Abstract
Current methods for identification of yeast from blood cultures may take several days after these microorganisms have been observed by Gram stain smears from positive blood cultures. We explored the use of a matrix-assisted laser desorption ionization (MALDI) Biotyper system in combination with Sepsityper specimen processing and Microflex analysis for improved detection and identification of yeast species directly from positive blood culture specimens demonstrating yeast-like organisms by Gram stain. The limit of detection of yeast species in blood culture medium was determined to be 5.9 × 105 CFU, with intra- and interstrain coefficients of variation of 1.8 to 3.6% and 2.9%, respectively. A total of 42 yeast-containing positive blood culture specimens were processed, and the identification results were compared to those obtained by routinely used phenotypic methods. Specimens with discrepant results between the Biotyper and phenotypic methods were identified on the basis of internal transcribed spacer region sequencing. The MALDI Biotyper system correctly identified the 42 specimens to species level, including 28 (66.7%) Candida albicans, 8 (19.0%) Candida parapsilosis, and 5 (11.9%) Candida tropicalis isolates and 1 (2.4%) Cryptococcus neoformans isolate. The entire procedure, from specimen extraction to final result reporting, can be completed within 1 h. Our data indicated that the Sepsityper specimen processing and Microflex analysis by the MALDI Biotyper system provide a rapid and reliable tool for yeast species identification directly from positive blood culture media.
INTRODUCTION
Rapid detection and identification of yeast species in blood specimens have been advocated in order to shorten the turnaround time for appropriate management of patients suffering from fungemia (25, 31, 33). While the automated, continuously monitoring blood culture system has reduced the delay for detecting the presence of blood-borne fungi, identification of such organisms still requires microscopic examination of the organism morphology after Gram staining as well as further identification after subculturing the organism onto solid medium.
Further identification of yeast-like fungal pathogens to species level is clinically important since intrinsic antibiotic resistance profiles vary among different yeast specimens (20). Current phenotypic methods for yeast identification, including colony morphology, germ tube test, urease activity, and the API 20C AUX strip (bioMeriuex, Durham, NC), take an additional 48 to 72 h to complete after the yeast-like fungal pathogens are observed in the blood culture media (33). Various molecular techniques, including real-time PCR, fluorescent in situ hybridization, and pyrosequencing, have been developed to speed the identification of blood-borne fungi (14, 23, 25), but they have not been implemented as routine techniques in the clinical microbiology laboratory.
Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) has emerged as a rapid and powerful tool for microbial species identification (1, 3, 6, 12, 24). One such system, the MALDI Biotyper system (Bruker Daltonics Inc., Billerica, MA), has been successfully used to rapidly identify yeast-like fungal pathogens after they grow on solid medium as pure colonies (2, 5, 17, 28, 32). Although direct identification from blood culture media has been successfully applied to bacterial pathogens (8, 9, 15, 22, 27), published studies are limited and have yielded variable results for direct identification of yeast-like fungal pathogens directly from positive blood culture medium (8, 9, 16).
Development and optimization of a protocol for specimen processing are critical for yeast-like fungal pathogen identification directly from positive blood culture medium specimens. Recently, a blood culture medium processing kit, the MALDI Sepsityper, has become commercially available (Bruker Daltonics). This kit contains all reagents and materials required for processing positive blood culture medium. This kit includes a dedicated lysis solution that disrupts blood cells but not bacteria and yeast cell walls, as well as a wash solution that conditions the sample for subsequent mass spectrometric analysis. In this study, we adapted and optimized it for yeast-containing positive blood culture medium processing and applied the extract for yeast identification by the Microflex instrument in the MALDI Biotyper system.
(This study was presented in part at the 3rd Mass Spectrometry Applications to the Clinical Laboratory Annual Conference & Exhibits [MSACL 2011], San Diego, CA, 5 to 9 February 2011.)
MATERIALS AND METHODS
Clinical specimen collection and phenotypic identification.
