Abstract
We compared the FilmArray RP (Idaho Technology, Inc., Salt Lake City, UT) and the xTAG RVP (Luminex Corporation, Toronto, Canada) multiplex respiratory virus PCR methods for the detection of respiratory viruses in a set of 200 patient specimens frozen at −70°C after standard viral culture and antigen detection methods were done. Both systems detected between 40 to 50% more viruses than traditional methods, primarily rhinoviruses and human metapneumovirus. The FilmArray RP detected significantly more total viruses either alone or as part of mixed infections than the xTAG RVP, as well as an additional 21.6% more respiratory syncytial viruses. The xTAG RVP requires 5 to 6 h with 2.5 to 3 h of hands-on time, while the FilmArray RP takes about an hour with 3 to 5 min of hands-on time, making it much easier to perform.
INTRODUCTION
Multiplex reverse transcriptase respiratory virus PCR has been shown to be more sensitive than standard respiratory virus culture, direct fluorescent-antigen, and direct enzyme-linked immunosorbent assay (ELISA) antigen detection methods (1, 2, 7–9, 13–14, 16, 20). Viral culture is labor-intensive, detects some viruses (e.g., rhinovirus and coronavirus) poorly, and requires 3 to 5 days to detect most agents. Consequently, results are generally not available early in the clinical decision-making process. Direct fluorescent-antibody assay (DFA) and chromatographic immunoassays are rapid enough to support real-time clinical decisions, but DFA is highly labor-intensive and chromatographic immunoassays are relatively insensitive. The FilmArray RP multiplex respiratory virus panel uses a pouch system that contains all reagents for the identification of 18 respiratory viruses and 3 bacterial respiratory pathogens in about 1 h after inoculation of a patient sample, obviating both labor and turnaround time (TAT) issues. We compared the performance of the FilmArray RP with that of the FDA-cleared Luminex xTAG RVP multiplex panel by using 200 retrospective clinical respiratory virus culture samples.
MATERIALS AND METHODS
Patient samples.
Patient specimens sent to the Shands at the University of Florida Hospital Clinical Virology laboratory between October 2008 and May 2010 were frozen at −70°C after standard viral culture was performed. There were 141 upper respiratory samples (nasopharyngeal [NP] swabs, n = 101; throat cultures, n = 25; miscellaneous, n = 15) and 59 lower respiratory tract specimens (bronchoalveolar lavage [BAL] fluid, n = 45; bronchial brushings, n = 2; endotracheal aspirates, n = 11; autopsy lung, n = 1). Sixty-one percent were from patients <18 years old. The study was approved by the University of Florida Institutional Review Board.
Viral culture and antigen detection.
One hundred eighty specimens were cultured using standard tube cultures and shell vials containing human diploid fibroblasts, monkey kidney cells, and A 549 cells (Diagnostic Hybrids, Athens, OH, and ViroMed Laboratories, Minnetonka, MN) at 33°C. Shell vials were stained on days 3 and 5 for influenza A virus, influenza B virus, respiratory syncytial virus (RSV), parainfluenza viruses 1, 2, and 3, and adenovirus using the Light Diagnostics (Temecula, CA) 7-way fluorescent-antibody screen and further identified with specific antisera if positive. Five samples (three influenza A virus and two RSV) were tested by direct antigen testing only (BinaxNOW; Binax, Inc., Scarborough, ME). Fifteen samples were tested by multiplex PCR only.
Multiplex respiratory virus PCR.
