Abstract
Several of the more recently proposed new species of Enterococcus are nearly identical based on 16S rRNA gene sequence analysis and phenotypic traits. In the present study, DNA-DNA reassociation experiments, in conjunction with sequencing of the 16S rRNA and rpoB genes, provided evidence that “Enterococcus sanguinicola” and Enterococcus thailandicus actually represent the same species. In contrast, Enterococcus caccae and Enterococcus silesiacus, two other species with nearly identical 16S rRNA gene sequences, were confirmed to be separate species.
TEXT
The number of species included in the genus Enterococcus has expanded considerably in the last two decades, and over 35 valid species are currently recognized (6) (http://www.bacterio.cict.fr). Several of them are associated with infections in humans, while other species have been isolated only from nonhuman sources to date. However, the differentiation of some of the enterococcal species based on comparative 16S rRNA gene sequencing and phenotypic testing alone is sometimes inconclusive for accurate identification. Strains of these Enterococcus species require a polyphasic approach using a combination of techniques, such as 16S rRNA gene sequencing, additional gene target sequencing, analysis of whole-cell protein profiles, phenotypic tests, and often DNA-DNA reassociation to conclusively determine their taxonomic status. In 2004, a single isolate, designated CDC PNS-E2, was determined to be a new Enterococcus species, and its phenotypic characteristics were described by our group (3). Formal species designation was not proposed at that time in deference to the recommendation in minute 10 of the July 2002 meeting of the International Committee on Systematics of Prokaryotes Subcommittee on the Taxonomy of Staphylococci and Streptococci (15), which refers to the description of new species based on a single isolate. Subsequently an additional strain was identified, and the epithet “Enterococcus sanguinicola” was proposed to accommodate these two strains recovered from clinically relevant human sources (2). This denomination was effectively but not validly published. Around this same time frame, a new species, designated Enterococcus thailandicus, comprising a single isolate from fermented sausage, was described (10). Although the source was not clinical, this isolate had phenotypic traits that were very similar and a 16S rRNA gene sequence that was identical to those of the E. sanguinicola isolates, indicating the need to further evaluate their taxonomic position. Two additional species of Enterococcus, Enterococcus caccae (4) and Enterococcus silesiacus (10), were also described within a short period of time and were indistinguishable by 16S rRNA gene sequence comparisons. However, there are several phenotypic differences between strains of these two taxa. The purpose of the present study was to reevaluate the taxonomic status of these proposed four species of Enterococcus.
Two E. sanguinicola strains, ATCC BAA-781T and CCUG 47884, isolated from blood cultures from two patients (2, 3), and E. caccae ATCC BAA-1240T, recovered from a stool sample obtained from a healthy individual (4), were obtained from the CDC Streptococcus Laboratory culture collection (Atlanta, GA). E. thailandicus (NRBC 101867T) isolated from fermented sausage (10) was obtained from the NITE Biological Research Center (NBRC) culture collection (Japan), and E. silesiacus (LMG 23085T), isolated from drinking water (9), was obtained from the BCCM/LMG bacterial collection (Belgium).
The strains were initially compared on the basis of their phenotypic characteristics by using conventional biochemical tests, as previously described (7, 12), and the API Rapid ID32 Strep system (bioMérieux, Inc., Durham, NC) according to the manufacturer's instructions. In addition, the AccuProbe Enterococcus identification test was performed as described by the manufacturer (Gen-Probe, Inc., San Diego, CA). Considering the Enterococcus identification scheme previously described (12), E. sanguinicola and E. thailandicus were placed in phenotypic group II (Table 1) based upon acid formation from mannitol and sorbose and hydrolysis of arginine. Group II currently includes 6 other valid species: E. casseliflavus, E. faecalis, E. faecium, E. gallinarum, E. haemoperoxidus, and E. mundtii. No biochemical reactions were observed to distinguish between E. sanguinicola and E. thailandicus. The two E. sanguinicola isolates were recovered from human blood indicating the potential to cause invasive infections in humans while the formal type strain E. thailandicus was isolated from a nonhuman source (fermented sausage). Interestingly, one of the E. sanguinicola isolates was vancomycin resistant, due to presence of the vanA gene. VanA is an important antimicrobial resistance marker with propensity to rapidly spread to other bacteria and it is often associated with E. faecalis and E. faecium, which belong to the same phenotypic group of enterococcal species. No additional conventional biochemical trait was found that could differentiate the E. sanguinicola strain from the E. thailandicus type strain, so the API Rapid ID32 Strep system was used to look for additional biochemical markers. The phenotypic profile obtained by this system for the 3 strains was classified as “doubtful profile” with the closest similarity to E. hirae with 67.7% of confidence.
