Abstract
Adamantane and oseltamivir resistance among influenza viruses is a major concern to public health officials. To determine the prevalence of antiviral-resistant influenza viruses in Guangdong, China, 244 seasonal A (H1N1) and 222 pandemic A (H1N1) 2009 viruses were screened for oseltamivir resistance by a fluorescence-based neuraminidase (NA) inhibition assay along with NA gene sequencing. Also, 147 seasonal A (H1N1) viruses were sequenced to detect adamantane resistance markers in M2. Adamantane-resistant seasonal A (H1N1) viruses clustering to clade 2C were dominant in 2008, followed by oseltamivir-resistant seasonal A (H1N1) viruses, clustering to clade 2B during January and May 2009. In June 2009, a lineage of double-resistant seasonal A (H1N1) viruses emerged, until it was replaced by the pandemic A (H1N1) 2009 viruses. The lineage most likely resulted from reassortment under the pressure of the overuse of adamantanes. As all viruses were resistant to at least one of the two types of antiviral agents, the need for close monitoring of the prevalence of antiviral resistance is stressed.
INTRODUCTION
Seasonal epidemics of influenza are responsible for substantial morbidity and mortality each year (23). Vaccination provides the primary means for the prevention of infection with influenza virus, but due to the continuous evolution of influenza viruses, vaccines must be updated periodically. In addition to vaccination, the use of antiviral agents plays an important role for prophylaxis and treatment. Currently, two classes of influenza antiviral drugs have been approved for treating influenza, i.e., M2 ion channel blockers, or adamantanes (rimantadine and amantadine), and neuraminidase (NA) inhibitors (NAIs; oseltamivir and zanamivir). Indeed, adamantanes are frequently used in China, because most remedies for common cold treatment contain adamantanes. It may have contributed to the emergence of resistant seasonal influenza virus strains during the 2005-2006 influenza season (18). All (8/8) of seasonal A (H3N2) and 72% (33/46) of seasonal A (H1N1) viruses were resistant to adamantanes in China during the 2005-2006 influenza season (4, 5, 9).
In contrast to adamantanes, NAIs are rarely used for treating influenza in China, as this is the drug of choice for treatment of severe cases. During the 2007-2008 influenza season, the emergence of oseltamivir-resistant seasonal A (H1N1) viruses (ORVs) in Europe was a point of considerable concern (12, 14). In the subsequent influenza season, ORVs spread worldwide. The WHO reported that the incidence of ORVs was virtually 100% in the United States, Eastern Mediterranean, Europe, and Southeast Asia during the 2008-2009 influenza season (16). Although the majority of ORVs remained sensitive to zanamivir and adamantanes, an alarming proportion (50/1,509) of both adamantane- and oseltamivir-resistant seasonal A (H1N1) viruses were found in Hong Kong, China (6, 8). In contrast, seasonal influenza B and A (H3N2) viruses remained sensitive to NAIs (3).
In March and early April 2009, pandemic A (H1N1) 2009 viruses emerged in Mexico and the United States and spread rapidly to many countries and regions, including China. This new reassortment virus was confirmed to be resistant to adamantanes. During the 2009-2010 influenza season, oseltamivir-resistant pandemic A (H1N1) 2009 viruses (pandemic ORVs) were reported sporadically, despite large-scale oseltamivir use (13, 26).
The global spread of antiviral-resistant influenza viruses stresses the need for monitoring of the susceptibility of circulating strains. This is particularly relevant in regions known to be hot spots for emergence of influenza virus variants (20). To characterize local patterns of emerging antiviral resistance, we studied the prevalence of adamantane- and oseltamivir-resistant seasonal A (H1N1) and pandemic A (H1N1) 2009 viruses in Guangdong Province, China.
MATERIALS AND METHODS
Compound.
Oseltamivir carboxylate Ro64-0802 (GS4071) was kindly provided by Hoffmann-La Roche.
Virus isolates.
