Abstract
This study aimed to determine the genetic diversity of clinical multidrug-resistant Pseudomonas aeruginosa. We used pulsed-field gel electrophoresis and multilocus sequence typing to analyze 187 strains isolated in different French hospitals. To illustrate the diversity of resistance mechanisms to antibiotics in a given clone, we identified β-lactamases with an extended spectrum by using phenotypic and genotypic methods. Typing results showed that the majority of our multidrug-resistant isolates belong to a few clonal types (ST235, ST111, and ST175) that are already spreading worldwide. These successful international clones sporadically produced extended-spectrum β-lactamase-encoding genes but mostly became extensively resistant to β-lactams after derepression of intrinsic resistance mechanisms (i.e., AmpC cephalosporinase). Our results indicate that cross-transmission plays a major role in the spread of multidrug-resistant P. aeruginosa in hospital settings.
INTRODUCTION
Pseudomonas aeruginosa is the third most important nosocomial pathogen in French hospitals, responsible for 10% of hospital-acquired infections (13). Additionally, the growing threat of antimicrobial resistance in P. aeruginosa is driven by its extraordinary capacity for developing resistance to almost any antibiotic by selection of mutations in chromosomal genes in conjunction with spread of horizontally acquired resistance (21, 30). Multidrug resistance in P. aeruginosa makes treatment of infections both difficult and expensive and can increase morbidity and mortality (16).
There appears to be a consensus that the P. aeruginosa population structure is nonclonal epidemic, that clinical isolates are indistinguishable from environmental isolates, and that there are no specific clones with a specific habitat or disease (25). However, some important controversial issues remain. First, since the 1980s, several studies have reported the emergence, spread, and persistence of multidrug-resistant clones in hospitals, evidencing a lack of genetic diversity (25). Second, cystic fibrosis (CF) clones have been reported worldwide, suggestive of an emergence of specific clones spreading within the CF population (25, 29).
The objective of this study was to assess the genetic diversity, by using pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST), of a large panel of multidrug-resistant (MDR) P. aeruginosa isolates from patients hospitalized in different French hospitals.
MATERIALS AND METHODS
Settings.
Five university hospitals (Besançon, Dijon, Nancy, Reims, and Strasbourg) of five administrative regions in eastern France took part in this survey over a 32-month period (from October 2007 to May 2010). These regions account for 8.5 million inhabitants, corresponding to 14% of the French population.
Bacterial strains.
All P. aeruginosa clinical and screening (if performed) isolates, except CF isolates, were collected and tested for antimicrobial resistance. The susceptibilities to ticarcillin, tazobactam/piperacillin, ceftazidime, cefepime, aztreonam, meropenem, imipenem, tobramycin, amikacin, gentamicin, and ciprofloxacin were assessed in each laboratory according to their routine testing methods. Nonduplicate (the first isolate of each patient) MDR P. aeruginosa (susceptible to ≤1 tested antibiotic) isolates were sent to the central laboratory, where the bacterial identification (using the biochemical tests of ID32 GN [bioMérieux, Marcy l'Étoile, France]) and resistance profile were confirmed. The susceptibilities to antibiotics were determined by the Kirby-Bauer disk method on Mueller-Hinton agar (Bio-Rad, Ivry sur Seine, France), as recommended by the Antibiogram Committee of the French Society for Microbiology (4). All the isolates from the collection were susceptible to colistin. A Sirscan automated image analyzer (I2A, Perols, France) was used to precisely measure inhibition diameters and to compile resistance data (23).
Genotyping. (i) PFGE.
The macrorestriction profile (determined with DraI) of total DNA from each isolate was determined by PFGE (CHEF DRIII; Bio-Rad), according to a method previously developed in our laboratory (2). We used the GelCompar software (Applied Maths, Kortrijk, Belgium) to establish a DNA similarity matrix. A dendrogram was constructed using the unweighted pair group method of the arithmetic average clustering method with the Dice coefficient. We ensured that the gels were comparable by including Staphylococcus aureus NCTC 8325 as a reference. PFGE results were interpreted according to international recommendations (28).
(ii) MLST.