The FilmArray RP detects the following agents: influenza A virus, influenza A virus subtype H1, influenza A virus subtype H3, influenza A virus subtype H1N1 swine-origin variant, influenza B virus, respiratory syncytial virus, human metapneumovirus, coronavirus NL63, coronavirus OC43, coronavirus 229E, coronavirus HKU1, adenovirus, parainfluenza virus 1, parainfluenza virus 2, parainfluenza virus 3, parainfluenza virus 4, bocavirus, rhinovirus/enterovirus, Bordetella pertussis, Mycoplasma pneumoniae, and Chlamydophila pneumoniae. The xTAG RVP detects influenza A virus, influenza A virus subtype H1, influenza A virus subtype H3, influenza B virus, respiratory syncytial virus, human metapneumovirus, adenovirus, parainfluenza 1 virus, parainfluenza 2 virus, parainfluenza 3 virus, and rhinovirus/enterovirus. Both assays include internal controls for amplification and extraction and were performed according to the manufacturer's instructions, following training by the respective companies. The FilmArray RP pouch contains dried reagents for all the steps needed for extraction, PCR amplification, and detection of the respiratory viruses listed above. As shown in Fig. 1, the pouch is rehydrated under negative pressure with 1 ml molecular reagent-grade water in the reagent port. Two hundred fifty microliters of sample is diluted into 0.5 ml sample buffer, of which 300 μl is injected into the sample port. The pouch is then placed in the FilmArray RP instrument and identified by bar code, and the assay is started. Results are available in about an hour. For the xTAG RVP, nucleic acid extraction with the addition of an extraction control was done with a Roche MagNA Pure compact instrument using a 200-μl sample eluted in 50 μl, of which 5 μl was used for the assay. The remaining extracted nucleic acid and aliquots of the original frozen specimen were stored at −70°C for further testing. The xTAG is currently FDA cleared only for nasopharyngeal swabs and extraction with the bioMérieux EasyMag, the bioMérieux MiniMag, and the Qiagen QIAamp MiniElute.
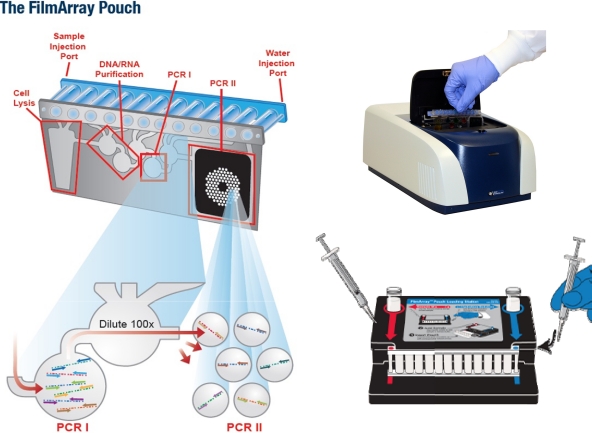
Fig. 1.
The dried reagents in the FilmArray RP pouch are reconstituted by the addition of 1 ml distilled water to the blue port (lower right of diagram), and the diluted sample is injected into the port shown in red. The pouch is then bar code read and placed in the FilmArray RP instrument (upper right). The steps inside the instrument are shown on the left: cell lysis, nucleic acid extraction, and washing, followed by multiplex RT-PCR I and then the specific-virus nested PCRs in PCR II.
Since the FilmArray RP requires approximately 1 h, we could test only 6 to 8 samples per day shift with 1 instrument. Therefore, the corresponding xTAG RVP assays were performed in batches of 20 to 24 within a 3-day time period, rotating the timing of the xTAG RVP so that it was equally distributed at the beginning, middle, and end of the period.
Resolution of discordant results.
“No agreement” was defined as one or more viruses being detected by one molecular method when no viruses were detected by the other method. “Essential agreement” was defined as at least one virus being the same with both methods if either method detected more than one virus. Viruses that were “equivocal” by xTAG RVP (i.e., mean fluorescence intensity [MFI] of 150 to 299) were considered positive for statistical purposes, since these results would be reported to physicians as such. Viruses not included in the xTAG RVP, i.e., coronavirus and bocavirus, that were identified by the FilmArray RP were not considered discordant between the two methods.
Discordant results were resolved by repeat testing in our laboratory of the original or remaining extracted samples by reverse transcription-PCR (RT-PCR) using primers (Invitrogen, Carlsbad, CA) and TaqMan probes (Biosearch Technologies, Inc., Novato, CA) based on published studies (10–12, 15). If probes were not available, the PCR products were confirmed by automated sequencing of PCR products at the University of Florida Biotechnology Core facility using an Applied Biosystems model 3130 Genetic Analyzer. To confirm the influenza virus, CDC primers were modified as follows and the product sequenced: sense, CAAGACCRATCCTGTCACCTCTGAC; antisense, GATCACTTGAATCGYTGCATCT. Coronavirus (10) and bocavirus (12) were confirmed using sequences from published methods as referenced.
Statistics.
The Kappa statistic was calculated from www.graphpad.com/quickcalcs/Kappa2.cfm. Sensitivity and specificity were calculated using the confirmed results as the gold standard at www.chestx-ray.com/statistics/twobytwo.html. McNemar's test was calculated at www.fon.hum.uva.nl/Service/Statistics/McNemars_test.html. These sites were accessible as of 30 March 2011.