The seasonal influenza A (H1N1) and pandemic influenza A (H1N1) 2009 viruses were obtained through the influenza surveillance network of Guangdong Province, which includes microbiological laboratories of municipal Centers for Disease Control and Prevention (CDC) throughout Guangdong province. The network included 8 representative municipal CDCs from January 2008 to August 2009, increasing to 21 municipal CDCs (all cities in Guangdong Province) after the outbreak of pandemic A (H1N1) 2009 virus. Each city had one or several sentinel practices in general hospitals. Sentinel practices collected samples from patients with influenza-like illness (ILI) and sent them to the municipal CDC for virus isolation in Madin-Darby canine kidney (MDCK) cells or embryonated chicken eggs. From all parts of the province, municipal CDCs submitted influenza virus isolates to the provincial CDC for (sub)typing using the hemagglutination inhibition assay.
NAI susceptibility.
A fluorescence-based NA enzyme inhibition assay using the fluorogenic substrate 2′-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid (4-MU-NANA; Sigma, St. Louis, MO) was conducted to measure influenza virus NA activity in the presence of oseltamivir. The NAI susceptibility of influenza virus was expressed as the concentration of NAI needed to inhibit the NA enzyme activity by 50% (IC50), as described previously (19). ORV was defined as influenza virus with an IC50 of >100 nmol/liter for oseltamivir (14). From a subset of viruses, the phenotypic oseltamivir susceptibility data were supplemented by NA sequence analysis. Briefly, viral RNA was extracted from culture isolates using a QIAamp viral RNA minikit (Qiagen, Hilden, Germany), according to the manufacturer's instructions. Reverse transcription-PCR (RT-PCR) amplification of the NA gene was conducted with a one-step RT-PCR kit (Qiagen, Hilden, Germany) using NA-specific primers, obtained from the influenza virus database from the Craig Venter Institute (http://gsc.jcvi.org/projects/msc/influenza/infl_a_virus/). Influenza viruses containing the previously reported oseltamivir resistance mutation H275Y in NA were considered oseltamivir resistant (10). Mutations in the NA gene are provided according to N1 numbering.
Box-and-whisker plot analysis was performed to identify potential outliers for NAI susceptibility (15). All viruses defined as resistant to oseltamivir (IC50 > 100 nmol/liter) were removed prior to analysis. IC50 values between 1.5 and 3 times the interquartile range (IQR) or more than 3 times the IQR outside the IQR were defined as mild and extreme outliers, respectively. All IC50 values were log transformed prior to analysis and backtransformed afterwards. Outliers for NAI susceptibility were subjected to NA gene sequencing for finding the H275Y mutation, as described above.
Adamantane susceptibility.
Adamantane susceptibility was examined by sequencing the M2 gene for well-characterized molecular markers known to confer resistance to adamantanes (4).
Phylogenetic analysis.
To study the genetic mechanism of emergence of antiviral resistance among seasonal A (H1N1) viruses in relation to the viral phylogeny, the hemagglutinin (HA) genes of ORVs and oseltamivir-susceptible A (H1N1) viruses (OSVs) were sequenced as described above, along with the NA and M2 genes for phylogenetic analysis. The phylogenetic trees were constructed by the neighbor-joining method with bootstrap analysis (n = 1,000) by MEGA (version 4.0) software (Center for Evolutionary Functional Genomics, Tempe, AZ) (24). Bootstrap values of ≥70% in the major branch were considered distinct clusters. The sequences of WHO-recommended vaccine strains and other reference strains included in the phylogenetic trees were downloaded from the NCBI Influenza Virus Resource (http://www.ncbi.nlm.nih.gov/genomes/FLU/FLU.html) (1).
Nucleotide sequence accession numbers.
The nucleotide sequence data obtained from this study were deposited into the GenBank database (accession nos. CY063704 to CY063742 and CY064249 to CY064389).
RESULTS
Influenza viruses collection.
