other (not classifiable) | 26 (17) | ||
| Comorbidity | |||
| Respiratory | |||
| Recurrent pulmonary disease | 28 (20) | 12 (7.5) | <0.01 |
| Recurrent ENT disease | 14 (10) | 31 (20) | <0.01 |
| Wheezing illness | 24 (17) | 23 (15) | 0.7 |
| Cystic fibrosis | 0 | 0 | |
| Prematurity | 39 (28) | 19 (12) | <0.01 |
URTI, upper respiratory tract infection; ENT disease, ear, nose, and throat disease; IQR, interquartile range.
NWs were performed by administering 1 to 2 ml sterile physiological saline into the nose, and nasopharyngeal secretions were aspirated through a small suction catheter that was inserted through the nostril to a depth of 3 to 4 cm. The suction catheter was connected to a mucus trap and fitted to a vacuum source for suction. Saline was not applied when apparent mucus was dripping from one of the nostrils. Samples were processed on the same day and stored at −80°C until further laboratory analysis. During the season of 2008-2009, parents of controls were contacted 1 week after sampling to inquire if the child had developed any respiratory or other complaints (fever, malaise, diarrhea, or vomiting). The study was approved by the hospital ethical board.
Virological assays.
Extraction of nucleic acids (NA) from 200 μl of the materials was performed by MagNA Pure (Roche Diagnostics, Penzberg, Germany) extraction using the total nucleic acid extraction kit (Roche Diagnostics, Penzberg, Germany). cDNA synthesis was performed as described earlier (1). The multiplex PCR is based on our previously described method (by Jansen et al. and Molenkamp et al. [10, 16]) and consists of four packages, as follows: package 1 contains influenza viruses A and B (InfA and InfB), enterovirus (EV), adenovirus (AdV), and the internal control (i.e., equine arteritis virus, which was added to the specimens prior to extraction); package 2 contains respiratory syncytial virus (RSV; A and B), rhinovirus (RV;A, B, and C), and hMPV; package 3 contains parainfluenza viruses (PIVs) 1, 2, 3, and 4 and parechovirus (PeV); package 4 containes hBoV and coronaviruses (hCoV; HKU1, NL63, 229E, and OC43). With every extraction and PCR, three controls were run: (i) a positive control, with plasmid containing the target sequence, (ii) an uninhibited internal control, equine arteritis virus, and (iii) a negative control, Tris-EDTA buffer containing carrier DNA.
PCRs were performed on a Roche LC480 platform (Roche Diagnostics, Penzberg, Germany). Reaction mixtures contained 10 μl of 2× Probes master mix (Roche Diagnostics, Penzberg, Germany), 900 nM (each) primer, 200 nM (each) probe, and 5 μl of cDNA in a total volume of 20 μl. Cycling conditions were as follows: 2 min at 50°C and 10 min at 95°C, followed by 45 cycles each consisting of 15 s at 95°C and 1 min at 60°C. Data were analyzed using the LC480 software (Roche Diagnostics, Penzberg, Germany).
Analytical sensitivity of the multiplex assay, expressed as the lower limit of detection, was between 40 and 50 copies/ml for every target. Dynamic ranges were determined to be linear between 500 and 108 copies/PCR reaction of target plasmid. Sensitivities and specificities of the PCRs were not determined, since a gold standard for viral respiratory infection has not been established and traditional methods like viral culture and antigen detection have moderate sensitivities and specificities compared to PCR (22, 23). Complete agreement (Cohen's kappa, 1.00) was seen between the multiplex PCR and the single-target PCR when samples spiked with plasmid were used. When testing clinical samples, good concordance was seen between multiplex and single-plex assays, except for clinical samples with EV (Cohen's kappa, 0.918) (10).
Values for viral copies/ml, calculated on the basis of standard curves, were used to estimate differences in quantities of respiratory viral nucleic acids between cases and controls. Standard curves were made using dilutions of cloned plasmids quantified by using Nanodrop diluted in TE (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]) supplemented with 20 μg/μl of calf thymus DNA (Sigma, Zwijndrecht, Netherlands). Considerable inhibition was defined as an inhibitory concentration value >2 times the standard deviation above the mean IC value in virus-negative samples; however, no samples were found to be considerably inhibited.