RESULTS
Both the FilmArray RP and the xTAG RVP detected significantly more viruses than standard culture and ELISA antigen methods. The great majority of these additional viruses were rhinovirus/enterovirus, which are poorly detected in culture, and human metapneumovirus, which was not specifically tested for in tissue culture (P < 0.00001 for culture versus both FilmArray RP and xTAG RVP). These data are shown in Table 1.
Table 1.
Viruses detected by FilmArray RP, xTAG RVP, and standard culture/antigen
| Virus | No. detected by: |
||
|---|---|---|---|
| Culture/antigen (n = 185)a | FilmArray RP (n = 200)b | xTAG RVP (n = 200)c | |
| Influenza A virus | 32 | 32 | 33 |
| Influenza B virus | 7 | 7 | 7 |
| RSV | 36 | 45 | 37 |
| Rhinovirus/enterovirus | 6 | 43 | 41 |
| Parainfluenza virus | 14 | 16 | 15 |
| Adenovirus | 11 | 10 | 10 |
| Metapneumovirus | 7 | 6 | |
| None (negative) | 82 | 62 | 68 |
| Total no. of viruses | 106 | 160 | 149 |
Culture, n = 180; influenza virus antigen, n = 3; RSV antigen, n = 2.
P < 0.00001 (chi-square test) for culture versus FilmArray RP.
P < 0.00001 (chi-square test) for culture versus xTAG RVP.
Complete agreement between the FilmArray RP and the xTAG RVP was found for 183/200 (91.5%) of specimens (kappa = 0.81; 95% confidence interval [CI], 0.73 to 0.90), and essential agreement was found for 192/200 (96%) (kappa = 0.91; 95% CI, 0.85 to 0.97). There were 15 viruses (in 13 specimens) found only in the FilmArray RP and 4 found only in the xTAG RVP (Table 2) (P = 0.01921). In all cases the discordant viruses detected by both systems were confirmed by independent PCR and sequencing (see Table 3 footnotes for details). With 8 samples, there was no agreement between the FilmArray RP and the xTAG RVP. The FilmArray RP was positive and the xTAG RVP was negative for 1 or more agents in 6 cases, while the xTAG RVP was positive with a negative FilmArray RP in 2 instances. Nine samples had multiple viruses, in which the FilmArray RP detected a total of 8 additional viruses, while the xTAG RVP detected 2 not found by the FilmArray RP.
Table 2.
Total numbers of viruses detecteda
| xTAG RVP result | No. with FilmArray RP result: |
|
|---|---|---|
| Positive | Negative | |
| Positive | 145 | 4 |
| Negative | 15 | 57 |
Excludes coronavirus and bocavirus; P = 0.01921 by McNemar test (http://www.fon.hum.uva.nl/Service/Statistics/McNemars_test.html).
Table 3.
Resolution of discordant results
| xTAG RVP result | FilmArray RP result | Resolution |
|---|---|---|
| Negative | RSV | RSVb |
| Negative | RSV | RSVc |
| Negative | RSV/rhinovirus | RSV/rhinovirusd |
| Negative | HMPa | HMPc |
| Negative | Rhinovirus | Rhinovirusc |
| Negative | RSV | RSVc |
| Rhinovirus | Negative | Rhinovirusc |
| Influenza A virus | Negative | Influenza A viruse |
| Adenovirus | Adenovirus/rhinovirus/RSV | Adenovirus/rhinovirusd/RSVc |
| Parainfluenza virus 2 | Parainfluenza virus 2/RSV | Parainfluenza virus 2/RSVc |
| Parainfluenza virus 2 | Parainfluenza virus 2/RSV | Parainfluenza virus 2/RSVb |
| Influenza A virus/rhinovirus | Influenza A virus | Influenza A virus/rhinovirusd |
| Influenza A virus | Influenza A virus/RSV | Influenza A virus/RSVf |
| Parainfluenza virus 1/adenovirus | Parainfluenza virus 1 | Parainfluenza virus 1/adenovirusg |
| RSV | RSV/rhinovirus | RSV/rhinovirusc |
| RSV | RSV/parainfluenza virus 2 | RSV/parainfluenza virus 2f |
| Influenza B virus/RSV/rhinovirus | Influenza B virus/RSV/rhinovirus/adenovirus | Influenza B virus/RSV/rhinovirus/adenovirusc |
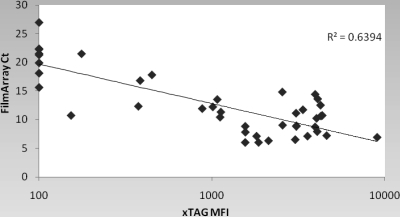
The FilmArray RP detected 8 RSVs while the corresponding xTAG RVP was negative (n = 4), or it was negative for RSV when one or more other viruses were detected by the xTAG RVP (P = 0.007813 by McNemar's test). Detection of RSV by the FilmArray RP but not the xTAG RVP suggests either that the FilmArray RP is more sensitive or that there could be sequence differences that are picked up in one method but not the other. In the former case, one would expect quantitation to show low titers on average, while in the latter, the quantity of virus should match that of the overall population. For this reason, we compared the semiquantitative cycle threshold (CT) available from the FilmArray RP with the MFI of the xTAG RVP for the RSV-positive specimens (Fig. 2). The FilmArray RP RSV-positive/xTAG RVP-negative specimens cluster in the high CT range, suggesting that the discrepancy in RSV detection in most samples may have been due to low levels of virus.