Through the influenza surveillance network in Guangdong Province, a total of 540 and 1,280 influenza isolates were collected in 2008 and 2009, respectively. Overall, seasonal A (H1N1) viruses accounted for 244 of 540 (45.2%) viruses, collected primarily from June to September 2008, and for 325 of 1,280 (25.4%) viruses, collected primarily from February to August 2009. The remaining viruses were seasonal A (H3N2) viruses, accounting for 119 of 540 (22.0%) and 396 of 1,280 (30.9%) viruses in 2008 and 2009, respectively, and influenza B viruses, accounting for 177 of 540 (32.8%) and 338 of 1,280 (26.4%) viruses in 2008 and 2009, respectively. Two hundred twenty-one (17.2%) influenza pandemic A (H1N1) 2009 viruses were primarily obtained from September 2009 onward (Table 1).
Table 1.
Numbers of influenza viruses isolated in Guangdong, China, during 2008 and 2009
| Yr | No. of influenza viruses by type (subtype) |
||||
|---|---|---|---|---|---|
| Total | Seasonal A (H1N1) | Seasonal A (H3N2) | Pandemic A (H1N1) 2009 | B | |
| 2008 | 540 | 244 | 119 | 0 | 177 |
| 2009 | 1,280 | 325 | 396 | 221 | 338 |
| Total | 1,820 | 569 | 515 | 221 | 515 |
NAI susceptibility.
The results for oseltamivir susceptibilities are summarized in Table 2. In total, 244 seasonal A (H1N1) viruses selected randomly were subject to NAI assay. Among them, 107 seasonal A (H1N1) viruses were oseltamivir resistant on the basis of an IC50 of >100 nmol/liter for oseltamivir. These ORVs had a median IC50 of 402 nmol/liter (range, 172 nmol/liter to 1148 nmol/liter). The remaining 137 viruses had a median IC50 of 2.83 nmol/liter (range, 0.153 nmol/liter to 10.8 nmol/liter), and no outliers were identified (mild threshold IC50s, >20.18 nmol/liter and <93.38 nmol/liter; extreme threshold IC50s, >93.38 nmol/liter). From a subset of 111 seasonal A (H1N1) viruses, the phenotypic susceptibility results were supplemented by sequencing data. The correlations were 100% between a sensitive phenotype (IC50 < 100 nmol/liter) and the presence of H275 (n = 33) and between a resistant phenotype (IC50 > 100 nmol/liter) and the presence of Y275 (n = 78).
Table 2.
Numbers of antiviral-resistant seasonal A (H1N1) viruses and pandemic A (H1N1) 2009 viruses
| Yr | Subtype | No. of viruses | No. of drug-resistant viruses/ no. of viruses tested (%) |
||
|---|---|---|---|---|---|
| Oseltamivir | Adamantane | Double resistant | |||
| 2008 | Seasonal A (H1N1) | 244 | 6/137 (4.4) | 59/64 (92.2) | 1/64 (1.6) |
| 2009 | Seasonal A (H1N1) | 325 | 101/107 (94.4) | 26/83 (31.3) | 23/83 (28.9) |
| Pandemic A (H1N1) 2009 | 221 | 0/221 (0) | NDa | ND | |
ND, not determined.
Of 221 pandemic A (H1N1) 2009 viruses tested, no virus had an IC50 of >100 nmol/liter for oseltamivir. Oseltamivir-susceptible pandemic A (H1N1) 2009 viruses had a median IC50 of 0.24 nmol/liter (range, 0.0169 nmol/liter to 1.66 nmol/liter), and seven outliers were identified (five mild outliers with IC50 thresholds of >0.71 nmol/liter and <1.59 nmol/liter; two extreme outliers with IC50 thresholds of >1.59 nmol/liter). However, no H275Y mutation was found in these outliers by sequencing.
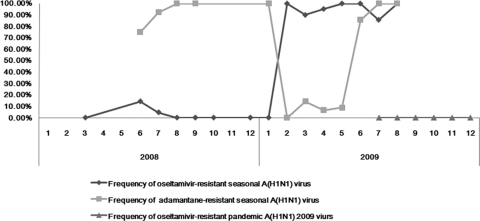
In this study, the earliest detection of ORVs was in Foshan City in June 2008. The proportions of the ORVs were 4.4% (6/137) and 94.4% (101/107) in 2008 and 2009, respectively. This high prevalence remained throughout the whole period of seasonal A (H1N1) viruses circulation, until they had been replaced by pandemic A (H1N1) 2009 viruses (Fig. 1).