MLST was performed according to the protocol by Curran et al. (5), as modified by van Mansfeld et al. (29). Nucleotide sequences were determined for internal fragments of the acsA, aroE, guaA, mutL, nuoD, ppsA, and trpE genes on both strands and compared to information on the P. aeruginosa MLST website (http://pubmlst.org/paeruginosa/) for assignment of allelic number and sequence type (ST). The Mega algorithm (http://www.megasoftware.net) was used to study the genetic diversity of the different populations of P. aeruginosa strains.
Determination of enzymatic resistance mechanisms to β-lactams.
All the isolates from the collection were screened for the production of β-lactamases with an extended spectrum (Ambler class A extended-spectrum β-lactamases [ESBLs], class D extended-spectrum oxacillinases [ES-OXAs], and class B metallo-β-lactamases [MBLs]) by using a double-disk synergy test (DDST). ESBLs and ES-OXAs were detected with a DDST using cefepime and ceftazidime as substrate and clavulanate and imipenem as inhibitors (12). MBLs were detected with imipenem- and EDTA-containing disks (19). A PCR-based approach was used to identify the genes encoding ESBLs, ES-OXAs, and MBLs in all DDST-positive isolates (n = 31). Whole-cell DNA suitable for PCR experiments was extracted and purified with the QiAmp DNA minikit (Qiagen, Courtabœuf, France). Specific primers were used to amplify the β-lactamase-encoding genes blaSHV, blaPER, blaVEB, blaGES, blaBEL, blaOXA-I group, blaOXA-II group, blaOXA-III group, blaOXA-9, and blaOXA-18 from the DNA (13). Positive PCR controls with appropriate strains were systematically run in parallel. Purified amplicons were sequenced on both strands using the dye terminator chemistry on an Applied Biosystems 3130 genetic analyzer (Applied Biosystems, Foster City, CA). The nucleotide sequences were compared and aligned with reference sequences by using the NCBI BLAST program.
RESULTS
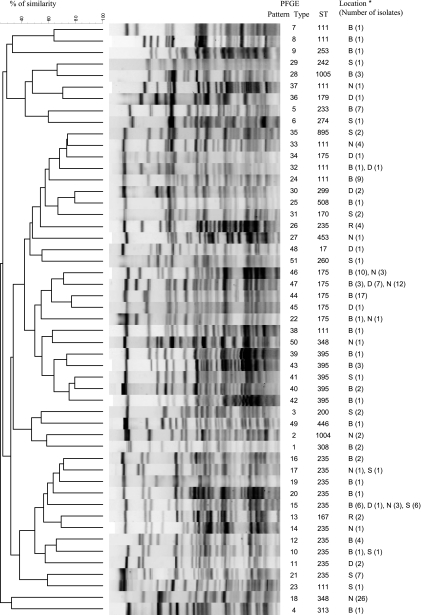
During the study period, 187 nonduplicate patients were colonized or infected with an MDR P. aeruginosa isolate (82 at Besançon, 56 at Nancy, 26 at Strasbourg, 17 at Dijon, and 6 at Reims). Ninety-one (48.7%) of these patients were hospitalized in intensive care units, whereas the others were distributed in medical units (n = 35; 18.7%), surgical units (n = 27; 14.4%), hematology units (n = 16; 8.6%), and chronic or long-term care units (n = 18; 9.6%). Among the 187 nonduplicate MDR P. aeruginosa isolates, 135 were nonsusceptible to all the tested antibiotics (Table 1). The 187 nonduplicate isolates yielded 51 unique PFGE patterns (designated pattern types PT1 to PT51) (Fig. 1 and Table 1). Most of the PTs were recovered from a single hospital (44/51), whereas 5 were recovered in two hospitals, 1 was recovered from three, and 1 was recovered in four hospitals.
Table 1.
