Abstract
Rapid identification of Cryptococcus neoformans var. grubii, Cryptococcus neoformans var. neoformans, and Cryptococcus gattii is imperative for facilitation of prompt treatment of cryptococcosis and for understanding the epidemiology of the disease. Our purpose was to evaluate a test algorithm incorporating commercial rapid biochemical tests, differential media, and DNA sequence analysis that will allow us to differentiate these taxa rapidly and accurately. We assessed 147 type, reference, and clinical isolates, including 6 other Cryptococcus spp. (10 isolates) and 14 other yeast species (24 isolates), using a 4-hour urea broth test (Remel), a 24-hour urea broth test (Becton Dickinson), a 4-hour caffeic acid disk test (Hardy Diagnostics and Remel), 40- to 44-hour growth assessment on l-canavanine glycine bromothymol blue (CGB) agar, and intergenic spacer (IGS) sequence analysis. All 123 Cryptococcus isolates hydrolyzed urea, along with 7 isolates of Rhodotorula and Trichosporon. Eighty-five of 86 C. neoformans (99%) and 26 of 27 C. gattii (96%) isolates had positive caffeic acid results, unlike the other cryptococci (0/10) and yeast species (0/24). Together, these two tests positively identified virtually all C. neoformans/C. gattii isolates (98%) within 4 h. CGB agar or IGS sequencing further differentiated these isolates within 48 h. On CGB, 25 of 27 (93%) C. gattii strains induced a blue color change, in contrast to 0 of 86 C. neoformans isolates. Neighbor-joining cluster analysis of IGS sequences differentiated C. neoformans var. grubii, C. neoformans var. neoformans, and C. gattii. Based on these results, we describe a rapid identification algorithm for use in a microbiology laboratory to distinguish clinically relevant Cryptococcus spp.
INTRODUCTION
Cryptococcus neoformans and Cryptococcus gattii are closely related species of basidiomycetous yeasts that cause potentially severe pulmonary and central nervous system (CNS) infections. C. neoformans primarily infects AIDS patients and other immunocompromised hosts, producing meningoencephalitis and other neurological complications (22). Globally, nearly 1 million persons with HIV develop cryptococcal meningitis each year. Depending on treatment, up to 70% will die within 3 months (26). Historically, human cases of cryptococcosis in temperate climates were primarily attributed to the two subspecies Cryptococcus neoformans var. grubii and C. neoformans var. neoformans owing to their worldwide distribution. Conversely, pulmonary and neurological infections of C. gattii generally occur at low incidence in immunocompetent hosts and were thought to be confined to tropical and subtropical regions where it is endemic (22). However, since 1999, hundreds of C. gattii infections have been identified in British Columbia, Canada, and the Northwestern United States. Characterized as an outbreak, these isolates signify a possible shift in the ecological distribution of C. gattii (5). Furthermore, macrophage intracellular proliferation studies and murine inhalation assays suggest that the outbreak strains of C. gattii from the Pacific Northwest are more virulent than nonoutbreak strains of C. gattii (2, 10, 23). With respect to patient treatment, differences in antifungal susceptibility patterns exist among the molecular subtypes of C. gattii and C. neoformans, in which molecular genotype VGII isolates, which comprise the majority (95%) of C. gattii clinical isolates from the Pacific Northwest of the United States and Canada (2, 17), are significantly less susceptible to fluconazole and other triazoles, as determined by the broth microdilution method, than other C. gattii and C. neoformans genotype isolates (3, 14, 28).
Given the severity of cryptococcosis, the changing ecology and virulence of C. gattii, and the differences in antifungal susceptibility, it is imperative that clinical laboratories implement testing algorithms that rapidly detect C. neoformans and C. gattii and distinguish them from other pathogenic Cryptococcus and yeast species. Both urea hydrolysis and detection of melanin production are key characteristics distinguishing C. neoformans and C. gattii from other yeast and Cryptococcus spp. However, the biochemical tests developed to detect these characteristics required either lengthy incubation times (2 to 7 days) (18) or tedious preparation of media and reagents (11, 16, 27, 29–31). l-Canavanine glycine bromothymol blue (CGB) agar has been reported to differentiate most C. gattii isolates from C. neoformans (18). Alternatively, DNA sequence analysis has been suggested to identify C. neoformans and C. gattii; however, the traditional fungal sequencing targets, such as the internal transcribed spacer (ITS) and the D2 region of the large ribosomal subunit, show >99% similarity between species and/or subspecies (13, 15, 18), suggesting that specialized DNA targets with more discriminatory power, such as the rRNA intergenic spacer (IGS) (1, 6, 7), may be more useful for differentiating isolates at the species and/or subspecies level in a clinical setting.