Fig. 2.
Relationship between the CT of the FilmArray RP and the MFI of the xTAG RVP. In the xTAG RVP, an MFI of ≥300 is considered positive and 150 to 299 is considered equivocal. For graphic purposes, specimens positive for RSV by the FilmArray RP but negative by the xTAG RVP are shown as having an xTAG RVP MFI of 100, below the positive range. There were 8 such specimens, but due to the closeness of some of the CTs, all 8 do not show up as individual points.
The sensitivity, specificity, negative predictive value (NPV), and positive predictive value (PPV) were calculated using the resolved results as the gold standard and are shown in Table 4.
Table 4.
Sensitivity and specificity of the FilmArray RP (FA) and xTAG RVP (xTAG) versus PCR-confirmed results
| Virus | No. detected |
Sensitivity (%) |
Specificity (%) |
PPV (%) |
NPV (%) |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FA+, xTAG+ | FA+, xTAG− | FA−, xTAG+ | FA−, xTAG− | FA | xTAG | FA | xTAG | FA | xTAG | FA | xTAG | |
| Influenza A virus | 32 | 0 | 1 | 167 | 97 | 100 | 100 | 100 | 100 | 100 | 99.4 | 100 |
| Influenza B virus | 7 | 0 | 0 | 193 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| RSV | 37 | 8 | 0 | 155 | 100 | 82.2 | 100 | 100 | 100 | 100 | 100 | 95.1 |
| Parainfluenza virus | 15 | 1 | 0 | 184 | 100 | 93.8 | 100 | 100 | 100 | 100 | 100 | 99.5 |
| Rhinovirus/enterovirus | 39 | 4 | 2 | 155 | 95.6 | 91.1 | 100 | 100 | 100 | 100 | 98.7 | 97.5 |
| Adenovirus | 9 | 0 | 1 | 190 | 90 | 100 | 100 | 100 | 100 | 100 | 99.5 | 100 |
| Metapneumovirus | 6 | 1 | 0 | 193 | 100 | 85.7 | 100 | 100 | 100 | 100 | 100 | 99.5 |
The FilmArray RP also detects coronavirus and bocavirus virus as well as Bordetella pertussis, Mycoplasma pneumoniae, and Chlamydophila pneumoniae. Coronavirus was detected in 3 specimens, which was confirmed by PCR (10), and bocavirus virus was detected in 6, all of which were also confirmed by PCR (12). B. pertussis was found in 3 patients, all of whose infections had been suspected and treated on clinical grounds. All three were confirmed by culture or molecular methods in other laboratories. No Mycoplasma or Chlamydophila was detected.
Both the FilmArray RP and the xTAG RVP performed well, with failure rates of 3% and 2%, respectively. In the FilmArray RP this rate was a result of failure of the vacuum seal, so that the vacuum did not function to rehydrate the pouch, and in the xTAG RVP, it was due to internal control failure.
DISCUSSION
Both the FilmArray RP and the xTAG RVP detected significantly more viruses than standard viral culture and antigen detection methods, primarily rhinoviruses/enteroviruses and human metapneumovirus. Numerous studies have also observed increased rates of virus detection when PCR is compared with culture and direct antigen detection methods, whether the PCRs are single or multiplexed (1, 2, 7–9, 13, 14, 16, 20).