MLST and PFGE types of 187 multidrug-resistant P. aeruginosa isolates from eastern France
| MLST | PT(s) (no. of isolates) | Antibiotica resistance profile (no. of isolates) | Total no. of isolates | Locationb (no. of isolates) | Yr of isolation |
|---|---|---|---|---|---|
| ST17 | PT48 (1) | TZP-ATM-CAZ-IPM-MEM-CIP-TOB-GEN | 1 | D (1) | 2007 |
| ST111 | PT32 (2) | ATM-CAZ-IPM-MEM-CIP-TM-GEN-AMK (1) | 20 | B (13), D (1), N (5), S (1) | 2007, 2008, 2009 |
| TZP-ATM-CAZ-IPM-MEM-CIP-TOB-GEN-AMK (1) | |||||
| PT23 (1) | TZP-CAZ-IPM-MEM-CIP-TOB-GEN-AMK (1) | ||||
| PT7 (1) | TZP-ATM-IPM-MEM-CIP-TOB-GEN-AMK (1) | ||||
| PT8 (1), PT24 (9), PT33 (4), PT37 (1), PT38 (1) | TZP-ATM-CAZ-IPM-MEM-CIP-TOB-GEN-AMK (16) | ||||
| ST167 | PT13 (2) | TZP-ATM-CAZ-IPM-MEM-CIP-TOB-GEN-AMK | 2 | R (2) | 2007 |
| ST170 | PT31 (2) | TZP-ATM-CAZ-IPM-MEM-CIP-TOB-GEN-AMK | 2 | S (2) | 2007, 2008 |
| ST175 | PT44 (17) | TZP-CAZ-IPM-MEM-CIP-TOB-GEN-AMK (3) | 56 | B (31), D (9), N (13), S (3) | 2007, 2008, 2009 |
| TZP-ATM-CAZ-IPM-MEM-CIP-TOB-GEN-AMK (14) | |||||
| PT47 (22) | TZP-ATM-CAZ-IPM-MEM-TOB-GEN-AMK (1) | ||||
| TZP-ATM-CAZ-IPM-MEM-CIP-TOB-GEN (3) | |||||
| TZP-ATM-CAZ-IPM-MEM-CIP-TOB-GEN-AMK (18) | |||||
| PT22 (2) | TZP-ATM-CAZ-IPM-MEM-CIP-TOB-GEN-AMK | ||||
| PT34 (1) | TZP-ATM-CAZ-IPM-MEM-CIP-TOB-GEN-AMK | ||||
| PT45 (1) | TZP-ATM-CAZ-IPM-MEM-Cip-TOB-GEN-AMK | ||||
| PT46 (13) | TZP-ATM-CAZ-IPM-MEM-CIP-TOB-GEN-AMK | ||||
| ST179 | PT36 (1) | TZP-ATM-CAZ-IPM-MEM-CIP-TOB-GEN-AMK | 1 | D (1) | 2007 |
| ST200 | PT3 (2) | TZP-ATM-CAZ-IPM-CIP-TOB-GEN-AMK (1) | 2 | S (2) | 2007 |
| TZP-ATM-CAZ-IPM-MEM-CIP-TOB-GEN-AMK (1) | |||||
| ST233 | PT5 (7) | TZP-ATM-CAZ-IPM-MEM-CIP-GEN-AMK (3) | 7 | B (7) | 2008 |
| TZP-ATM-CAZ-IPM-MEM-CIP-TOB-AMK (1) | |||||
| TZP-ATM-CAZ-IPM-MEM-CIP-TOB-GEN (1) | |||||
| TZP-ATM-CAZ-IPM-MEM-CIP-TOB-GEN-AMK (2) | |||||
| ST235 | PT12 (5) | ATM-CAZ-IPM-MEM-CIP-TOB-GEN-AMK (4) | 42 | B (15), D (4), N (4), R (4), S (15) | 2007, 2008, 2009 |
| TZP-ATM-CAZ-MEM-CIP-TOB-GEN-AMK (1) | |||||
| PT15 (15) | TZP-CAZ-IPM-MEM-CIP-TOB-GEN-AMK (1) | ||||
| TZP-ATM-CAZ-IPM-CIP-TOB-GEN-AMK (2) | |||||
| TZP-ATM-CAZ-IPM-MEM-CIP-GEN-AMK (1) | |||||
| TZP-ATM-CAZ-IPM-MEM-CIP-TOB-AMK (1) | |||||
| TZP-ATM-CAZ-IPM-MEM-CIP-TOB-GEN-AMK (10) | |||||
| PT21 (7) | TZP-CAZ-IPM-MEM-CIP-TOB-GEN-AMK (1) | ||||
| TZP-ATM-CAZ-MEM-CIP-TOB-GEN-AMK (1) | |||||
| TZP-ATM-CAZ-IPM-MEM-CIP-GEN-AMK (1) | |||||
| TZP-ATM-CAZ-IPM-MEM-CIP-TOB-GEN-AMK (4) | |||||
| PT10 (2) | TZP-ATM-CAZ-IPM-MEM-CIP-GEN-AMK (1) | ||||
| TZP-ATM-CAZ-IPM-MEM-CIP-TOB-GEN-AMK (1) | |||||