Our goal was to develop a rapid, accurate, and comprehensive testing algorithm incorporating biochemical tests and/or DNA sequence analysis (i) to distinguish C. neoformans and C. gattii from other Cryptococcus and yeast species and (ii) to differentiate C. neoformans and C. gattii at the species or subspecies level depending on the required level of discrimination. To this end, we utilized rapid commercial preparations of tests to detect urea hydrolysis (urease test) and melanin production (caffeic acid disk test) in order to positively identify C. neoformans/C. gattii isolates in 4 h with minimal preparatory work. Within 48 h, isolates were further differentiated at the species level as C. neoformans or C. gattii by CGB agar. Alternatively, highly discriminatory subspecies identification of C. neoformans var. grubii, C. neoformans var. neoformans, and C. gattii was achieved within 48 h using IGS sequence analysis. When implemented in a clinical laboratory, this algorithm provides rapid and accurate species identification to aid both patient diagnosis and epidemiological study.
MATERIALS AND METHODS
Strains.
A total of 147 yeast isolates were tested, including 123 Cryptococcus strains and 24 non-Cryptococcus yeasts. Of the Cryptococcus isolates, 47 were isolated from clinical samples submitted to the Mycology Laboratory of the Ontario Public Health Laboratory from 2007 to 2010. Another 49 were obtained from the Fungal Testing Laboratory, Department of Pathology, University of Texas Health Sciences Center, and an additional 15 were from the British Columbia Centre for Disease Control. Twelve type and reference strains (24) were also included in the analysis (Table 1). Of the 24 non-Cryptococcus yeast isolates, 10 were clinical isolates from the Ontario Public Health Laboratory and 14 were type strains (Table 1).
Table 1.
Study isolates
| Yeast group | No. of type, reference, and clinical isolatesa | Type or reference strain(s)b |
|---|---|---|
| Cryptococcus | ||
| Cryptococcus albidus | 1 | ATCC 10666T |
| Cryptococcus gattii | 27 | CBS 6289,c ATCC MYA-4561,c CBS 6955,c ATCC MYA-4563c |
| Cryptococcus laurentii | 1 | ATCC 18803T |
| Cryptococcus luteolus | 1 | JCM 3689T |
| Cryptococcusneoformans var. grubii | 82 | ATCC MYA-4564,c ATCC MYA-4565c |
| Cryptococcusneoformans var. neoformans | 4 | ATCC MYA-4567c |
| Cryptococcus magnus | 4 | CBS 140T (AF190008.1d) |
| Cryptococcus terreus | 1 | CBS 1895T |
| Cryptococcusunigutulatus | 2 | JCM 3685T |
| Non-Cryptococcus | ||
| Candida albicans | 2 | NRRL Y-12983T |
| Candida dubliniensis | 1 | NRRL Y-17841T |
| Candida glabrata | 3 | NRRL Y-65T |
| Candida guilliermondii | 1 | NRRL Y-2075T |
| Candida krusei | 2 | NRRL Y-5396T |
| Candida lusitaniae | 1 | NRRL Y-11827T |
| Candida parapsilosis | 2 | DSM 5784T |
| Candida tropicalis | 2 | DSM 11953T |
| Geotrichum candidum | 1 | JCM 6359T |
| Rhodotorula mucilaginosa | 2 | JCM 8115T |
| Saccharomyces cerevisiae | 2 | JCM 7255T |
| Trichosporon asahii | 3 | JCM 2466T |
| Trichosporon asteroides | 1 | JCM 2937T |
| Trichosporon inkin | 1 | JCM 9195T |
Identification based on >99% ITS and/or >98% IGS sequence identity to a type or reference strain.
Superscript “T” indicates type strain.