Fig. 1.
Trend of antiviral-resistant seasonal A (H1N1) viruses and pandemic A (H1N1) 2009 viruses by month during 2008 and 2009.
Adamantane susceptibility.
Results for adamantane susceptibility on the basis of M2 sequence analysis are summarized in Table 2. Among 244 viruses tested for NAI susceptibility, 147 seasonal A (H1N1) viruses, 64 viruses in 2008 and 83 viruses in 2009, were also used for M2 gene sequencing. Sequence analysis showed that 59 and 26 viruses in 2008 and 2009, respectively, carried the serine-to-asparagine amino acid substitution at residue 31 (S31N) in the M2 gene. No other known adamantane resistance substitutions were found.
Noticeably, among 21 ORVs isolated from June to August 2009, 20 were resistant to both oseltamivir and adamantanes. These double-resistant viruses were the majority strain isolated in June, July, and August 2009, until they were replaced by pandemic A (H1N1) 2009 viruses.
Phylogenetic analysis.
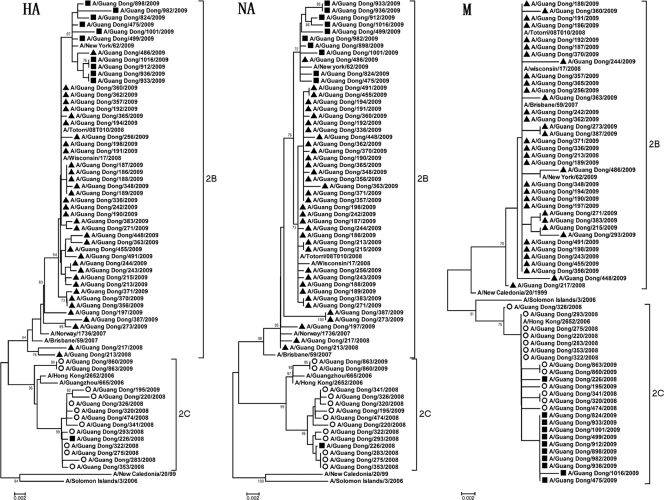
To further study the genetic mechanism of the emergence of antiviral resistance among seasonal A (H1N1) viruses, phylogenetic analysis was performed on the HA, NA, and M2 genes on 13/137 (9.5%) OSVs and 47/107 (43.9%) ORVs from both seasons (Fig. 2).
Fig. 2.
Phylogenetic trees constructed on the basis of the HA, NA, and M2 genes of 60 seasonal A (H1N1) viruses by the neighbor-joining method. Bootstrap values of more than 70% are shown. Closed triangles, oseltamivir-resistant viruses; open circles, adamantane-resistant viruses; closed squares, double-resistant viruses.
On the basis of HA clustering, most viruses during 2008 clustered within clade 2C, represented by A/Hong Kong/2652/2006, an adamantane-resistant virus, whereas most viruses during 2009 clustered within clade 2B, represented by A/Brisbane/59/2007, the WHO-recommended vaccine strain used in the Northern Hemisphere in the 2008-2009 season. All but one of the ORVs belonging to clade 2B possessed H275Y and D354G substitutions in the NA protein (25). Phylogenetic analysis of HA genes showed that 44 ORVs detected during 2009 were genetically close to the ORVs from Japan and the United States (i.e., A/Tottori/08T010/2008 and A/Wisconsin/17/2008, respectively), but 2 ORVs isolated during 2008 were distinct from ORVs isolated during 2009. In total, 36 of 46 ORVs remained sensitive to adamantanes, with M2 genes clustered to clade 2B. However, nine viruses isolated during 2009 were resistant to both oseltamivir and adamantanes; they carried adamantane-resistance-conferring M2 genes clustered in clade 2C. All double-resistant viruses during 2009 sharing the V131A substitution in the HA protein formed a subclade, including adamantane-susceptible ORVs with the M2 gene clustered in clade 2C isolated domestically and from the United States. One double-resistant virus (A/Guang Dong/226/2008) isolated in July 2008 had HA, NA, and M genes similar to those of OSVs from 2008 in clade 2C but carried the H275Y substitution in the NA protein.