Positive blood culture media randomly collected from the Bactec FX blood culture system (Becton Dickinson Diagnostic Instrument Systems, Sparks, MD) which demonstrated yeast-like fungal pathogens by Gram stain in the Clinical Microbiology Laboratory at the Vanderbilt University Medical Center (VUMC) during the entire year of 2009 were included in the study. The research project was approved by the VUMC Institutional Research Board. The positive blood culture contents were subcultured onto 5% sheep blood agar plates, and the plates were incubated in a 35°C atmosphere for 24 to 48 h. Phenotypic identification and differentiation were performed by routine phenotypic methods, including colony morphology, germ tube test, urease activity, and an API 20C AUX strip (bioMérieux) (21, 26).
Specimen processing for MALDI-TOF MS analysis.
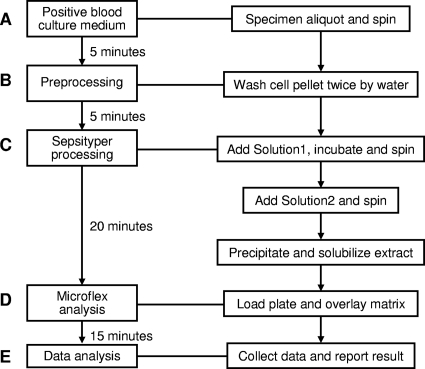
Yeast-containing blood culture medium was processed using a modification of the product of the MALDI Sepsityper kit (270170; Bruker Daltoniks GmbH). Prior to the Sepsityper procedure, two brief washing/centrifugation steps were done to remove red blood cells and proteins from the blood culture broths. In brief, 1 ml of the blood culture fluid was centrifuged at 8,000 × g for 2 min, and the cell pellet was washed twice with 1 ml of water. The pellet was then resuspended in 1 ml water by vortexing, and 200 μl Sepsityper lysis solution (solution 1) was added. After incubation for 2 min at room temperature, the yeast cell pellet was obtained by centrifugation and resuspended into 1 ml Sepsityper washing solution (solution 2) by vortexing. The cell pellet was obtained by centrifugation and resuspended into 300 μl water by vortexing. An additional 900 μl absolute ethanol was added into the suspension, the contents were mixed, and a cell pellet was obtained by centrifugation. The initial supernatant was discarded completely, and the cell pellet was air dried and eluted into 50 μl of 70% formic acid and 50 μl of acetonitrile (Fig. 1). After a second centrifugation, the final supernatant was used for analysis.
Fig. 1.
Flowchart of the MALDI Biotyper system for yeast identification directly from positive blood culture media.
MALDI-TOF mass spectrometry measurement.
One microliter of extract was added to the target plate, and 1 μl of matrix solution (Bruker Daltonics) was then overlaid on the spot. The mixture was allowed to dry in air at room temperature and crystallized (Fig. 1). MALDI-TOF MS spectra measurement was performed on a Microflex LT instrument (Bruker Daltonics) according to the manufacturer's instructions. Spectra were collected using FlexControl (version 3.0) software in the linear positive mode in the mass range of 2,000 to 20,000 m/z (laser frequency, 20 Hz; ion source 1 voltage, 20 kV; ion source 2 voltage, 18.4 kV; lens voltage, 9.1 kV). The standard Biotyper AutoX method was used for automated measurement (12, 19). Before the specimens were processed, the Bruker Daltonics bacterial test standard (BTS; no. 255343) was measured for calibration of the instrument.
MALDI Biotyper identifications.
Automated data analysis and identification of raw spectra were performed by the MALDI Biotyper (version 2.0) software (Bruker Daltonics) using a library of 3,476 entries and default settings (13, 18, 19). The current database covers 110 different yeast species, of which 66 are Candida species. An identification log(score) value ranging from 0 to 3 was given for each specimen. The score indicated the pattern-matching extent, according to the specifications of the MALDI Biotyper system (12, 17, 28). Score values of 0 to 1.699 generally indicated no reliable identification; score values of 1.7 to 1.999 indicated probable genus identification; score values of 2.0 to 2.299 indicated secure genus identification, probable species identification; and score values of 2.300 to 3.000 indicated highly probable species identification.