The FilmArray RP appeared to detect more total viruses and more RSVs than the xTAG RVP; in 6 instances (4 RSV and 2 rhinovirus) the xTAG RVP was negative, while in the other 9 instances, the xTAG RVP was positive for at least 1 of the 2 or 3 viruses found in the FilmArray RP. Pabbaraju et al. (17) also noted a greater number of positive RSV results using an in-house PCR than found by the xTAG RVP (92 versus 78, in 9 of which RSV was a single positive result). Although the experiments were not definitive, most of the discordant RSVs in our study had low titers in the FilmArray RP, suggesting a higher sensitivity in the FilmArray RP rather than lack of amplification due to RSV sequence differences detected by the FilmArray RP but not by the xTAG RVP. Were the latter the case, one would expect the distribution of CTs in the FilmArray RP to be similar to that of the entire group, which they clearly are not (Fig. 2). Pabbaraju et al. (17) also observed that their discordant RSVs were of low titer as well.
Mixed viral infections were common and were found in 22/138 (15.9%) positive results in the FilmArray RP and in 16/132 (12.1%) in the xTAG RVP, in line with what others have reported. We did not have clinical information to look into whether mixed infections were of greater severity than single infections as has been reported by others (3, 4, 18, 19, 21). The xTAG RVP does not report quantitative results, but “mean fluorescence intensity” (MFI) is available. The research version of the FilmArray RP has a cycle threshold (CT), although this number only indirectly measures the initial amount of virus present and is not planned for the clinical product. However, as shown in Fig. 2 for RSV, the xTAG RVP MFI and the FilmArray RP CT do show a general correlation, but the range is somewhat less than in typical single real-time PCR assays. RSV titers in infants have been reported to be high on admission and to fall approximately 2 log10 units over a 7- to 8-day hospitalization (5) as symptoms improve. This pattern was also observed for infection with human rhinovirus (HRV) as a single agent, but when either RSV or HRV was found in low titer on admission as part of a dual infection, its titer tended to remain unchanged over the course of hospitalization, while that of the higher-titer coinfecting virus fell as it did with single-virus infections (6). It will be important for future studies to define the natural course of viral titers in both single and multiple viral infections when antiviral treatment is given. In view of the extensive clinical experience with the responses of HIV and HCV viral loads to antiviral treatment, it is likely that clinicians will want to apply this paradigm to the treatment of respiratory viral disease.
The xTAG RVP requires 5 to 6 h to complete, including separate reverse transcriptase, PCR, and linking steps to attach the PCR product to the Luminex beads before the results can be read. Logistically, the process takes an entire workday or has to be split into 2 half-day sessions. The test also requires approximately 2.5 to 3 h of actual hands-on time, as well as meticulous technique to prevent cross contamination in the open system. We found it necessary to aliquot reconstituted reagents and store them at −70°C between runs to maintain reagent quality and reliability. In contrast, the FilmArray RP requires 3 to 5 min of hands-on time to inoculate the pouch, read a bar code, and load the instrument. Results are available in about an hour, but only one test at a time can be run as the system is presently configured. The list price for the FilmArray RP is $129/test, but this includes essentially everything necessary to perform the assay. The xTAG RVP lists for $120/test, but the extraction kits and positive-control material have to be purchased separately. The ease of use (literally 3 to 5 min of hands-on time) and rapid turnaround time (TAT) were remarkable features of the FilmArray RP. However, since only one sample can be processed at a time, in its current format, the FilmArray RP may be suitable for low- to moderate-volume laboratories but would have only an adjunctive role in high-volume laboratories.
ACKNOWLEDGMENTS
This study was supported in part by a grant from Idaho Technology, Inc., Salt Lake City, UT, and by the Department of Pathology, Immunology and Laboratory Medicine, University of Florida, Gainesville, FL.
The support of the staff of the Shands at the University of Florida Hospital Clinical Virology laboratory is gratefully acknowledged.