Table 1.
Phenotypic characteristics of Enterococcus sanguinicola, Enterococcus thailandicus, Enterococcus caccae, Enterococcus silesiacus, and physiologically related species of Enterococcus
| Species | Phenotypic characteristica |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAN | SOR | ARG | ARA | SBL | RAF | TEL | MOT | PIG | SUC | PYU | MGP | TRE | XYL | |
| Group II | ||||||||||||||
| E. faecium | +d | − | + | + | V | V | − | − | − | +d | − | − | + | −d |
| E. casseliflavus | + | − | +d | + | V | + | −d | +d | +d | + | V | + | + | + |
| E. gallinarum | + | − | +d | + | − | + | − | +d | − | + | − | + | + | + |
| E. mundtii | + | − | + | + | V | + | − | − | + | + | − | − | + | + |
| E. faecalis | +d | − | +d | − | + | − | + | − | − | +d | + | − | + | −d |
| E. haemoperoxidusb | +c | − | +c | − | − | − | − | − | − | + | − | + | + | − |
| E. sanguinicola | + | − | + | − | − | − | Ve | − | − | + | − | V | + | − |
| E. thailandicusb | + | − | + | − | − | − | − | − | − | + | − | − | + | − |
| Group IV | ||||||||||||||
| E. aquimarinusb | − | − | − | + | − | + | − | − | − | + | − | + | + | + |
| E. phoeniculicolab | − | − | − | + | − | + | − | − | − | + | − | + | + | + |
| E. cecorumb | − | − | − | − | − | + | − | − | − | + | + | − | + | − |
| E. sulfureus | − | − | − | − | − | + | − | − | + | + | − | + | + | − |
| E. asinib | − | − | − | − | − | − | − | − | − | + | − | − | + | + |
| E. caccae | − | − | − | − | − | − | − | − | − | + | + | +c | + | − |
| E. silesiacusb | − | − | − | − | − | − | − | − | − | + | − | +c | + | + |
| E. termitisb | − | − | − | − | − | − | − | − | − | + | + | + | + | + |
Based on the conventional identification scheme proposed by Teixeira et al. (12). Abbreviations and symbols: MAN, mannitol; SOR, sorbose; ARG, arginine; ARA, arabinose; SBL, sorbitol; RAF, raffinose; TEL, 0.04% tellurite; MOT, motility; PIG, pigment; SUC, sucrose; PYU, pyruvate; MGP, methyl-α-d-glucopyranoside; TRE, trehalose; XYL, xylose; +, 90% or more of the strains are positive; −, 90% or more of the strains are negative; V, variable (11 to 89% of the strains are positive). Data for species other than E. sanguinicola, E. thailandicus, E. caccae, and E. silesiacus were taken from the work of Teixeira et al. (12).
Phenotypic characteristics based on data from type strains.
Late positive (3 days of incubation or longer).
Occasional exceptions occur (<3% of strains show atypical reactions).
Weak reaction.
E. caccae and E. silesiacus were placed in phenotypic Group IV using the same Enterococcus identification scheme (Table 1). Group IV also includes 6 additional species: E. aquimarinus, E. asini, E. cecorum, E. phoeniculicola, E. sulfureus, and E. termitis. The type strains of E. caccae and E. silesiacus were distinguished from one another based on the following conventional biochemical tests: pyruvate utilization and production of acids from melibiose and xylose. In addition, results for the Accuprobe Enterococcus test were also useful in distinguishing between these two strains. E. caccae tests pyruvate positive, melibiose negative, xylose negative, and probe positive, while E. silesiacus tests pyruvate negative, melibiose positive, xylose positive, and probe negative. E. caccae is lactose negative in the API Rapid ID32 Strep system; however, lactose is positive in the longer incubated conventional test and will probably not be a reliable criterion when additional strains are isolated and tested. Moreover, E. caccae tests saccharose/ sucrose positive in the API Rapid ID32 Strep system, while E. silesiacus tests negative. However, both are positive by the conventional biochemical method, so these two differences would probably not be reliable for differentiation. Acid production from tagatose may also be useful in differentiating these two species. The API Rapid ID32 Strep system misidentified E. caccae isolates as E. durans with a confidence level of 87% and misidentified E. silesiacus as E. durans with a confidence level of 99.9%.