Limit of detection (LOD) and reproducibility.
A positive blood culture specimen (specimen 2-4), which was vortexed vigorously until all yeast cells separated singly, as seen by Gram stain, was 10-fold serially diluted with a pooled negative blood culture specimen. Each dilution was performed in triplicate for MALDI Biotyper identification and CFU determination. For MALDI Biotyper identification, three scores for each sample were obtained for both intra- and interspecimen reproducibility determination. The numbers of CFU were determined by a quantitative subculture of each diluted sample onto 5% sheep blood agar plates, which were incubated at 35°C for 48 h. Variations in identification log(score) values between three measurement spots and between three preparations were used to determine the intra- and interspecimen reproducibilities, respectively.
28S rRNA gene sequencing.
Specimens with discrepant identification results between the phenotypic and MALDI Biotyper methods were further identified by sequencing analysis. A primer set (28S-Haynes-40F [5′-GCA TAT CAA TAA GCG GAG GA-3′] and 28S-Haynes-654R [5′-GGT CCG TGT TTC AAG ACG-3′]) was used to target the conserved sequences in the V3 region of the 28S rRNA gene (7). A loopful of the purified yeast isolate was put into 1 ml of distilled water and heated at 95°C for 10 min, the suspension was centrifuged, and 1 μl of supernatant was used for PCR amplification. PCR amplification was carried out as previously described (11, 29). Nucleotide sequences were determined bidirectionally and were analyzed with BLAST (Basic Local Alignment Search Tool) on the NCBI website.
RESULTS
Concentrations of yeast cells of five positive blood culture media ranged from 1.2 × 106 to 8.5 × 106 CFU/ml. Direct application of the MALDI Sepsityper kit for specimen processing failed to yield reliable identification (data not shown). However, the two additional preprocessing washing steps added (Fig. 1, step B) prior to using the MALDI Sepsityper kit resulted in satisfactory extraction results. Sepsityper processing resulted in reliable satisfactory identification results for all 42 clinical positive blood culture medium specimens, with identification log(score) values being >1.9 (Tables 1 and 2). The LOD was determined with one positive clinical specimen (identified as Candida parapsilosis), which was serially diluted with pooled negative blood culture medium. The yeast load of this specimen was determined to be 5.9 × 106 CFU/ml by quantitative subculture. The LOD of the MALDI Biotyper system for reliable yeast identification was 5.9 × 105 CFU/ml when it was used directly on positive blood culture medium. The intraspecimen coefficients of variation (CVs) of the identification log(score) values were 1.83 to 3.55%, and the interspecimen CV was 2.91% for the original, undiluted specimen (Table 1).
Table 1.
Limit of detection and reproducibility of MALDI Biotyper system for yeast identification directly from a positive blood culture specimen
| Sample 2-4 dilution | No. of CFU/ml | Identification | Score |
% CV | |||
|---|---|---|---|---|---|---|---|
| Specimen 1 | Specimen 2 | Specimen 3 | Mean ± SD | ||||
| Undiluted | 5.9 × 106 | Reliable identification | 2.230 | 2.112 | 2.211 | 2.184 ± 0.063 | 2.90 |
| 2.234 | 2.245 | 2.170 | 2.216 ± 0.041 | 1.83 | |||
| 2.204 | 2.354 | 2.229 | 2.262 ± 0.080 | 3.55 | |||
| 1:10 | 5.9 × 105 | Reliable identification | 1.985 | 2.087 | 2.013 | 2.028 ± 0.053 | 2.60 |
| 2.160 | 2.057 | 2.079 | 2.099 ± 0.054 | 2.58 | |||
| 1.942 | 1.984 | 2.006 | 1.977 ± 0.033 | 1.64 | |||
| 1:100 | 5.9 × 104 | Not reliable identification | 1.328 | 1.230 | 1.276 | 1.278 ± 0.049 | 3.84 |
| 1.174 | 1.381 | 1.273 | 1.276 ± 0.104 | 8.11 | |||
| 1.382 | 1.361 | 1.370 | 1.371 ± 0.011 | 0.77 | |||
| 1:1,000 | 5.9 × 103 | Not reliable identification | 1.446 | 1.540 | 1.483 | 1.489 ± 0.047 | 3.18 |
| 1.231 | 1.264 | 1.245 | 1.246 ± 0.017 | 1.33 | |||
| 1.212 | 1.363 | 1.288 | 1.288 ± 0.076 | 5.86 | |||
Table 2.