On the other hand, 13 OSVs were grouped in clade 2C on the basis of the analysis of the HA, NA, and M genes. Most OSVs (10 in 2008 and 1 at the beginning of 2009) had HA, NA and M2 genes similar to those of adamantane-resistant viruses which were prevalent during the 2007-2008 influenza season (i.e., A/Hong Kong/2652/2006). Two OSVs isolated in 2009 were similar to adamantane-resistant viruses isolated domestically in 2006 (i.e., A/Guangzhou/665/2006).
DISCUSSION
Here, we present a comprehensive overview of the antiviral resistance patterns of influenza viruses identified through systematic surveillance in Guangdong Province, China. In the present study, all viruses tested were resistant to at least one of the two types of antiviral agents. Generally, seasonal A (H1N1) viruses isolated during 2008 were resistant to adamantanes, whereas those isolated during 2009 were resistant to oseltamivir. Of concern is the observed emergence of a double-resistant lineage circulating during June, July, and August 2009, when it was replaced with the pandemic A (H1N1) 2009 viruses. Conversely, influenza viruses resistant to zanamivir were rarely reported (21). We also tested the zanamivir susceptibility of a few influenza viruses in Guangdong and identified no viruses resistant to zanamivir.
In previous studies, the prevalence of adamantane-resistant seasonal A (H1N1) virus (72%) was lower than that of seasonal A (H3N2) virus (100%) in China during the 2005-06 influenza season (5, 9). However, our study showed that almost all influenza A (H1N1) viruses (92.9%) isolated during 2008 in Guangdong were resistant to adamantanes, which was a rate higher than that reported previously. In China, adamantanes are available in over-the-counter formulations and are included in various cold remedies that do not need a prescription. Thus, drug selective pressure might have led to the emergence and subsequent spread of adamantane resistance among influenza viruses in Guangdong, given that resistant viruses typically emerge readily in treated patients and are transmissible (18).
In 2008, we identified two ORVs and one double-resistant virus (A/Guang Dong/226/2008), all of which were isolated from Foshan City in 2008. Although we do not have information about the use of oseltamivir, it is conceivable that this sporadic dual resistance was therapy induced, as reported previously (7, 25). The two ORVs detected in 2008 were distinct from the lineage that became dominant during 2009, again, possibly reflecting sporadic cases of resistance that were therapy induced. Previous studies suggested that influenza A (H1N1) virus with the H275Y mutation most often showed impaired replicative ability in cell culture and attenuated infectivity and transmissibility in animal models (15).
In contrast to the sporadic detections of ORV in 2008, we found a high prevalence of oseltamivir-resistant seasonal A (H1N1) viruses (94.4%) during 2009, similar to the observations in other parts of the world since their emergence in Norway in 2007 (11). Oseltamivir is rarely used to treat influenza virus infection in Guangdong, and all ORVs from Guangdong clustered phylogenetically with the Northern European lineage carrying a D354G substitution on the NA protein (2, 27) and with ORVs from the United States and Japan which were isolated before the emergence of ORVs in Guangdong. Thus, the emergence of ORVs in 2009 in Guangdong most likely reflects seeding of strains through importation, as Guangdong is highly connected to the rest of the world though travel and trade (20).
The most remarkable finding was the emergence of a double-resistant lineage of influenza A (H1N1) virus in 2009, which had HA and NA genes similar to those of domestic ORVs in clade 2B but had M2 genes similar to those of domestic OSVs in clade 2C, suggesting that they arose from reassortment. We are not aware of similar findings elsewhere in the region, suggesting that the reassortment occurred while strains were cocirculating in Guangdong. This could also explain the sporadic findings by the NAI surveillance in Hong Kong (7, 8, 22). This subclade might have been therapy induced, considering the frequent use of adamantanes in China. In addition, the double-resistant virus emerged at a time when the pandemic A (H1N1) 2009 virus outbreak may have resulted in an increased use of adamantanes in some populations. The predominance of double-resistant virus indicated that it can efficiently compete with adamantane-susceptible ORV. Further studies are required to investigate possible mechanisms underlying the fitness improvement of these recombinant viruses, but our findings stress the need for systematic surveillance of resistance, particularly in regions known to be at risk for reassortment due to local epidemiological factors (20). The cocirculation of these double-resistant seasonal influenza viruses and the pandemic A (H1N1) 2009 strains in September in Guangdong exemplifies this risk.