| PT16 (2) | TZP-ATM-CAZ-IPM-MEM-CIP-GEN-AMK (1) | ||||
| TZP-ATM-CAZ-IPM-MEM-CIP-TOB-AMK (1) | |||||
| PT17 (2) | TZP-ATM-CAZ-IPM-MEM-CIP-TOB-AMK (1) | ||||
| TZP-ATM-CAZ-IPM-MEM-CIP-TOB-GEN-AMK (1) | |||||
| PT26 (4) | TZP-ATM-CAZ-IPM-MEM-CIP-TOB-AMK (1) | ||||
| TZP-ATM-CAZ-IPM-MEM-CIP-TOB-GEN-AMK (3) | |||||
| PT11 (2), PT14 (1), PT19 (1), PT20 (1) | TZP-ATM-CAZ-IPM-MEM-CIP-TOB-GEN-AMK (5) | ||||
| ST242 | PT29 (1) | TZP-ATM-CAZ-IPM-MEM-CIP-TOB-AMK | 1 | S (1) | 2008 |
| ST253 | PT9 (1) | TZP-ATM-CAZ-IPM-MEM-TOB-GEN-AMK | 1 | B (1) | 2009 |
| ST260 | PT51 (1) | TZP-ATM-CAZ-IPM-CIP-TOB-GEN-AMK | 1 | S (1) | 2007 |
| ST274 | PT6 (1) | TZP-ATM-CAZ-IPM-MEM-CIP-TOB-GEN-AMK | 1 | S (1) | 2008 |
| ST299 | PT30 (2) | TZP-ATM-CAZ-IPM-MEM-CIP-TOB-AMK (1) | 2 | D (2) | 2007 |
| TZP-ATM-CAZ-IPM-MEM-CIP-TOB-GEN-AMK (1) | |||||
| ST308 | PT1 (2) | TZP-ATM-CAZ-IPM-MEM-TOB-GEN-AMK (1) | 2 | B (2) | 2008, 2009 |
| TZP-ATM-CAZ-IPM-MEM-CIP-TOB-GEN-AMK (1) | |||||
| ST313 | PT4 (1) | TZP-ATM-CAZ-IPM-MEM-CIP-TOB-AMK | 1 | B (1) | 2008 |
| ST348 | PT18 (26) | TZP-ATM-CAZ-IPM-MEM-CIP-TOB-GEN (1) | 27 | N (27) | 2007, 2008 |
| TZP-ATM-CAZ-IPM-MEM-CIP-TOB-GEN-AMK (25) | |||||
| PT50 (1) | TZP-ATM-CAZ-IPM-MEM-CIP-TOB-GEN (1) | ||||
| ST395 | PT39 (2) | TZP-ATM-CAZ-IPM-MEM-CIP-TOB-AMK (1) | 8 | B (7), S (1) | 2007, 2008, 2009 |
| TZP-ATM-CAZ-IPM-MEM-CIP-TOB-GEN-AMK (1) | |||||
| PT40 (1) | TZP-ATM-CAZ-IPM-MEM-CIP-TOB-GEN-AMK | ||||
| PT41 (1) | TZP-ATM-CAZ-IPM-MEM-CIP-TOB-GEN-AMK | ||||
| PT42 (1) | TZP-ATM-CAZ-IPM-MEM-CIP-TOB-GEN-AMK | ||||
| PT43 (3) | TZP-ATM-CAZ-IPM-MEM-TOB-GEN-AMK (1) | ||||
| TZP-ATM-CAZ-IPM-MEM-CIP-TOB-AMK (1) | |||||
| TZP-ATM-CAZ-IPM-MEM-CIP-TOB-GEN (1) | |||||
| ST446 | PT49 (1) | TZP-ATM-CAZ-IPM-MEM-CIP-TOB-GEN-AMK | 1 | B (1) | 2008 |
| ST453 | PT27 (1) | TZP-ATM-CAZ-IPM-MEM-CIP-TOB-GEN-AMK | 1 | N (1) | 2008 |
| ST508 | PT25 (1) | TZP-ATM-CAZ-IPM-MEM-CIP-TOB-AMK | 1 | B (1) | 2007 |
| ST895 | PT35 (2) | TZP-ATM-CAZ-IPM-MEM-CIP-TOB-AMK (1) | 2 | S (2) | 2008 |
| TZP-ATM-CAZ-IPM-MEM-CIP-TOB-GEN-AMK (1) | |||||
| ST1004 | PT2 (2) | TZP-ATM-CAZ-IPM-MEM-CIP-TOB-GEN-AMK | 2 | N (2) | 2007, 2008 |
| ST1005 | PT28 (3) | TZP-ATM-Caz-IPM-MEM-CIP-GEN-AMK (1) | 3 | B (3) | 2008, 2009 |
| TZP-ATM-CAZ-IPM-MEM-CIP-TOB-GEN-AMK (2) |
Antibiotics tested: ticarcillin (TIC), tazobactam/piperacillin (TZP), ceftazidime (CAZ), cefepime (FEP), aztreonam (ATM), meropenem (MEM), imipenem (IPM), tobramycin (TOB), amikacin (AMK), gentamicin (GEN), and ciprofloxacin (CIP). All isolates were nonsusceptible to TIC and FEP.