Performance of MALDI Biotyper system for yeast identification on 42 yeast pathogens in blood
| No. of isolates tested | Phenotypic identification | Biotyper identification |
|
|---|---|---|---|
| Positive blood culture | Pure colony | ||
| 28 | C. albicans | C. albicans | C. albicans |
| 8 | C. parapsilosis | C. parapsilosis | C. parapsilosis |
| 4 | C. tropicalis | C. tropicalis | C. tropicalis |
| 1 | C. neoformans | C. neoformans | C. neoformans |
| 1a | C. lusitaniae | C. tropicalis | C. tropicalis |
The isolate was identified to be C. tropicalis by 28S rRNA gene sequencing.
A total of 42 yeast-containing positive blood culture medium specimens were collected during the study period. Current phenotypic methods, including colony morphology, germ tube test, urease activity analysis, and an API 20C AUX strip, identified these isolates to two genera and five species, including Candida albicans (n = 28, 66.7%), C. parapsilosis (n = 8, 19.0%), Candida tropicalis (n = 4, 9.5%), Cryptococcus neoformans (n = 1, 2.4%), and Candida lusitaniae 1 (2.4%). The isolate identified as C. lusitaniae (isolate 43-154) had a poor match, with an identification ratio of 64% by the API 20C AUX strip. The MALDI Biotyper system was able to provide reliable identification to the species level for all 42 specimens (Table 2). The identification results were identical to those of phenotypic methods for all except one specimen, in which C. tropicalis was identified by the MALDI Biotyper system and C. lusitaniae was identified by phenotypic methods. MALDI Biotyper analysis on pure isolates obtained by subsequent subculture gave results identical to those obtained when it was used directly on positive blood culture media (Table 2).
The specimen with discrepant identification results (specimen 43-154) between the conventional methods and the MALDI Biotyper system was further analyzed by 28S rRNA gene sequencing. The nucleotide sequence of a partial 603-bp sequence of the 28S rRNA gene (GenBank accession number HQ214057) was identical to that of a clinical C. tropicalis isolate identified and reported previously (11).
The entire procedure, from specimen extraction to final result reporting, was completed within 1 h.
DISCUSSION
In this pilot study, we explore the use of the MALDI Biotyper system, which combines a blood culture medium specimen processing kit (MALDI Sepsityper), a MALDI-TOF MS analyzer (Microflex), and dedicated software (MALDI Biotyper, version 2.0), for rapid identification of yeast-like pathogens directly from positive blood culture medium. With two additional preprocessing washing steps, the system reached a limit of detection of 105 CFU/ml with satisfactory intra- and interspecimen reproducibilities. The entire procedure, from specimen extraction to final result reporting, can be completed within 1 h, providing another reliable tool for rapid yeast identification directly from positive blood culture media.
Yeast-like fungal pathogens are among the commonest etiological agents of invasive fungal infections and are also seen in nosocomial bloodstream infections. Invasive fungal infections and fungal nosocomial bloodstream infections are life-threatening, with overall and specific mortality rates of 60 and 49%, respectively (20). The selection of an effective treatment for candidemia depends on the infecting Candida species. There are more than 20 Candida species, each with different antifungal susceptibility profiles (20, 30). At present, species determination requires from 2 to 5 days after collection of blood specimens for culture from patients. MALDI-TOF MS-based identification has the potential to shorten the time to results, thereby providing a substantial reduction in the delay before initiation of or adjustment to an effective therapy to improve therapeutic efficacy, minimize adverse effects, reduce costs, and lessen the risk of resistance development (6, 24).