ATCC, American Type Culture Collection, Manassas, VA; CBS, Centraalbureau voor Schimmelcultures, Utrecht, Netherlands; JCM, Japanese Collection of Microorganisms, Saitama, Japan; NRRL, United States Department of Agriculture Agricultural Research Service, Peoria, IL; DSM, Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany.
Meyer et al. (16).
GenBank accession number.
Biochemical testing.
Urease tests were performed on 24- or 48-hour inhibitory mold agar (IMA; Becton Dickinson [BD], Sparks, MD) or Sabouraud dextrose agar (BD) plus 0.005% chloramphenicol (SAB+C) cultures using Remel rapid urea broth (Remel, Lenexa) or BD BBL urease test broth according to the manufacturer's instructions. Briefly, 3 ml of broth was heavily inoculated with yeast culture and incubated at 37°C. Tubes were inspected at 4 and 24 h (Remel) or 24 and 48 h (BD) for pink color development, indicating positive urea hydrolysis.
Caffeic acid disk tests were performed by inoculating disks from Remel or Hardy Diagnostics (Santa Maria, CA) with cultures previously grown for 72 hours on 1% cornmeal agar plus 0.01% Tween 80 (CMA). (Inoculation of the disks with CMA cultures was recommended by both manufacturers since cultures grown on media containing glucose yield false-negative results.) Disks were incubated at 37°C on CMA medium for 4 h before they were assessed for brown pigment, indicating a positive result.
l-Canavanine glycine bromothymol blue agar (CGB) was prepared as previously described (19). Isolates were grown on CGB at 27°C and examined at 40 to 44 h (most isolates did not grow at 24 h). Development of a bright blue color was indicative of a positive result. If the medium remained yellow-green in color, the result was interpreted as negative. Prolonged incubation of cultures beyond 48 h resulted in false-positive interpretation due to blue color production by non-C. gattii cryptococci.
DNA sequence analysis.
As a gold standard, all isolates were identified by DNA sequence analysis of the rRNA internal transcribed spacer (ITS) and the rRNA intergenic spacer (IGS) for C. neoformans var. grubii, C. neoformans var. neoformans, and C. gattii. Products were amplified using Phire polymerase (New England BioLabs, Ipswich, MA) and the primer pairs comprising ITS-1 (5′TCCGTAGGTGAACCTGCGG3′)/ITS4 (5′TCCTCCGCTTATTGATATGC) and IGSF (5′ATCCTTTGCAGACGACTTGA3′)/IGSR (5′GTGATCAGTGCATTGCATGA3′) (24). ITS PCRs were cycled at 98°C for 30 s, followed by 35 cycles at 98°C for 5 s, 56°C for 5 s, and 72°C for 20 s, followed by 72°C for 1 min, while the IGS PCR conditions were 98°C for 30 s, followed by 35 cycles at 98°C for 5 s, 60°C for 5 s, and 72°C for 20 s, followed by 72°C for 1 min. PCR products were sequenced using BigDye v1.1 and a model 3130xl genetic analyzer (Applied Biosystems, Foster City, CA). Forward and reverse fragments were aligned and trimmed in BioNumerics v6.0.1 (Applied Maths, Austin, TX). Identities of clinical isolates were assigned based on a >99% match to the ITS sequence of a type strain (Table 1) or a >98% match to the IGS sequence of a Cryptococcus reference strain (Table 1). ITS and IGS sequences of all type and reference strains were determined during the course of this study except for the ITS sequence of Cryptococcus magnus type strain CBS 140, which was obtained from GenBank (accession no. AF190008.1). Using BioNumerics v6.0.1, IGS sequences were aligned and trimmed to defined start and end positions, yielding fragments of 728 to 761 bp. An unrooted neighbor-joining (NJ) tree was generated in accordance with Kimura-2 parameter (K2P) correction. Bootstrap analysis (500 replicates) was used to access the robustness of the clusters.