Footnotes
Published ahead of print on 20 April 2011.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. Aguilar J. C., et al. 2000. Detection and identification of human parainfluenza viruses 1, 2, 3, and 4 in clinical samples of pediatric patients by multiplex reverse transcription-PCR. J. Clin. Microbiol. 38:1191–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Balada-Llasat J. M., Larue H., Kelly C., Rigali L., Pancholi P. 2010. Evaluation of commercial ResPlex II v2.0, MultiCode ®-PLx, and xTAG RVP ® respiratory viral panels for the diagnosis of respiratory viral infections in adults. J. Clin. Virol. S1386–6532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cilla G., et al. 2008. Viruses in community-acquired pneumonia in children aged less than 3 years old: high rate of viral coinfection. J. Med. Virol. 80:1843–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Freymuth F., et al. 2006. Comparison of multiplex PCR assays and conventional techniques for the diagnostic of respiratory virus infections in children admitted to hospital with an acute respiratory illness. J. Med. Virol. 78:1498–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gerna G., et al. 2008. Correlation of viral load as determined by real-time RT-PCR and clinical characteristics of respiratory syncytial virus lower respiratory tract infections in early infancy. J. Clin. Virol. 41:45–48 [DOI] [PubMed] [Google Scholar]
- 6. Gerna G., et al. 2009. Correlation of rhinovirus load in the respiratory tract and clinical symptoms in hospitalized immunocompetent and immunocompromised patients. J. Med. Virol. 81:1498–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Greenberg S. B. 2007. Rhinovirus and coronavirus infections. Semin. Respir. Crit. Care Med. 28:182–192 [DOI] [PubMed] [Google Scholar]
- 8. Kim S. R., Ki C. S., Lee N. Y. 2009. Rapid detection and identification of 12 respiratory viruses using a dual priming oligonucleotide system-based multiplex PCR assay. J. Virol. Methods 156:111–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liao R. S., Tomalty L. L., Majury A., Zoutman D. E. 2009. Comparison of viral isolation and multiplex real-time reverse transcription-PCR for confirmation of respiratory syncytial virus and influenza virus detection by antigen immunoassays. J. Clin. Microbiol. 47:527–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lieberman D., et al. 2009. Identification of respiratory viruses in adults: nasopharyngeal versus oropharyngeal sampling. J. Clin. Microbiol. 47:3439–3443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lönnrot M., et al. 1999. Diagnosis of enterovirus and rhinovirus infections by RT-PCR and time-resolved fluorometry with lanthanide chelate labeled probes. J. Med. Virol. 59:378–384 [PubMed] [Google Scholar]
- 12. Lu X., et al. 2006. Real-time PCR assays for detection of bocavirus in human specimens. J. Clin. Microbiol. 44:3231–3235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mahony J., et al. 2007. Development of a respiratory virus panel test for detection of twenty human respiratory viruses by use of multiplex PCR and a fluid microbead-based assay. J. Clin. Microbiol. 45:2965–2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marshall D. J., et al. 2007. Evaluation of a multiplexed PCR assay for detection of respiratory viral pathogens in a public health laboratory setting. J. Clin. Microbiol. 45:3875–3882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mojica M. F., Escobar M. F., Escalante M. P., Jaramillo C. A., el Pilar Delgado M. Detección y tipificación del virus sincitial respiratorio mediante la técnica RT-PCR anidada en pacientes con infección respiratoria aguda. Rev. Inst. Nal. Enf. Resp. Mex. 21:92–98 [Google Scholar]
- 16. Nolte F. S., et al. 2007. MultiCode-PLx system for multiplexed detection of seventeen respiratory viruses. J. Clin. Microbiol. 45:2779–2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pabbaraju K., Tokaryk K. L., Wong S., Fox J. D. 2008. Comparison of the Luminex xTAG RVP respiratory viral panel with in-house nucleic acid amplification tests for diagnosis of respiratory virus infections. J. Clin. Microbiol. 46:3056–3062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Paranhos-Baccala G., et al. 2008. Mixed respiratory virus infections. J. Clin. Virol. 43:407–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Richard N., et al. 2008. The impact of dual viral infection in infants admitted to a pediatric intensive care unit associated with severe bronchiolitis. Pediatr. Infect. Dis. J. 27:213–217 [DOI] [PubMed] [Google Scholar]
- 20. Schindera C., et al. 2010. Immunofluorescence versus xTAG RVP multiplex PCR for the detection of respiratory picornavirus infections in children. J. Clin. Virol. 48:223–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Utokaparch S., et al. 2010. The relationship between respiratory viral loads and diagnosis in children presenting to a pediatric hospital emergency department. Pediatr. Infect. Dis. J. [DOI] [PubMed] [Google Scholar]