Previous published efforts on identification of yeast specimens by the MALDI-TOF MS-based procedures directly from yeast-containing positive blood culture specimens have yielded different findings (8, 9, 16). In this situation, the yeast-like fungi grew in liquid medium of complex composition due to the presence of both medium proteins and blood cells. In particular, the hemoglobin, proteins and peptides derived from leukocytes, and serum proteins must be discarded, as those yield strong signals in the mass spectra which hamper the interpretation of specific peaks. Development of a standardized and efficient specimen processing protocol to remove interference materials and yield clear microbial protein components is critical (10). In this study, we adapted and optimized a commercial kit, MALDI Sepsityper, for positive blood culture medium specimen processing. In our experience, the Sepsityper kit has the following characteristics: (i) identification of microorganisms directly from positive blood culture bottles is easy and rapid (less than 30 min); (ii) the sample preparation protocol is simple and uses only 1 ml sample material, few centrifugation steps, and only one tube for each analysis; (iii) identification of yeasts is reliable by the MALDI Biotyper system; and (iv) all reagents and consumables required for processing blood culture fluid are supplied in the kit ready to use. We added two washing steps to the specimen procedure prior to the MALDI Sepsityper analysis step to remove red blood cells and proteins. This modification made the MALDI Sepsityper an efficient tool for processing positive blood culture media containing yeast-like pathogens.
In contrast to published protocols, which employed differential centrifugation for the isolation of microorganisms in a positive blood culture and usually failed in identification of yeast, this method destroys the human cells (white and red blood cells) by lysis. The lysis solution used in this study is designed to lyse only human blood cells and not microorganisms. Therefore, in the high-speed centrifugation steps, only microorganisms, which are not affected by the lysis step because of their robust cell walls, are precipitated. Thereby, interference of white blood cells, which are sedimented similarly to yeast cells and therefore cannot be separated from them by differential centrifugation, is prevented. We also applied two additional washing steps prior to the Sepsityper procedure, which resulted in more reproducible and satisfactory results on the basis of the identification of organisms in a total of 42 clinical specimens.
The sensitivity of MALDI-TOF MS to identify organisms in blood clearly depends on the inoculum (9, 10). Spiking experiments with Staphylococcus aureus and Escherichia coli indicated that organisms present at 107 to 108 CFU/ml were correctly identified, whereas 106 CFU/ml yielded signals that were indistinguishable from the background for the negative controls, indicating that it was necessary to deposit at least 104 to 105 CFU on the MALDI target plate in order to obtain an identifying spectrum (4). Additionally, the inoculum must contain sufficient microorganisms to overcome the background peaks derived from blood during measurement. In our experiment, we reached a limit of detection of 105 CFU/ml on a clinical specimen with C. parapsilosis, which is more sensitive than that previously reported. This may be attributed to the adaptation and employment of the MALDI Sepsityper kit for processing of 1 ml of a positive blood specimen. The Sepsityper kit itself is much more effective at removing the interfering human cells (red and white blood cells) from the complex mixture and enriches microorganisms in a greater ratio than other methods. In addition, both intra- and interstrain identification log(score) value CVs were small, suggesting that the Sepsityper kit for sample preparation possessed satisfactory reproducibility results similar to those achieved with bacterial and yeast pathogen identification methods (18, 28).
In summary, the direct MS fingerprinting method with specimen processing by the modified MALDI Sepsityper kit offers a favorable combination of easy sample handling, accuracy, reproducibility, short turnaround time, and modest reagent costs. While our pilot study results are promising, more yeast-containing positive blood culture medium specimens covering a variety of genera and species need to be tested to fully evaluate this potentially revolutionary microbial identification tool.