RESULTS
When evaluated using Remel rapid urea broth and BD urease test broth, isolates of Cryptococcus, Trichosporon, and Rhodotorula were found to turn the medium bright pink, while other species of yeast produced a negative reaction. The reactions of different isolates within a given genus were more consistent when the isolates were grown on IMA than when they were grown on SAB+C and were found to be more consistent when the isolates were tested using the Remel rapid urea broth than when they were tested using the BD urease test broth (Table 2). While most positive results were obtained in 4 (Remel) or 24 (BD) hours, negative reactions could not be confirmed until 24 (Remel) or 48 (BD) hours, due to a small number of weakly positive isolates (IMA and Remel, 2.3%; IMA and BD, 1%; SAB+C and Remel, 4%; SAB+C and BD, 1%). All but one (85/86 [99%]) of the C. neoformans isolates and one (26/27 [99%]) of the C. gattii isolates induced a brown color, indicative of a positive reaction, on the caffeic acid disks. All other species of Cryptococcus and other yeast species had a negative reaction (Table 2). Nearly all (25/27 [93%]) of the C. gattii isolates produced a bright blue color after 40 to 44 h on CGB agar. All 86 of the C. neoformans isolates had a negative reaction on CGB. As well, isolates of Trichosporon asahii (3/3 [100%]), Trichosporon asteroides (1/1 [100%]), Trichosporon inkin (1/1 [100%]), Cryptococcus laurentii (1/1 [100%]), Cryptococcus luteolus (1/1 [100%]), Candida parapsilosis (2/2 [100%]), and Geotrichum candidum (1/1 [100%]) also produced a positive reaction on CGB agar.
Table 2.
Results for the urease broth, caffeic acid disk, and CGB agar biochemical tests
| Identificationa | No. of isolates | No. of isolates with positive reaction/total no. of isolates (%) |
||||||
|---|---|---|---|---|---|---|---|---|
| Urease broth test using: |
Caffeic acid disk test |
CGB agar | ||||||
| Remel IMA | Remel SAB+C | BD IMA | BD SAB+C | Remel | Hardy Diagnostics | |||
| C. neoformans | 86 | 86/86 (100) | 86/86 (100) | 86/86 (100) | 85/86 (99) | 85/86 (99) | 85/86 (99) | 0/86 (0) |
| C. gattii | 27 | 27/27 (100) | 27/27 (100) | 27/27 (100) | 27/27 (100) | 26/27 (96) | 26/27 (96) | 25/27 (93) |
| Other cryptococci | 10 | 10/10 (100) | 9/10 (90) | 7/10 (70) | 7/10 (70) | 0/10 (0) | 0/10 (0) | 2/10 (20) |
| Trichosporon | 5 | 5/5 (100) | 5/5 (100) | 3/5 (60) | 3/5 (60) | 0/5 (0) | 0/5 (0) | 5/5 (100) |
| Rhodotorula | 2 | 2/2 (100) | 2/2 (100) | 2/2 (100) | 2/2 (100) | 0/2 (0) | 0/2 (0) | 0/2 (0) |
| Geotrichum | 1 | 0/1 (0) | 0/1 (0) | 0/1 (0) | 0/1 (0) | 0/1 (0) | 0/1 (0) | 1/1 (100) |
| Saccharomyces | 2 | 0/2 (0) | 0/2 (0) | 0/2 (0) | 0/2 (0) | 0/2 (0) | 0/2 (0) | 0/2 (0) |
| Candida | 14 | 0/14 (0) | 0/14 (0) | 0/14 (0) | 0/14 (0) | 0/14 (0) | 0/14 (0) | 2/14 (14) |
Identification based on >99% ITS and/or >98% IGS sequence identity to a type or reference strain.
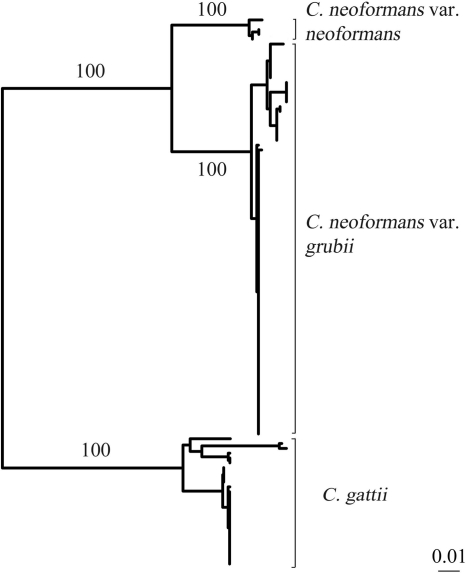
As a gold standard, all clinical isolates were identified by DNA sequence analysis. Except for C. neoformans and C. gattii, ITS sequence analysis clearly distinguished all yeast species. Clinical isolates demonstrated >99% similarity to type strains, with between-species divergence of >5.8%. With >99% between-species similarity in the ITS region, C. neoformans var. grubii, C. neoformans var. neoformans, and C. gattii were distinguished using IGS sequencing. Neighbor-joining cluster analysis clearly differentiated C. neoformans var. grubii (82 isolates), C. neoformans var. neoformans (4 isolates), and C. gattii (27 isolates), with 100% bootstrap support for each of the three clusters (Fig. 1 and Table 1). Isolates showed >98.9% similarity to a reference strain (24) and >4% divergence between species and subspecies.