B, Besancon University Hospital; D, Dijon University Hospital; N, Nancy University Hospital; S, Strasbourg University Hospital; R, Reims University Hospital.
Fig. 1.
Dendrogram of the percent similarity between DraI-digested genomic DNAs from multidrug-resistant P. aeruginosa isolates. B, Besancon University Hospital; D, Dijon University Hospital; N, Nancy University Hospital; S, Strasbourg University Hospital; R, Reims University Hospital.
Considering that PFGE typing is more discriminatory than MLST, we assumed that all the isolates with a given PT shared the same ST. Consequently, MLST was performed on 1 randomly selected isolate of each PT. The 51 PTs clustered into 24 STs (Table 1). Of these, 2 STs were not previously submitted to the P. aeruginosa MLST database (ST1004 and ST1005). ST235, ST111, ST175, and ST395 were the most frequent, being detected in 11, 8, 6, and 5 PTs, respectively, and by deduction in 42, 20, 56, and 8 isolates. ST348 was found in 2 PTs, and the remaining 20 STs were found only once (Table 1). ST235 was present at the five participating hospitals, whereas ST111 and ST175 were recovered in four hospitals.
Finally, the 24 STs could be clustered by analysis with the eBURST program into 23 different clonal complexes (CCs; ST308 and ST1004 belonging to CC308).
One isolate from PT16 (ST235) produced the ESBL PER-1. Four and 26 isolates produced the ES-OXAs OXA-28 and OXA-19, respectively. None of the isolates from our series produced an MBL. The remaining 156 isolates were considered AmpC overproducers. The four OXA-28-producing isolates were recovered in the same hospital and clustered in the same PT (PT12, ST235), which was shared by 42 isolates. Similarly, the 26 OXA-19-producing isolates were gathered in the same PT (PT18, ST348) shared by 27 isolates.
DISCUSSION
Several molecular typing schemes have been proposed for P. aeruginosa to determine the relatedness of nosocomial pathogens. PFGE, which is recognized as the most discriminatory method for molecular typing, has been a clinically valuable tool for evaluating short-term epidemiological P. aeruginosa infections (14). Due to the dynamic genome of P. aeruginosa, there has been debate regarding the usefulness of PFGE for evaluating the long-term global epidemiology of this bacterium. The observed extensive genomic and phenotypic diversities between P. aeruginosa strains may originate from the frequent recombinations and transfers of mobile genetic elements in this species. Consequently, there was a need for other less ambiguous methods, such as MLST, which is more appropriate for exploring long-term evolutionary relationships in P. aeruginosa populations (5). Besides, MLST is now documented as the current standard for investigating the population structure of a bacterial species (22).