Statistics.
Differences in copies/ml were assessed using the Mann-Whitney U test. Independent categorical variables were analyzed using a chi-square test or a Fisher exact test. These statistics were performed using SPSS 16.0 software. Receiver operating characteristic (ROC) analysis and graph design were performed using GraphPad Prism 4 software.
RESULTS
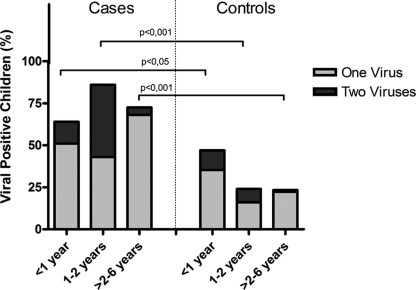
Characteristics of patients and controls are displayed in Table 1. Cases were younger than controls: the median age of cases was 0.9 years, versus 2.9 years for controls (P < 0.001). At least one virus was detected in 72% (102/141) of the cases. The presence of multiple viral species was demonstrated in 27 out of these 102 cases (26%). In the controls at least one virus could be detected in 27% (42/157), of which 12% (5/42) represented infections with multiple viral species (Table 2). To correct for the different age distribution between cases and controls, virus prevalence was stratified in three age groups (0 to 1 year, 1 to 2 years, and 2 to 6 years old). These results are displayed in Table 2. The greatest prevalence was observed in cases in the age group 1 to 2 years old, with the presence of at least one virus in 89% of cases. A strikingly high prevalence of 44% was observed in asymptomatic children in the age group <1 year old (Fig. 1). The detection of a virus did correlate with the presence of common cold symptoms among household members, attendance at a day care center, smoking parents, or any respiratory tract disease in the past (data not shown).
Table 2.
Viruses detected per age group, categorized by single or double infection
| Virus(es) | No. (%) in which virus was detected, by age (n), among cases and controls |
|||||||
|---|---|---|---|---|---|---|---|---|
| Cases |
Controls |
|||||||
| <1 yr (72) | 1–2 yrs (28) | >2–6 yrs (41) | Total (141) | <1 yr (30) | 1–2 yrs (23) | >2–6 yrs (97) | Total (157) | |
| InfA | 1 (1.4) | 3 (7.3) | 4 (2.8) | 2 (2.0) | 2 (1.2) | |||
| InfA + RV | 1 (3.6) | 1 (0.7) | ||||||
| InfA + hMPV | 1 (1.4) | 1 (0.7) | ||||||
| InfA + hCoV | 1 (2.9) | 1 (0.6) | ||||||
| InfB | 1 (2.4) | 2 (1.4) | 1 (2.9) | 1 (1.0) | 2 (1.2) | |||
| EV | ||||||||
| AdV | 3 | 2 (4.8) | 5 (3.5) | 1 (2.9) | 1 (0.6) | |||
| AdV + RSV | 1 (1.4) | 2 (7.2) | 1 (2.4) | 4 (2.8) | ||||
| AdV + hCoV | 1 (3.6) | 1 (0.7) | ||||||
| AdV + RV | 2 (7.2) | 2 (1.4) | ||||||
| RSV | 18 (25) | 5 (18) | 7 (17) | 30 (21) | 1 (2.9) | 1 (1.0) | 2 (1.2) | |
| RSV + RV | 1 (3.6) | 1 (0.7) | ||||||
| RSV + hCoV | 3 (11) | 3 (2.0) | ||||||
| RSV + hBoV | 1 (1.4) | 1 (3.6) | 2 (1.4) | |||||
| RV | 11 (15) | 5 (18) | 6 (15) | 22 (16) | 4 (12) | 3 (12) | 13 (14) | 20 (14) |
| RV + EV | 1 (1.4) | 1 (0.7) | ||||||
| RV + hMPV | 1 (2.4) | 1 (0.7) | ||||||
| RV + hBoV | 1 (1.4) | 2 (7.2) | 3 (2.1) | 1 (4.0) | 1 (0.6) | |||
| RV + PIV2 | 1 (0.7) | |||||||
| RV + EV + hBoV | 1 (1.0) | 1 (0.6) | ||||||