Fig. 1.
NJ tree constructed using IGS sequences (bp) from 82 isolates of C neoformans var. grubii, 4 isolates of C. neoformans var. neoformans, and 27 isolates of C. gattii. Bootstrap values (500 replicates) are indicated. Bar, 1% nucleotide substitutions.
DISCUSSION
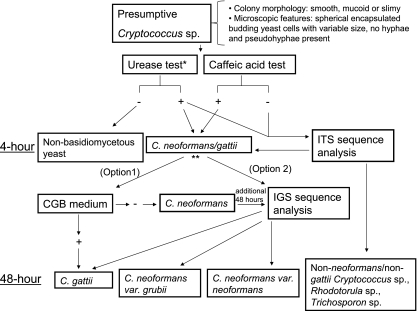
As a severe fungal infection of the CNS leading to meningoencephalitis and neurological complications, successful treatment of cryptococcosis depends on rapid and accurate identification of the causative agents, C. neoformans and C. gattii. Although cryptococcosis infections in temperate regions were previously attributed to C. neoformans, the recent outbreak of C. gattii in the Pacific Northwest region of North America suggests a recent ecological expansion of C. gattii from tropical and subtropical regions where C. gattii was previously endemic into temperate climates (17). Additional differences in host preference (22), virulence (2, 10), and antifungal drug susceptibility patterns (3, 14, 28) necessitate that clinical laboratories implement testing algorithms that differentiate these species in order to aid patient treatment and advance the understanding of the disease and its epidemiology. Here, we describe a testing algorithm (Fig. 2) using two rapid commercial biochemical tests that distinguishes the majority (98%) of C. neoformans/C. gattii isolates from other Cryptococcus and yeast species in 4 h. Growth on CGB selective medium and/or IGS sequencing further differentiates C. neoformans/C. gattii isolates at the species or subspecies level within 48 h. This testing algorithm represents a rapid and accurate method for identifying clinically relevant isolates of Cryptococcus.
Fig. 2.
Clinical laboratory testing algorithm for rapidly identifying Cryptococcus spp. *, urease test. Optimal test performance is achieved by testing culture isolates grown on IMA agar and using Remel rapid urea broth. **, C. neoformans/C. gattii isolates can be further differentiated using either CGB medium (option 1) or IGS sequence analysis (option 2).
Using various test broth formulations, studies have shown that urea hydrolysis with medium alkalinization is a pivotal, accurate biochemical test in the dichotomous classification of yeasts, with positive results distinguishing the genera Cryptococcus, Rhodotorula, and Trichosporon (18, 27, 31). When coupled with the detection of melanin production through brown pigmentation of caffeic acid agar medium (11, 16, 29) or caffeic acid-ferric citrate-infused paper disks (30), C. neoformans/C. gattii are easily distinguished from other Cryptococcus and yeast species. In this study, we utilized rapid commercial forms of these two biochemical tests, which are advantageous over formulations that require >24 h of incubation for results or require tedious preliminary preparation of media and reagents. All Cryptococcus, Rhodotorula, and Trichosporon isolates were differentiated from other yeasts by use of the two commercial urea broth tests. However, growth on IMA yielded the most-consistent results, and the Remel rapid urea broth provided more-accurate and -rapid results (4 h) than the BD urease test broth (24 h). With the use of the caffeic acid disk test, all but 2 of the C. neoformans/C. gattii isolates (98%) were differentiated from other Cryptococcus spp., Rhodotorula, and Trichosporon. Although the performances of the caffeic acid disks from Remel and Hardy Diagnostics were identical, the disks from Hardy Diagnostics were significantly less expensive ($1.70/disk) than those from Remel ($3.00/disk) based on the list price indicated on the website. With the use of the Remel rapid urease test and the caffeic acid disk test as outlined in Fig. 2, most C. neoformans/C. gattii isolates were identified in 4 h. Since the caffeic acid disk test produced two false negatives (1.8% [2/113]), also noted by Klein et al. (18), the testing algorithm was designed to refer these isolates (positive for the urease test and negative for the caffeic acid disk test) for ITS sequencing (Fig. 2). ITS sequencing clearly distinguished these false negatives as C. neoformans/C. gattii and identified other Cryptococcus, Trichosporon, and Rhodotorula species as expected (4).