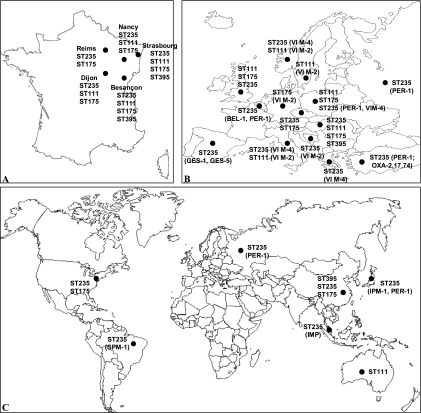
The present study was designed to determine whether particular epidemic clones were associated with multidrug resistance among hospital P. aeruginosa isolates. We used a two-step typing scheme, with PFGE typing of all isolates and MLST for 1 randomly selected isolate of each pulsotype. In contrast with other recent studies focusing mostly on particular resistance mechanisms (i.e., ESBLs or MBLs) (8, 10, 11, 15, 17, 27), we attempted to determine the genetic diversity of all MDR P. aeruginosa isolates recovered during a long period (32 months) in a large population (all the university hospitals of the northeast part of France). Our results show that the majority of multidrug-resistant isolates belong to a few clonal types: ST235, ST111, and ST175. ST235 was the founder ST of the clonal complex CC235 (previously described as CC11/BG11). CC235 has been long known as responsible for outbreaks throughout Europe, Asia, and South America (8, 11, 15, 17, 18, 20, 27) while armed with various ESBLs (such as PER-1, GES-1, and BEL-1) as well as MBLs (such as VIM-1, VIM-4, and IMP-1) (Fig. 2). Interestingly, of the 42 ST235 isolates of our series, 1 produced the ESBL PER-1, 4 produced the ES-OXA OXA-28, and none produced MBL (26). ST111 has been also, to a lesser extent, reported as an MBL-producing epidemic lineage (6), as has ST175 (7) (Fig. 2). We observed differences in genomic variabilities of the STs. The internationally distributed STs (111, 175, 235, and 395) were represented by diverse PTs in our series (8, 6, 11, and 6, respectively), indicating that these clones had spread for several years in our hospitals. On the other hand, ST348 with 27 isolates had only two PTs, indicating a localized outbreak over a limited period of time. This outbreak has been described elsewhere (3). It is noteworthy that all these international clones as well as ST227 (the colistin-only-susceptible Brazilian clone [9]), including most of the MDR P. aeruginosa strains, have not been recovered in large CF P. aeruginosa strain collections (29, 31). Well-characterized CF epidemic strains (e.g., Liverpool-ST146 or Dutch-ST406) are also absent from the collections of non-CF MDR P. aeruginosa (29, 31).
Fig. 2.
Distribution of epidemic lineages of P. aeruginosa in eastern France (A) (this study), Europe (B), and worldwide (C). For maps B and C, the β-lactamase content is specified in brackets when available, according to references 11, 14, 16, 18 to 23, 25, 26, and 31 and the MLST database (http://pubmlst.org/paeruginosa).
As genetic capitalism predicts (1), the most successful clones are also more likely to acquire MDR determinants. Subsequently, they are selected more easily under antibiotic pressure, allowing their spread in hospitals. Indeed, this study and recent reports have demonstrated the presence of widespread MDR P. aeruginosa clones in hospitals. These clones are responsible for an important proportion (more than 50% in our survey) of multidrug resistance within the species. Taking resistance to β-lactams as an example, these MDR clones were repeatedly reported to carry extended-spectrum β-lactamase-encoding genes located in diverse horizontally acquired elements (integrons, transposons, and plasmids) (6–8, 10, 11, 15, 20, 27). The present study is the first, to our knowledge, to provide evidence that these successful MDR clones may become resistant to β-lactams after chromosomal mutations and without requiring the transfer of foreign DNA.
Some limitations of our study should be addressed. First, the MDR isolates were not equally distributed across the five participating centers, due to local differences in resistance levels, as previously observed in French hospitals (13). Second, we did not type wild-type or non-MDR P. aeruginosa isolates to determine the distribution of the epidemic clones within these populations. However, a nationwide study in the Czech Republic demonstrated a nonrandom distribution of genomic types between susceptible and MDR isolates of P. aeruginosa, with most of the latter associated with three clonal lineages (ST235, ST175, and ST132) (24).
In conclusion, and despite its limitations, our study demonstrates a nonrandom distribution of genomic types among MDR P. aeruginosa isolates. These findings are in accordance with all published studies on this topic. However, they should be considered with caution, since the MLST database may be not representative of the whole P. aeruginosa population. Moreover, we showed that these epidemic clones can become MDR after chromosomal mutations without transfer of foreign DNA, although previous studies reported their association with ESBL- or MBL-encoding genes. Understanding a species population structure and the evolutionary paths is necessary to decide control strategies. Given the possible life-threatening consequences of P. aeruginosa infection and the continued emergence of MDR strains that hamper effective antimicrobial therapy, it is clear that strategies to reduce the likelihood of antimicrobial resistance has become a key issue. Our results indicate that cross-transmission plays a major role in the hospital spread of MDR P. aeruginosa. While the wise use of antibiotics is essential, we suggest that priority be given to improvement of standard hygiene procedures.