Our study shows that the antiviral resistance profile of influenza viruses is a dynamic process: resistance patterns may change when a new predominant virus replaces the previous subtype or by reassortment among viruses from different lineages. The detection of double-resistant seasonal A (H1N1) viruses also stresses the importance of restricted use of antivirals for influenza treatment. We will continue to closely monitor antiviral drug resistance among circulating viruses, including pandemic and seasonal influenza viruses, to track the evolutionary mechanisms of antiviral resistance and provide treatment suggestions for clinics.
ACKNOWLEDGMENTS
We thank all members of the Working Group for Influenza Surveillance Network in Guandong Province. Members are Jianhong Tang (Shaoguan CDC), Jiemin Lin (Shantou CDC), Jian Zhang (Huizhou CDC), Hua Chen (Zhaoqing CDC), Qinghua Ou (Yunfu CDC), Guodong Liao (Maoming CDC), Jialin Chen (Zhanjiang CDC), Zhiqing Chen (Meizhou CDC), Xiaochang Xu (Yangjiang CDC), Songyan Lin (Chaozhou CDC), Yanping Hong (Jieyang CDC), Weijun Chen (Shanwei CDC), Pingyuan Wang (Qingyuan CDC), Fengguang Huang (Heyuan CDC), Junhe Liang (Jiangmen CDC), Yiyun Chen (Guangzhou CDC), Xiaowen Cheng (Shenzhen CDC), Hongxia Li (Zhuhai CDC), Hong Liang (Zhongshan CDC), Suyi Zhu (Foshan CDC), and Yong Huang (Dongguan CDC).
This study was supported by an Emergency Response Grant ([2008]1216), the Science and Technology Planning Project of Guangdong Province (2009A020101006), and the Medical Science Foundation of Guangdong Province (A2010066), People's Republic of China.
We declare that none of us has any conflict of interest.
Footnotes
Published ahead of print on 18 May 2011.
REFERENCES
- 1. Bao Y., et al. 2008. The influenza virus resource at the National Center for Biotechnology Information. J. Virol. 82:596–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baranovich T., et al. 2010. Emergence of H274Y oseltamivir-resistant A(H1N1) influenza viruses in Japan during the 2008-2009 season. J. Clin. Virol. 47:23–28 [DOI] [PubMed] [Google Scholar]
- 3. Bauer K., Richter M., Wutzler P., Schmidtke M. 2009. Different neuraminidase inhibitor susceptibilities of human H1N1, H1N2, and H3N2 influenza A viruses isolated in Germany from 2001 to 2005/2006. Antiviral Res. 82:34–41 [DOI] [PubMed] [Google Scholar]
- 4. Bright R. A., et al. 2005. Incidence of adamantane resistance among influenza A (H3N2) viruses isolated worldwide from 1994 to 2005: a cause for concern. Lancet 366:1175–1181 [DOI] [PubMed] [Google Scholar]
- 5. Bright R. A., Shay D. K., Shu B., Cox N. J., Klimov A. I. 2006. Adamantane resistance among influenza A viruses isolated early during the 2005-2006 influenza season in the United States. JAMA 295:891–894 [DOI] [PubMed] [Google Scholar]
- 6. Chen H., et al. 2009. Oseltamivir-resistant influenza A pandemic (H1N1) 2009 virus, Hong Kong, China. Emerg. Infect. Dis. 15:1970–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cheng P. K., et al. 2009. Oseltamivir- and amantadine-resistant influenza viruses A (H1N1). Emerg. Infect. Dis. 15:966–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cheng P. K., et al. Oseltamivir- and amantadine-resistant influenza virus A (H1N1). Emerg. Infect. Dis. 16:155–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Deyde V. M., et al. 2007. Surveillance of resistance to adamantanes among influenza A (H3N2) and A (H1N1) viruses isolated worldwide. J. Infect. Dis. 196:249–257 [DOI] [PubMed] [Google Scholar]