The test algorithm presented in Fig. 2 provides two options for further differentiating the C. neoformans/C. gattii isolates identified using the urease and caffeic acid disk tests. As option 1, CGB selective medium was used to distinguish C. gattii from C. neoformans based on the former's ability to grow in the presence of l-canavanine and metabolize glycine in the process, thereby raising the pH and causing the bromothymol blue indicator to turn the agar blue (25). Similar to results reported by Klein et al. (18), none of the C. neoformans isolates in this study grew on CGB agar; however, two of the C. gattii isolates (7%) failed to produce a positive reaction (Table 2). Thus, the use of GCB medium is an inexpensive and feasible way to distinguish C. neoformans from C. gattii in 48 h. CGB-negative isolates can be referred for IGS sequence analysis to identify the small number of CGB-nonreactive C. gattii isolates in an additional 48 h (Fig. 2). Besides C. gattii, seven other yeast species (10 isolates) of Trichosporon spp., Geotrichum spp., and C. parapsilosis turned the CGB medium blue, also noted by Klein et al. (18). However, these species are distinguishable from C. gattii based on microscopic examination and biochemical analysis. In addition to the fact that these species are negative for the urease (except for Trichosporon sp.) and caffeic acid disk tests, the presence of arthroconidia distinguishes Trichosporon and Geotrichum from Cryptococcus while well-developed pseudohyphae differentiate C. parapsilosis from Cryptococcus (21). Thus, microscopic examination and the urease and caffeic acid disk tests are crucial in conjunction with CGB medium in order to positively identify C. gattii (Fig. 2).
For the second option, IGS sequence analysis was used to distinguish C. neoformans var. grubii, C. neoformans var. neoformans, and C. gattii as previously reported (1, 6, 7). NJ cluster analysis of IGS sequences clearly differentiated C. gattii, C. neoformans var. grubii, and C. neoformans var. neoformans with 100% bootstrap support (Fig. 1). Although more labor-intensive and costly, DNA sequencing is rapidly becoming a common procedure in most clinical laboratories because is has greater discriminatory power than that generally provided by differential media and biochemical tests. In laboratories where DNA sequencing is routinely available, IGS sequence analysis represents the optimal method for identification of Cryptococcus because it identifies all C. gattii isolates, including CGB-nonreactive isolates, and differentiates C. neoformans at the subspecies level. While there are no clinical data suggesting any treatment or outcome differences for C. neoformans var. grubii versus C. neoformans var. neoformans infections, there are epidemiological differences that suggest that it is prudent for clinical reference laboratories to track the species distribution. For instance, although the majority of clinical infections worldwide are caused by C. neoformans var. grubii, C. neoformans var. neoformans infections are more prevalent in certain geographic locations (India, France, Italy, and Denmark) (8, 12, 20) and are associated with infections in the elderly, the presence of skin lesions, and corticosteroid use (9).