ACKNOWLEDGMENTS
We are grateful to the following biologists for their participation in this survey: Christophe De Champs (Reims), Alain Lozniewski (Nancy), Catherine Neuwirth (Dijon), and Benoit Jaulhac (Strasbourg). We are also grateful to Fabrice Poncet from the Institut Fédératif de Recherche IFR133, Besançon, France, for his expertise in DNA sequencing.
This work was funded by grants from the French Ministry of Health in the framework of a clinical research program 2007.
We do not have any conflicts of interest to declare.
Footnotes
Published ahead of print on 18 May 2011.
REFERENCES
- 1. Baquero F. 2004. From pieces to patterns: evolutionary engineering in bacterial pathogens. Nat. Rev. Microbiol. 2:510–518 [DOI] [PubMed] [Google Scholar]
- 2. Bertrand X., et al. 2001. Endemicity, molecular diversity and colonisation routes of Pseudomonas aeruginosa in intensive care units. Intensive Care Med. 27:1263–1268 [DOI] [PubMed] [Google Scholar]
- 3. Cholley P., et al. 2010. Hospital outbreak of Pseudomonas aeruginosa producing extended-spectrum oxacillinase OXA-19. J. Med. Microbiol. 59:866–869 [DOI] [PubMed] [Google Scholar]
- 4. Comité de l'Antibiogramme de la Société Française de Microbiologie 2007. Recommandations 2007. Société Française de Microbiologie, Paris, France [Google Scholar]
- 5. Curran B., Jonas D., Grundmann H., Pitt T., Dowson C. G. 2004. Development of a multilocus sequence typing scheme for the opportunistic pathogen Pseudomonas aeruginosa. J. Clin. Microbiol. 42:5644–5649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Edalucci E., et al. 2008. Acquisition of different carbapenem resistance mechanisms by an epidemic clonal lineage of Pseudomonas aeruginosa. Clin. Microbiol. Infect. 14:88–90 [DOI] [PubMed] [Google Scholar]
- 7. Elias J., et al. 2010. Nosocomial outbreak of VIM-2 metallo-beta-lactamase producing Pseudomonas aeruginosa associated with retrograde urography. Clin. Microbiol. Infect. 16:1494–1500 [DOI] [PubMed] [Google Scholar]
- 8. Empel J., et al. 2007. Outbreak of Pseudomonas aeruginosa infections with PER-1 extended-spectrum beta-lactamase in Warsaw, Poland: further evidence for an international clonal complex. J. Clin. Microbiol. 45:2829–2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fonseca E. L., Freitas Fdos S., Vicente A. C. 2010. The colistin-only-sensitive Brazilian Pseudomonas aeruginosa clone SP (sequence type 277) is spread worldwide. Antimicrob. Agents Chemother. 54:2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Giske C. G., et al. 2006. Establishing clonal relationships between VIM-1-like metallo-beta-lactamase-producing Pseudomonas aeruginosa strains from four European countries by multilocus sequence typing. J. Clin. Microbiol. 44:4309–4315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Glupczynski Y., et al. 2010. Detection and characterization of class A extended-spectrum beta-lactamase-producing Pseudomonas aeruginosa isolates in Belgian hospitals. J. Antimicrob. Chemother. 65:866–871 [DOI] [PubMed] [Google Scholar]
- 12. Hocquet D., Dehecq B., Bertrand X., Plesiat P. 30 March 2011. A strain-tailored double-disc synergy test detects extended-spectrum oxacillinases in Pseudomonas aeruginosa. J. Clin. Microbiol. doi:10.1128/JCM.02585-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hocquet D., et al. 2010. Nationwide investigation of extended-spectrum beta-lactamases, metallo-beta-lactamases, and extended-spectrum oxacillinases produced by ceftazidime-resistant Pseudomonas aeruginosa strains in France. Antimicrob. Agents Chemother. 54:3512–3515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Johnson J. K., Arduino S. M., Stine O. C., Johnson J. A., Harris A. D. 2007. Multilocus sequence typing compared to pulsed-field gel electrophoresis for molecular typing of Pseudomonas aeruginosa. J. Clin. Microbiol. 45:3707–3712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Juan C., et al. 2010. Metallo-beta-lactamase-producing Pseudomonas putida as a reservoir of multidrug resistance elements that can be transferred to successful Pseudomonas aeruginosa clones. J. Antimicrob. Chemother. 65:474–478 [DOI] [PubMed] [Google Scholar]