| RV + RSV + hCoV | 1 (2.9) | 1 (0.6) | ||||||
| RV + hCoV | 1 (2.9) | 1 (0.6) | ||||||
| hMPV | 3 (4.1) | 1 (3.6) | 3 (7.3) | 7 (4.9) | ||||
| hMPV + hBoV | 1 (1.4) | 1 (0.7) | ||||||
| PIV1 | ||||||||
| PIV2 | ||||||||
| PIV3 | 2 (2.4) | 2 (1.4) | ||||||
| PIV4 | ||||||||
| PeV | 2 (2.4) | 2 (1.4) | ||||||
| hCoV | 2 (2.4) | 1 (2.4) | 3 (2.1) | 3 (8.8) | 2 (8.0) | 3 (3.0) | 8 (5.1) | |
| hCoV + hBoV | 1 (1.4) | 1 (0.7) | ||||||
| hBoV | 1 (1.4) | 1 (3.6) | 1 (2.4) | 3 (2.1) | 2 (5.8) | 2 (1.2) | ||
| No virus detected | 21 (29) | 3 (11) | 15 (37) | 39 (28) | 19 (56) | 19 (76) | 77 (79) | 115 (73) |
| Total | 72 (100) | 28 (100) | 41 (100) | 141 (100) | 34 (100) | 25 (100) | 98 (100) | 157 (100) |
Fig. 1.
Viral prevalence in cases and controls, stratified by age. The percentage represents the amount of positive cases for the total group and for each specific age group. Data for single infections and double infections (i.e., two or more virus species detected) are shown.
A total of 130 viruses were detected in 102 cases, with RSV and RV the main pathogens, representing 31% (40/130) and 25% (32/130), respectively, of all viruses detected (Table 2). In controls, RV (53%; 24/45) and hCoV (24%; 11/45) were the most common pathogens detected.
To address differences in virus quantities, the viral copies/ml values for cases and controls were compared. Overall, asymptomatic children had a median of 2,755 copies/ml (interquartile range [IQR], 821 to 18,055), which was significantly lower than the median of 46,252 copies/ml (IQR, 4,147 to 381,898) in symptomatic children (P < 0.01). After stratifying for age, differences in viral copies/ml remained significant for all age categories. The median viral loads for the different age groups were as follows. In the age group for 0 to 1 year, asymptomatic children had 2,856 copies/ml (IQR, 748 to 15,364), versus the symptomatic group, in which we found 63,045 copies/ml (IQR, 4,376 to 417,094; P = 0.02). In the age group for 1 to 2 years old, asymptomatic chidlren had 3,319 copies/ml (IQR, 1,458 to 28,821), versus the symptomatic group, with 95,117 copies/ml (IQR, 2,608 to 471,918; P = 0.03). In the age group for 2 to 6 years old, asymptomatic children had 2,295 copies/ml (IQR, 785 to 30,573), versus the symptomatic group, with 31,068 copies/ml (IQR, 2,163 to 297,703; P = 0.01).
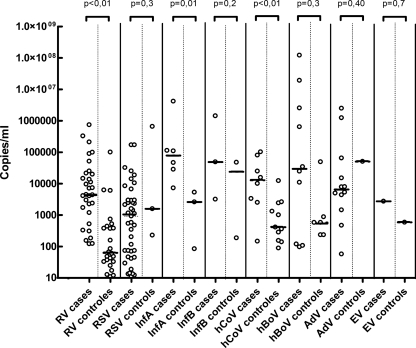
Analyses for separate viral species showed significantly lower median viral loads in asymptomatic children for RV (P < 0.01), InfA (P = 0.01), and hCoV (P < 0.01) (Fig. 2). These results did not change when we limited the analyses to single infections (data not shown). For RV these results were also not changed by age stratification, as median loads for controls were significantly lower than for the cases in each age category. The sample size was too small to repeat the age-stratified analysis for other viral species.