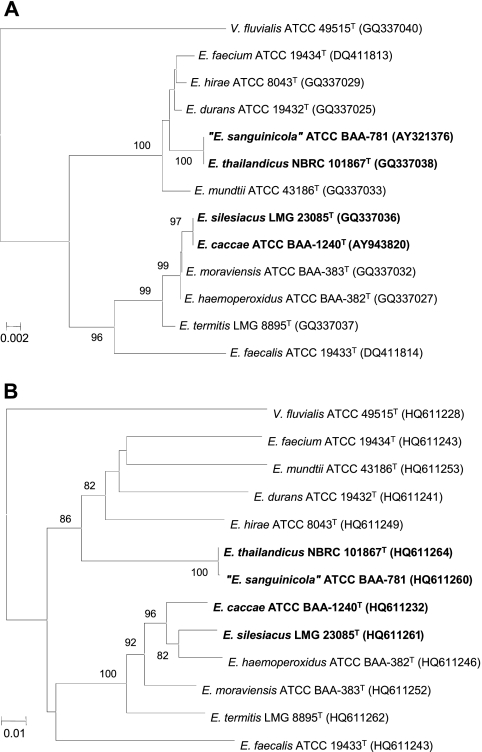
The phylogenetic positions of the isolates were initially determined by comparative analysis of the 16S rRNA gene sequences, as previously described (3). Sequencing reaction products were purified using Centri-Sep plates (Princeton Separations, Princeton, NJ). Reaction products were electrophoresed on an ABI 3130 or 3730 genetic analyzer using the POP-7 polymer (Applied Biosystems). Chromatograms were assembled and analyzed in the Seqmerge software program (Wisconsin Package, version 10.3; Accelrys, Inc., San Diego, CA). The sequences were aligned using the CLUSTAL-W algorithm (13) and trimmed to a 1,493-bp consensus, and a neighbor-joining tree was created (Fig. 1 A) using MEGA 4.0 (8). The 16S rRNA gene sequence analysis (1,483 bp) comparing E. thailandicus to E. sanguinicola showed identical sequences. The 16S RNA gene sequence analysis (1,483 bp) comparing E. silesiacus to E. caccae also showed complete sequence identity. These findings, along with the limited number of strains available for compilation of biochemical data, indicated the need for analysis of additional genetic targets and DNA-DNA hybridization experiments.
Fig. 1.
Phylogenetic trees based on comparative analysis of the 16S rRNA (A) or rpoB (B) gene sequences, showing the relationships among the type strains of Enterococcus sanguinicola, Enterococcus thailandicus, Enterococcus caccae, Enterococcus silesiacus, and other selected species of Enterococcus. GenBank accession numbers are given in parentheses. The neighbor-joining method was used to create the dendrograms, and 1,000 resamplings at the nodes are displayed as percentages. Scale bars indicate numbers of substitutions per nucleotide position. V. fluvialis, Vagococcus fluvialis.
A ∼1,300-bp portion of the RNA polymerase beta subunit (rpoB) gene from each isolate was amplified with HotStarTaq polymerase (Qiagen, Valencia, CA) using the primers UnivrpoB3F (5′-ATGGGNDCGNAAYATGCA) and UnivrpoB23R (5′-GAYATGGAYGTNTGYGC) in a 50-μl PCR. The thermal cycling conditions were as follows: 95°C for 5 min; 15 cycles at 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min, with a decrease in the annealing temperature by 1°C at each cycle; 25 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 1 min; 72°C for 10 min; and a final hold at 4°C. The PCR amplicons were purified through a Nucleospin column (Clontech) and eluted in 120 μl PCR-grade water. Cycle sequencing was performed as described for the 16S rRNA gene above using the primers UnivrpoB3F, UnivrpoB23R, UnivrpoBseq1 (5′-GGNGAYAARNTNKSNRR), and UnivrpoBseq2 (5′-YYNSMNANYTTRTCNCC). Comparative analysis of an 1,177-bp section of the rpoB gene of the E. sanguinicola and E. thailandicus strains showed a similarity of 99.9%, while rpoB sequences of E. caccae and E. silesiacus revealed 96.7% sequence identity. Considering the criterion of similarity greater than 97% as indicative of species-level relatedness (5), such findings suggested that E. sanguinicola and E. thailandicus may constitute a single taxon, while E. caccae and E. silesiacus should be considered separate species.