- 16. Kerr K. G., Snelling A. M. 2009. Pseudomonas aeruginosa: a formidable and ever-present adversary. J. Hosp. Infect. 73:338–344 [DOI] [PubMed] [Google Scholar]
- 17. Koh T. H., et al. 2010. Multilocus sequence types of carbapenem-resistant Pseudomonas aeruginosa in Singapore carrying metallo-beta-lactamase genes, including the novel blaIMP-26 gene. J. Clin. Microbiol. 48:2563–2564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kouda S., et al. 2009. Increased prevalence and clonal dissemination of multidrug-resistant Pseudomonas aeruginosa with the blaIMP-1 gene cassette in Hiroshima. J. Antimicrob. Chemother. 64:46–51 [DOI] [PubMed] [Google Scholar]
- 19. Lee K., Lim Y. S., Yong D., Yum J. H., Chong Y. 2003. Evaluation of the Hodge test and the imipenem-EDTA double-disk synergy test for differentiating metallo-beta-lactamase-producing isolates of Pseudomonas spp. and Acinetobacter spp. J. Clin. Microbiol. 41:4623–4629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Libisch B., et al. 2008. Identification of PER-1 extended-spectrum beta-lactamase producing Pseudomonas aeruginosa clinical isolates of the international clonal complex CC11 from Hungary and Serbia. FEMS Immunol. Med. Microbiol. 54:330–338 [DOI] [PubMed] [Google Scholar]
- 21. Livermore D. M. 2002. Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: our worst nightmare? Clin. Infect. Dis. 34:634–640 [DOI] [PubMed] [Google Scholar]
- 22. Maiden M. C. 2006. Multilocus sequence typing of bacteria. Annu. Rev. Microbiol. 60:561–588 [DOI] [PubMed] [Google Scholar]
- 23. Medeiros A. A., Crellin J. 2000. Evaluation of the Sirscan automated zone reader in a clinical microbiology laboratory. J. Clin. Microbiol. 38:1688–1693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nemec A., Krizova L., Maixnerova M., Musilek M. Multidrug-resistant epidemic clones among bloodstream isolates of Pseudomonas aeruginosa in the Czech Republic. Res. Microbiol. 161:234–242 [DOI] [PubMed] [Google Scholar]
- 25. Pirnay J. P., et al. 2009. Pseudomonas aeruginosa population structure revisited. PLoS One 4:e7740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Poirel L., Girlich D., Naas T., Nordmann P. 2001. OXA-28, an extended-spectrum variant of OXA-10 beta-lactamase from Pseudomonas aeruginosa and its plasmid- and integron-located gene. Antimicrob. Agents Chemother. 45:447–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Samuelsen O., et al. Molecular epidemiology of metallo-beta-lactamase-producing Pseudomonas aeruginosa isolates from Norway and Sweden shows import of international clones and local clonal expansion. Antimicrob. Agents Chemother. 54:346–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tenover F. C., et al. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. van Mansfeld R., et al. 2009. Pseudomonas aeruginosa genotype prevalence in Dutch cystic fibrosis patients and age dependency of colonization by various P. aeruginosa sequence types. J. Clin. Microbiol. 47:4096–4101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vettoretti L., et al. 2009. Emergence of extensive-drug-resistant Pseudomonas aeruginosa in a French university hospital. Eur. J. Clin. Microbiol. Infect. Dis. 28:1217–1222 [DOI] [PubMed] [Google Scholar]
- 31. Waine D. J., Honeybourne D., Smith E. G., Whitehouse J. L., Dowson C. G. 2009. Cross-sectional and longitudinal multilocus sequence typing of Pseudomonas aeruginosa in cystic fibrosis sputum samples. J. Clin. Microbiol. 47:3444–3448 [DOI] [PMC free article] [PubMed] [Google Scholar]