Fig. 2.
Crossing point values for each positive sample. White diamonds denote single infections, and black diamonds denote positive samples as part of a double or triple infection. Horizontal lines represent the median. PIV 1, PIV2, PIV3, PIV4, and hMPV are not shown in this figure because no positive samples were found among controls for these species.
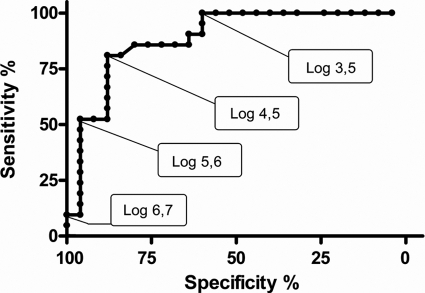
For all viruses, except InfA, overlap was seen between viral loads of cases and controls. To assess the effect of viral load cutoff values on the sensitivity and specificity for diagnosing disease, a ROC analysis was performed for RV (Fig. 3), for which sufficient infections in cases and controls were available. To prevent possible difficulties in interpretation, double infections were excluded. The ROC analysis showed that the specificity of clinically relevant positive PCR results (i.e., the presence of both virus and clinical symptoms) for RV single infections rapidly declined when loads were lower than approximately 30,000 copies/ml (104.5 copies/ml) and lost all correlation to clinical symptoms at 103.5 copies/ml.
Fig. 3.
ROC curve for RV-positive samples, excluding double infections. The curve shows the relationship between sensitivity and specificity when different cutoffs in crossing point values for association with clinical illness were applied.
During the season of 2008-2009, the parents of 57 controls were contacted by telephone 7 days after the visit to determine if clinical symptoms had developed during the week after sampling. Of the controls in whom at least one virus could be detected, 37% (7/19) developed symptoms during the week after sampling, while in those without detectable viruses, 43% (14/32) developed symptoms (P = 0.2). There was no correlation between specific viral species and development of symptoms, nor were there correlations found between the levels of viral load and the development of symptoms during the following week (data not shown).
DISCUSSION
With the availability of very sensitive molecular techniques to detect viral pathogens, questions can be addressed about the causality between the presence of microbial nucleic acids and a clinical syndrome in individual patients. To aid a better understanding of the significance of a positive PCR test and to improve the interpretation of diagnostic results, it is imperative to evaluate the detection of pathogens in asymptomatic controls, in addition to persons with clinical disease. To our knowledge this is the first prospective study using real-time PCR that has compared the presence of a comprehensive panel of respiratory viruses between young children with respiratory illness and those without symptoms.
Our study showed that the prevalence of respiratory viruses is high in asymptomatic children (27%), particularly in infants (44%). In previous studies respiratory viruses were detected at varying frequencies in asymptomatic children, depending on the different virus species tested and the age of the patient population studied. For example, Winther et al. (29) found that picornaviruses were found in only 5% of asymptomatic episodes, while Garcia-Garcia et al. (8) found a 20% carriage rate of hBoV, RV, and AdV in their control group. Studies using a large panel of respiratory viruses have reported higher prevalences; Kusel et al. (15) reported a prevalence of 25% in prospectively followed infants during periods without symptoms, and Jartti et al. (12) found at least one virus in 45% of asymptomatic infants younger than 1 year old.