- 10. Gubareva L. V., Kaiser L., Matrosovich M. N., Soo-Hoo Y., Hayden F. G. 2001. Selection of influenza virus mutants in experimentally infected volunteers treated with oseltamivir. J. Infect. Dis. 183:523–531 [DOI] [PubMed] [Google Scholar]
- 11. Kramarz P., Monnet D., Nicoll A., Yilmaz C., Ciancio B. 2009. Use of oseltamivir in 12 European countries between 2002 and 2007—lack of association with the appearance of oseltamivir-resistant influenza A(H1N1) viruses. Euro Surveill. 14(5):pii=19112. [DOI] [PubMed] [Google Scholar]
- 12. Lackenby A., et al. 2008. Emergence of resistance to oseltamivir among influenza A(H1N1) viruses in Europe. Euro Surveill. 13(5):pii=8026. [DOI] [PubMed] [Google Scholar]
- 13. Leung T. W. C., Tai A. L. S., Cheng P. K. C., Kong M. S. Y., Lim W. 2009. Detection of an oseltamivir-resistant pandemic influenza A/H1N1 virus in Hong Kong. J. Clin. Virol. 46:298–299 [DOI] [PubMed] [Google Scholar]
- 14. Meijer A., Lackenby A., Hungnes O., Lina B., van-der-Werf S., Schweiger B., Opp M., Paget J., van-de-Kassteele J., Hay A., Zambon M. 2009. Oseltamivir-resistant influenza virus A (H1N1), Europe, 2007-08 season. Emerg. Infect. Dis. 15:552–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Monto A. S., et al. 2006. Detection of influenza viruses resistant to neuraminidase inhibitors in global surveillance during the first 3 years of their use. Antimicrob. Agents Chemother. 50:2395–2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Moscona A. 2009. Global transmission of oseltamivir-resistant influenza. N. Engl. J. Med. 360:953–956 [DOI] [PubMed] [Google Scholar]
- 17. Reference deleted.
- 18. Nelson M. I., Simonsen L., Viboud C., Miller M. A., Holmes E. C. 2009. The origin and global emergence of adamantane resistant A/H3N2 influenza viruses. Virology 388:270–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Potier M., Mameli L., Belisle M., Dallaire L., Melancon S. B. 1979. Fluorometric assay of neuraminidase with a sodium (4-methylumbelliferyl-α-DN-acetylneuraminate) substrate. Anal. Biochem. 94:287–296 [DOI] [PubMed] [Google Scholar]
- 20. Russell C. A., et al. 2008. The global circulation of seasonal influenza A (H3N2) viruses. Science 320:340–346 [DOI] [PubMed] [Google Scholar]
- 21. Sheu T. G., et al. 2008. Surveillance for neuraminidase inhibitor resistance among human influenza A and B viruses circulating worldwide from 2004 to 2008. Antimicrob. Agents Chemother. 52:3284–3292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sheu T. G., et al. Dual resistance to adamantanes and oseltamivir among seasonal influenza A(H1N1) viruses: 2008-2010. J. Infect. Dis. 203:13–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Simonsen L. 1999. The global impact of influenza on morbidity and mortality. Vaccine 17:S3–S10 [DOI] [PubMed] [Google Scholar]
- 24. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596. [DOI] [PubMed] [Google Scholar]
- 25. Ujike M., et al. 2010. Oseltamivir-resistant influenza viruses A (H1N1) during 2007-2009 influenza seasons, Japan. Emerg. Infect. Dis. 16:926–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang B., et al. 2010. Detection of the rapid emergence of the H275Y mutation associated with oseltamivir resistance in severe pandemic influenza virus A/H1N1 09 infections. Antiviral Res. 87:16–21 [DOI] [PubMed] [Google Scholar]
- 27. Zaraket H., et al. 2010. Genetic makeup of amantadine-resistant and oseltamivir-resistant human influenza A/H1N1 viruses. J. Clin. Microbiol. 48:1085–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]