C. neoformans and C. gattii cannot be differentiated by routine methods used in clinical laboratories for yeast identification, such as API 20C AUX (bioMérieux, Durham, NC), Vitek (bioMérieux), and MicroScan (Siemens, West Sacramento, CA). Serotyping distinguishes C. neoformans (serotypes A and D) from C. gattii (serotypes B and C); however, the only commercial kit for serotyping cryptococci (Crypto-Check kit; Iatron, Inc., Tokyo, Japan) was discontinued. Multilocus sequence analysis identifies C. neoformans and C. gattii and delineates these species into eight molecular types (24); however, this method is costly and time-consuming, and this level of discrimination is usually unnecessary in a clinical laboratory. DNA sequencing of the D2 region of the 28S large ribosomal subunit also distinguishes C. neoformans and C. gattii (18) but may not be able to reliably distinguish C. neoformans var. grubii and C. neoformans var. neoformans (13). Similarly, we found that DNA sequencing of the ITS region differentiates C. gattii from the other two groups but has poor discrimination (≥99.5% similarity) between C. neoformans var. neoformans and C. neoformans var. grubii.
The testing algorithm presented here (Fig. 2) provides an easy, rapid, and accurate method of identifying Cryptococcus species from among other yeasts in a clinical laboratory. C. neoformans/C. gattii are distinguished from other species in 4 h and then further differentiated at the species or subspecies level in 48 h. Depending on the required level of discrimination, cost constraints, workflow, and expertise of personnel, the testing algorithm provides laboratories with the option of CGB medium for biochemical differentiation of C. gattii and C. neoformans or IGS sequence-based distinction of C. neoformans var. grubii, C. neoformans var. neoformans, and C. gattii. Rapid and accurate identification will aid patient treatment and epidemiological understanding of cryptococcosis.
Footnotes
Published ahead of print on 18 May 2011.
REFERENCES
- 1. Bovers M., et al. 2007. Identification of genotypically diverse Cryptococcus neoformans and Cryptococcus gattii isolates by Luminex xMAP technology. J. Clin. Microbiol. 45:1874–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Byrnes E. J., III, et al. 2010. Emergence and pathogenicity of highly virulent Cryptococcus gattii genotypes in the northwest United States. PLoS Pathog. 6:e1000850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chong H. S., Dagg R., Malik R., Chen S., Carter D. 2010. In vitro susceptibility of the yeast pathogen Cryptococcus to fluconazole and other azoles varies with molecular genotype. J. Clin. Microbiol. 48:4115–4120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ciardo D. E., Schar G., Bottger E. C., Altwegg M., Bosshard P. P. 2006. Internal transcribed spacer sequencing versus biochemical profiling for identification of medically important yeasts. J. Clin. Microbiol. 44:77–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Datta K., et al. 2009. Spread of Cryptococcus gattii into Pacific Northwest region of the United States. Emerg. Infect. Dis. 15:1185–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Diaz M. R., Boekhout T., Kiesling T., Fell J. W. 2005. Comparative analysis of the intergenic spacer regions and population structure of the species complex of the pathogenic yeast Cryptococcus neoformans. FEMS Yeast Res. 5:1129–1140 [DOI] [PubMed] [Google Scholar]
- 7. Diaz M. R., Fell J. W. 2005. Use of a suspension array for rapid identification of the varieties and genotypes of the Cryptococcus neoformans species complex. J. Clin. Microbiol. 43:3662–3672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dromer F., Mathoulin S., Dupont B., Laporte A. 1996. Epidemiology of cryptococcosis in France: a 9-year survey (1985–1993). French Cryptococcosis Study Group. Clin. Infect. Dis. 23:82–90 [DOI] [PubMed] [Google Scholar]
- 9. Dromer F., Mathoulin S., Dupont B., Letenneur L., Ronin O. 1996. Individual and environmental factors associated with infection due to Cryptococcus neoformans serotype D. French Cryptococcosis Study Group. Clin. Infect. Dis. 23:91–96 [DOI] [PubMed] [Google Scholar]
- 10. Fraser J. A., et al. 2005. Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature 437:1360–1364 [DOI] [PubMed] [Google Scholar]