The prevalence of specific viral species differed considerably between symptomatic and asymptomatic children. Among symptomatic patients, RSV was the most prevalent virus, but this virus was rarely detected among controls. This is in agreement with other studies showing that RSV is infrequently detected in asymptomatic individuals (5, 8, 14, 25), suggesting that RSV infection is usually associated with clinical illness and should be regarded as the causative pathogen when detected in a patient with respiratory symptoms. The very low prevalence of hMPV and AdV in controls compared to relatively high prevalences in cases in our study suggests that the same may be true for these viruses. Previous studies have shown that hMPV is common in asymptomatic adults but indeed is rare in asymptomatic young children (26, 27). However, for AdV, asymptomatic carriage in children has been reported to be as high as 30% (20). The low prevalence among controls in our study may be explained by the sampling method or by adenovirus serotype (2). Our study and other studies showed a prevalence of AdV of ≤4% in asymptomatic individuals when using NW (8), while a study using throat swabs revealed a prevalence of 33% (20). This difference may be related to the latent presence of AdV DNA in human tonsil tissues (9) and indicates the importance of taking into account specimen type for virological diagnosis. Further studies comparing different specimen types and adenovirus serotypes in symptomatic and asymptomatic individuals are clearly needed.
In contrast to RSV, hMPV, and AdV, viruses such as RV, hCoV, and hBoV were frequently found in asymptomatic children. This suggests that a causal inference based on the detection of these viruses in symptomatic patients should be made with caution. Assessment of viral load could potentially aid the interpretation of positive test results. Indeed, virus loads were higher in cases than in controls. The magnitude of these differences varied between different virus species. For RV, InfA, and hCoV, median viral loads differed significantly between cases and controls. A similar trend was observed for InfB and hBoV, but this trend failed to reach statistical significance, perhaps due to the limited numbers of positive samples for these species. Few studies have compared quantitative PCR results between symptomatic and asymptomatic children. For RV, Peltola et al. did not find statistical differences in RV load in symptomatic versus asymptomatic persons (18). However, the methodology of this study differed from our study, as it compared both adults and children and analyzed samples taken at different time points of infection.
When differences in viral load between symptomatic and asymptomatic children are present, it should be feasible to define cutoff levels for the various viruses to aid in the clinical interpretation of test results. InfA was the only virus for which no overlap was noted between viruses in cases versus controls. One could speculate, therefore, that cutoff values for influenza virus could easily be determined, but larger sample sizes of InfA-positive cases and controls are clearly needed for this. For viruses that show overlap of viral loads amid cases and controls, ROC curves are useful to define cutoff values, as they demonstrate the effects of different cutoff levels on test specificity and sensitivity. By using the depicted ROC curve for RV, one could inform a clinician that for RV loads of ≥104.5 copies/ml, this virus is very likely to be the cause of the presenting illness, while doubts concerning the etiology may remain for cases with loads of <104.5 copies/ml.
There are obvious challenges in determining clinically relevant cutoff values for respiratory viruses in different patient groups, as these will likely be influenced by factors such as the timing of sampling during the course of illness and the presence of comorbidities. In addition, challenges exist with regard to sample quality and standardization of sampling methods. For example, sampling of the upper respiratory tract will always show some degree of interpatient variation. We used NWs, which will results in variations based on dilution of sampling. However, dilution of NWs is likely never greater than 10-fold, based on the amount of mucus excreted in asymptomatic individuals (4). Therefore, semiquantitation is still applicable in samples that can reach up to 1010 viral particles per milliliter (6).
Our observations point to the need for a more nuanced look at diagnostic results and the potential value of quantitative results in interpreting the clinical relevance of test results. Larger samples sizes in a variety of different patient groups are clearly needed to overcome the above-mentioned challenges and to design reliable ROC curves for infections with viruses that are also found frequently among controls.
Regarding the presence of a respiratory virus in the upper respiratory tract in the absence of symptoms, this observation may be explained by several causes. First, detectable virus could represent the period of incubation before the onset of symptoms. In our study we did not find a higher frequency of symptom development during the following week in asymptomatic children who were virus positive compared to those in whom no virus could be detected. However, reliable analyses could not be performed for the less-prevalent viruses in our study, like InfA, InfB, and hBoV. Second, the detection of virus could represent postinfectious shedding. Duration of viral shedding differs greatly between different studies, varying between several days from the start of symptoms to several weeks after resolution of symptoms. Duration of shedding likely depends on virus species, clinical background of the patient, and the detection method used (11, 19). In a prospective birth cohort study, about 25% of infants shed respiratory viruses (RV, hMPV, and hCoV or mixed infection) for 3 weeks or more after clinical illness (19). We included young children as controls if they had been symptom-free for at least 7 days prior to inclusion; hence, it remains possible that a proportion of detectable viruses represented long-term postinfectious shedding. A third explanation could be subclinical infections. Studies of adult volunteers challenged with InfA or RSV showed that 23% to 33% do not develop clinically manifest symptoms (3, 28). Although such human volunteer studies have not been performed in children, it seems likely that asymptomatic infections also occur in children, especially in cases of less pathogenic viruses, like RV and hCoV.