Acknowledgements
We thank Susan Sefers, Joni Williams, Bunny Ambrose, Jasper Benton, Rusty Bowden, Donna Brewer, Beth Brown, Sonia Cerruti, Emily Cauanaugh, Kathy Ewing, Pam Foster, Rene Gerald, Tonia Goodman, Mary Hedges, Monna Jedd, Lindsay Johnson, Kim Klocek, Sue May, Amy Montgomery, Kim Neville, Carla Nicholson, and Jennifer Steinhauer for helping collect clinical specimens.
This study was partially supported by Vanderbilt CTSA grant UL1 RR024975 from NCRR/NIH and an instrument lease agreement between Vanderbilt University Medical Center and Bruker Daltonics.
Footnotes
Published ahead of print on 4 May 2011.
REFERENCES
- 1. Alispahic M., et al. 2010. Species-specific identification and differentiation of Arcobacter, Helicobacter and Campylobacter by full-spectral matrix-associated laser desorption/ionization time of flight mass spectrometry analysis. J. Med. Microbiol. 59:295–301 [DOI] [PubMed] [Google Scholar]
- 2. Bader O., et al. 14 October 2010. Improved clinical laboratory identification of human pathogenic yeasts by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Microbiol. Infect. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 3. Cherkaoui A., et al. 2010. Comparison of two matrix-assisted laser desorption ionization-time of flight mass spectrometry methods with conventional phenotypic identification for routine identification of bacteria to the species level. J. Clin. Microbiol. 48:1169–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Christner M., et al. 2010. Rapid identification of bacteria from positive blood culture bottles by use of matrix-assisted laser desorption-ionization time of flight mass spectrometry fingerprinting. J. Clin. Microbiol. 48:1584–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dhiman N., Hall L., Wohlfiel S. L., Buckwalter S. P., Wengenack N. L. 2011. Performance and cost analysis of matrix-assisted laser desorption ionization-time of flight mass spectrometry for routine identification of yeast. J. Clin. Microbiol. 49:1614–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Drancourt M. 2010. Detection of microorganisms in blood specimens using matrix-assisted laser desorption ionization time-of-flight mass spectrometry: a review. Clin. Microbiol. Infect. 16:1620–1625 [DOI] [PubMed] [Google Scholar]
- 7. Fell J. W. 1993. Rapid identification of yeast species using three primers in a polymerase chain reaction. Mol. Mar. Biol. Biotechnol. 2:174–180 [PubMed] [Google Scholar]
- 8. Ferreira L., et al. 12 August 2010. Rapid method for direct identification of bacteria in urine and blood culture samples by matrix-assisted laser desorption ionization time-of-flight mass spectrometry: intact cell vs. extraction method. Clin. Microbiol. Infect. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 9. Ferroni A., et al. 2010. Real-time identification of bacteria and Candida species in positive blood culture broths by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 48:1542–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Giebel R., et al. 2010. Microbial fingerprinting using matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) applications and challenges. Adv. Appl. Microbiol. 71:149–184 [DOI] [PubMed] [Google Scholar]
- 11. Hall L., Wohlfiel S., Roberts G. D. 2003. Experience with the MicroSeq D2 large-subunit ribosomal DNA sequencing kit for identification of commonly encountered, clinically important yeast species. J. Clin. Microbiol. 41:5099–5102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. He Y., Li H., Lu X., Stratton C. W., Tang Y. W. 2010. Mass spectrometry Biotyper system identifies enteric bacterial pathogens directly from colonies grown on selective stool culture media. J. Clin. Microbiol. 48:3888–3892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ilina E. N., et al. 2009. Direct bacterial profiling by matrix-assisted laser desorption-ionization time-of-flight mass spectrometry for identification of pathogenic Neisseria. J. Mol. Diagn. 11:75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jordan J. A., Jones-Laughner J., Durso M. B. 2009. Utility of pyrosequencing in identifying bacteria directly from positive blood culture bottles. J. Clin. Microbiol. 47:368–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. La Scola B., Raoult D. 2009. Direct identification of bacteria in positive blood culture bottles by matrix-assisted laser desorption ionisation time-of-flight mass spectrometry. PLoS One 4:e8041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Marinach-Patrice C., et al. 2010. Rapid species diagnosis for invasive candidiasis using mass spectrometry. PLoS One 5:e8862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Marklein G., et al. 2009. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for fast and reliable identification of clinical yeast isolates. J. Clin. Microbiol. 47:2912–2917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mellmann A., et al. 2009. High interlaboratory reproducibility of matrix-assisted laser desorption ionization-time of flight mass spectrometry-based species identification of nonfermenting bacteria. J. Clin. Microbiol. 47:3732–3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mellmann A., et al. 2008. Evaluation of matrix-assisted laser desorption ionization-time-of-flight mass spectrometry in comparison to 16S rRNA gene sequencing for species identification of nonfermenting bacteria. J. Clin. Microbiol. 46:1946–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pappas P. G., et al. 2009. Clinical practice guidelines for the management of candidiasis: 2009 update by the Infectious Diseases Society of America. Clin. Infect. Dis. 48:503–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pincus D. H., et al. 1999. Rapid identification of Candida dubliniensis with commercial yeast identification systems. J. Clin. Microbiol. 37:3533–3539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Prod'hom G., Bizzini A., Durussel C., Bille J., Greub G. 2010. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for direct bacterial identification from positive blood culture pellets. J. Clin. Microbiol. 48:1481–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Selvarangan R., Bui U., Limaye A. P., Cookson B. T. 2003. Rapid identification of commonly encountered Candida species directly from blood culture bottles. J. Clin. Microbiol. 41:5660–5664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Seng P., et al. 2009. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Infect. Dis. 49:543–551 [DOI] [PubMed] [Google Scholar]
- 25. Shepard J. R., et al. 2008. Multicenter evaluation of the Candida albicans/Candida glabrata peptide nucleic acid fluorescent in situ hybridization method for simultaneous dual-color identification of C. albicans and C. glabrata directly from blood culture bottles. J. Clin. Microbiol. 46:50–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Smith M. B., Dunklee D., Vu H., Woods G. L. 1999. Comparative performance of the RapID Yeast Plus System and the API 20C AUX clinical yeast system. J. Clin. Microbiol. 37:2697–2698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stevenson L. G., Drake S. K., Murray P. R. 2010. Rapid identification of bacteria in positive blood culture broths by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 48:444–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stevenson L. G., Drake S. K., Shea Y. R., Zelazny A. M., Murray P. R. 2010. Evaluation of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of clinically important yeast species. J. Clin. Microbiol. 48:3482–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tang Y. W., et al. 1998. Comparison of phenotypic and genotypic techniques for identification of unusual aerobic pathogenic gram-negative bacilli. J. Clin. Microbiol. 36:3674–3679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tortorano A. M., et al. 2004. Epidemiology of candidaemia in Europe: results of 28-month European Confederation of Medical Mycology (ECMM) hospital-based surveillance study. Eur. J. Clin. Microbiol. Infect. Dis. 23:317–322 [DOI] [PubMed] [Google Scholar]
- 31. Trenholme G. M., et al. 1989. Clinical impact of rapid identification and susceptibility testing of bacterial blood culture isolates. J. Clin. Microbiol. 27:1342–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. van Veen S. Q., Claas E. C., Kuijper E. J. 2010. High-throughput identification of bacteria and yeast by matrix-assisted laser desorption ionization-time of flight mass spectrometry in conventional medical microbiology laboratories. J. Clin. Microbiol. 48:900–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Weinstein M. P., et al. 1997. The clinical significance of positive blood cultures in the 1990s: a prospective comprehensive evaluation of the microbiology, epidemiology, and outcome of bacteremia and fungemia in adults. Clin. Infect. Dis. 24:584–602 [DOI] [PubMed] [Google Scholar]