- 11. Gokulshankar S., Babu K., Valli S., Ranjitsingh A. J., Ranjith M. S. Cowitch seed agar medium—a simple new medium for identification and melanin production of Cryptococcus neoformans. Mycoses, in press [DOI] [PubMed]
- 12. Gokulshankar S., Ranganathan S., Ranjith M. S., Ranjithsingh A. J. 2004. Prevalence, serotypes and mating patterns of Cryptococcus neoformans in the pellets of different avifauna in Madras, India. Mycoses 47:310–314 [DOI] [PubMed] [Google Scholar]
- 13. Gueho E., Improvisi L., Christen R., de Hoog G. S. 1993. Phylogenetic relationships of Cryptococcus neoformans and some related basidiomycetous yeasts determined from partial large subunit rRNA sequences. Antonie Van Leeuwenhoek 63:175–189 [DOI] [PubMed] [Google Scholar]
- 14. Iqbal N., et al. 2010. Correlation of genotype and in vitro susceptibilities of Cryptococcus gattii strains from the Pacific Northwest of the United States. J. Clin. Microbiol. 48:539–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Katsu M., et al. 2004. The internal transcribed spacers and 5.8S rRNA gene show extensive diversity among isolates of the Cryptococcus neoformans species complex. FEMS Yeast Res. 4:377–388 [DOI] [PubMed] [Google Scholar]
- 16. Kaufmann C. S., Merz W. G. 1982. Two rapid pigmentation tests for identification of Cryptococcus neoformans. J. Clin. Microbiol. 15:339–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kidd S. E., et al. 2007. Characterization of environmental sources of the human and animal pathogen Cryptococcus gattii in British Columbia, Canada, and the Pacific Northwest of the United States. Appl. Environ. Microbiol. 73:1433–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Klein K. R., et al. 2009. Identification of Cryptococcus gattii by use of L-canavanine glycine bromothymol blue medium and DNA sequencing. J. Clin. Microbiol. 47:3669–3672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kwon-Chung J., Polacheck I., Bennett J. 1982. Improved diagnostic medium for separation of Cryptococcus neoformans var. neoformans (serotypes A and D) and Cryptococcus neoformans var. gattii (serotypes B and C). J. Clin. Microbiol. 15:535–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kwon-Chung K. J., Bennett J. E. 1984. Epidemiologic differences between the two varieties of Cryptococcus neoformans. Am. J. Epidemiol. 120:123–130 [DOI] [PubMed] [Google Scholar]
- 21. Larone D. H. 1995. Medically important fungi: a guide to identification. ASM Press, Washington, DC [Google Scholar]
- 22. Lin X., Heitman J. 2006. The biology of the Cryptococcus neoformans species complex. Annu. Rev. Microbiol. 60:69–105 [DOI] [PubMed] [Google Scholar]
- 23. Ma H., et al. 2009. The fatal fungal outbreak on Vancouver Island is characterized by enhanced intracellular parasitism driven by mitochondrial regulation. Proc. Natl. Acad. Sci. U. S. A. 106:12980–12985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Meyer W., et al. 2009. Consensus multi-locus sequence typing scheme for Cryptococcus neoformans and Cryptococcus gattii. Med. Mycol. 47:561–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Min K. H., Kwon-Chung K. J. 1986. The biochemical basis for the distinction between the two Cryptococcus neoformans varieties with CGB medium. Zentralbl. Bakteriol. Mikrobiol. Hyg. A 261:471–480 [DOI] [PubMed] [Google Scholar]
- 26. Park B. J., et al. 2009. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS 23:525–530 [DOI] [PubMed] [Google Scholar]
- 27. Roberts G. D., Horstmeier C. D., Land G. A., Foxworth J. H. 1978. Rapid urea broth test for yeasts. J. Clin. Microbiol. 7:584–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Trilles L., Fernandez-Torres B., Lazera Mdos S., Wanke B., Guarro J. 2004. In vitro antifungal susceptibility of Cryptococcus gattii. J. Clin. Microbiol. 42:4815–4817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vidotto V., et al. 2004. A new caffeic acid minimal synthetic medium for the rapid identification of Cryptococcus neoformans isolates. Rev. Iberoam. Micol. 21:87–89 [PubMed] [Google Scholar]
- 30. Wang H. S., Zeimis R. T., Roberts G. D. 1977. Evaluation of a caffeic acid-ferric citrate test for rapid identification of Cryptococcus neoformans. J. Clin. Microbiol. 6:445–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zimmer B. L., Roberts G. D. 1979. Rapid selective urease test for presumptive identification of Cryptococcus neoformans. J. Clin. Microbiol. 10:380–381 [DOI] [PMC free article] [PubMed] [Google Scholar]