The most important limitation of our report is the cross-sectional character of the study. Since controls were sampled once and clinical follow-up was done in only a proportion of controls, it is difficult to determine the exact explanation for positivity in individual participants (e.g., post-viral shedding, subclinical infection, or incubation before symptomatic infection). Larger longitudinal studies are needed to fully address the causes and implications of respiratory virus detection in asymptomatic persons. Such studies will also help to define cutoff values for specific viruses to aid in diagnostic interpretation. Our data indicate that defining such cutoff levels may be feasible and represent the next necessary step toward reliable laboratory diagnosis of viral respiratory infections.
ACKNOWLEDGMENT
We have no potential conflicts of interest with this work.
Footnotes
Published ahead of print on 4 May 2011.
REFERENCES
- 1. Beld M., et al. 2004. Highly sensitive assay for detection of enterovirus in clinical specimens by reverse transcription-PCR with an armored RNA internal control. J. Clin. Microbiol. 42:3059–3064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brandt C. D., et al. 1969. Infections in 18,000 infants and children in a controlled study of respiratory tract disease. I. Adenovirus pathogenicity in relation to serologic type and illness syndrome. Am. J. Epidemiol. 90:484–500 [DOI] [PubMed] [Google Scholar]
- 3. Carrat F., et al. 2008. Time lines of infection and disease in human influenza: a review of volunteer challenge studies. Am. J. Epidemiol. 167:775–785 [DOI] [PubMed] [Google Scholar]
- 4. Eichner H., Behbehani A. A., Hochstrasser K. 1983. Diagnostic value of nasal secretions, current state: normal values. Laryngol. Rhinol. Otol. (Stuttg.) 52:561–565 (In German.) [PubMed] [Google Scholar]
- 5. Falsey A. R., Criddle M. C., Walsh E. E. 2006. Detection of respiratory syncytial virus and human metapneumovirus by reverse transcription polymerase chain reaction in adults with and without respiratory illness. J. Clin. Virol. 35:46–50 [DOI] [PubMed] [Google Scholar]
- 6. Franz A., et al. 2010. Correlation of viral load of respiratory pathogens and co-infections with disease severity in children hospitalized for lower respiratory tract infection. J. Clin. Virol. 48:239–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Freymuth F., et al. 2006. Comparison of multiplex PCR assays and conventional techniques for the diagnostic of respiratory virus infections in children admitted to hospital with an acute respiratory illness. J. Med. Virol. 78:1498–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Garcia-Garcia M. L., et al. 2008. Human bocavirus detection in nasopharyngeal aspirates of children without clinical symptoms of respiratory infection. Pediatr. Infect. Dis. J. 27:358–360 [DOI] [PubMed] [Google Scholar]
- 9. Garnett C. T., et al. 2009. Latent species C adenoviruses in human tonsil tissues. J. Virol. 83:2417–2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jansen R. R., et al. 13 May 2011, posting date. Development and evaluation of a four-tube real-time multiplex PCR assay covering fourteen respiratory viruses, and comparison to its corresponding single target counterparts. J. Clin. Virol. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jansen R. R., et al. 2010. Quantitation of respiratory viruses in relation to clinical course in children with acute respiratory tract infections. Pediatr. Infect. Dis. J. 29:82–84 [DOI] [PubMed] [Google Scholar]
- 12. Jartti T., et al. 2008. Serial viral infections in infants with recurrent respiratory illnesses. Eur. Respir. J. 32:314–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Johnston S. L., et al. 1993. Use of polymerase chain reaction for diagnosis of picornavirus infection in subjects with and without respiratory symptoms. J. Clin. Microbiol. 31:111–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kumar S., et al. 2008. Detection of 11 common viral and bacterial pathogens causing community-acquired pneumonia or sepsis in asymptomatic patients by using a multiplex reverse transcription-PCR assay with manual (enzyme hybridization) or automated (electronic microarray) detection. J. Clin. Microbiol. 46:3063–3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kusel M. M., et al. 2006. Role of respiratory viruses in acute upper and lower respiratory tract illness in the first year of life: a birth cohort study. Pediatr. Infect. Dis. J. 25:680–686 [DOI] [PubMed] [Google Scholar]
- 16. Molenkamp R., van der H. A., Schinkel J., Beld M. 2007. Simultaneous detection of five different DNA targets by real-time Taqman PCR using the Roche LightCycler480: application in viral molecular diagnostics. J. Virol. Methods 141:205–211 [DOI] [PubMed] [Google Scholar]
- 17. Nokso-Koivisto J., Kinnari T. J., Lindahl P., Hovi T., Pitkaranta A. 2002. Human picornavirus and coronavirus RNA in nasopharynx of children without concurrent respiratory symptoms. J. Med. Virol. 66:417–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Peltola V., et al. 2008. Rhinovirus transmission within families with children: incidence of symptomatic and asymptomatic infections. J. Infect. Dis. 197:382–389 [DOI] [PubMed] [Google Scholar]
- 19. Regamey N., et al. 2008. Viral etiology of acute respiratory infections with cough in infancy: a community-based birth cohort study. Pediatr. Infect. Dis. J. 27:100–105 [DOI] [PubMed] [Google Scholar]
- 20. Sackesen C., Pinar A., Sekerel B. E., Akyon Y., Saraclar Y. 2005. Use of polymerase chain reaction for detection of adenovirus in children with or without wheezing. Turk. J. Pediatr. 47:227–231 [PubMed] [Google Scholar]
- 21. Thavagnanam S., et al. 2010. Respiratory viral infection in lower airways of asymptomatic children. Acta Paediatr. 99:394–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. van de Pol A. C., et al. 2007. Increased detection of respiratory syncytial virus, influenza viruses, parainfluenza viruses, and adenoviruses with real-time PCR in samples from patients with respiratory symptoms. J. Clin. Microbiol. 45:2260–2262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. van Elden L. J., et al. 2008. Enhanced severity of virus associated lower respiratory tract disease in asthma patients may not be associated with delayed viral clearance and increased viral load in the upper respiratory tract. J. Clin. Virol. 41:116–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. van Elden L. J., et al. 2002. Polymerase chain reaction is more sensitive than viral culture and antigen testing for the detection of respiratory viruses in adults with hematological cancer and pneumonia. Clin. Infect. Dis. 34:177–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. von Linstow M. L., et al. 2008. Acute respiratory symptoms and general illness during the first year of life: a population-based birth cohort study. Pediatr. Pulmonol. 43:584–593 [DOI] [PubMed] [Google Scholar]
- 26. von Linstow M. L., et al. 2004. Human metapneumovirus and respiratory syncytial virus in hospitalized danish children with acute respiratory tract infection. Scand. J. Infect. Dis. 36:578–584 [DOI] [PubMed] [Google Scholar]
- 27. Walsh E. E., Peterson D. R., Falsey A. R. 2008. Human metapneumovirus infections in adults: another piece of the puzzle. Arch. Intern. Med. 168:2489–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wilson R., et al. 1987. Upper respiratory tract viral infection and mucociliary clearance. Eur. J. Respir. Dis. 70:272–279 [PubMed] [Google Scholar]
- 29. Winther B., Hayden F. G., Hendley J. O. 2006. Picornavirus infections in children diagnosed by RT-PCR during longitudinal surveillance with weekly sampling: association with symptomatic illness and effect of season. J. Med. Virol. 78:644–650 [DOI] [PubMed] [Google Scholar]