For DNA-DNA reassociation studies, bacterial cells were grown in flasks containing 2 liters of Todd-Hewitt broth for 18 to 24 h and harvested by centrifugation. The cells were lysed, and DNA was extracted as previously described (11). The techniques and reassociation of DNA using the hydroxyapatite method were as previously described (1). The temperatures used for DNA reassociation were 55°C (optimal conditions) and 70°C (stringent conditions). The percent divergence (% D) was calculated, considering that a decrease of 1°C in thermal stability of a heterologous DNA duplex compared with that of the homologous duplex correlates to 1% unpaired DNA bases. DNA-DNA reassociation is considered the “gold standard” for species relatedness. The three criteria for defining a species include relatedness of greater than 70% under optimal conditions, relatedness of greater than 60% under stringent conditions, and divergence of less than 5% (14). The relative binding ratio at the optimal and stringent temperatures for the two E. sanguinicola blood isolates and the E. thailandicus sausage isolate was greater than 70%, and the divergence was less than 1% (Table 2), which meets the criteria for the definition of species-level relatedness (14). Therefore, results of DNA-DNA reassociation experiments between the two E. sanguinicola strains and the E. thailandicus strain confirm that these three strains actually belong to the same taxon. According to the rules of nomenclature, since E. thailandicus is the earliest formally published and valid epithet (10), it should be considered the senior subjective synonym of E. sanguinicola and must be used to name this taxon. Therefore, E. thailandicus should include strains previously described as CDC PNS-E2 (3) and later designated E. sanguinicola (2). Clinical laboratories should be aware on the occurrence of this additional enterococcal species potentially associated with vanA gene acquisition and transmission.
Table 2.
Levels of DNA relatedness between Enterococcus sanguinicola and Enterococcus thailandicus and between Enterococcus caccae and Enterococcus silesiacus
| Source of unlabeled DNA | Result with labeled DNA froma: |
|||||
|---|---|---|---|---|---|---|
|
E. sanguinicola ATCC BAA781T |
E. caccae ATCC BAA-1240T |
|||||
| RBR at 55°C | % D | RBR at 70°C | RBR at 55°C | % D | RBR at 70°C | |
| E. sanguinicola ATCC BAA-781T | 100 | 0.0 | 100 | NP | NP | NP |
| E. sanguinicola CCUG 47884 | 75 | 0.0 | 84 | NP | NP | NP |
| E. thailandicus NRBC 101867T | 73 | 0.5 | 77 | NP | NP | NP |
| E. caccae ATCC BAA-1240T | NPb | NP | NP | 100 | 0.0 | 100 |
| E. silesiacus LMG 23085T | NP | NP | NP | 40 | 11.0 | NP |
RBR, relative binding ratio; % D, percentage of divergence, calculated to the nearest 0.5%.
NP, not performed.
In contrast, results of DNA-DNA reassociation experiments with strains of E. caccae and E. silesiacus confirmed that they are two valid species even though 16S rRNA gene sequencing could not resolve them. The relative binding ratio at the optimal temperatures for the E. caccae and E. silesiacus type strains was 40%, and the divergence was 11%, indicating that these strains represent distinct species (Table 2). Our observations indicate that potentially conventional biochemical tests, such as pyruvate utilization and production of acids from melibiose and xylose, may be used to reliably distinguish these species. In addition, E. caccae tests positive using the Accuprobe Enterococcus genetic probe, while E. silesiacus tests negative.
Results of DNA-DNA reassociation experiments corroborate the rpoB sequencing data and indicate that analysis of the rpoB gene sequence is a reliable component for use in distinguishing isolates of the enterococcal species included in the present study.
On the basis of our findings, isolates previously proposed as E. sanguinicola should be assigned to the species E. thailandicus, while E. caccae and E. silesiacus should remain as distinct species. In addition to clarifying the taxonomic status of these proposed new species of Enterococcus, the results of the present study highlight the need to use more than a single gene sequence for resolution of enterococcal species. For that, rpoB gene sequencing may represent a useful additional tool. As more closely related enterococcal species are being recognized, it is also important to test enterococcal isolates from different sources for a wide variety of biochemical characteristics in order to prepare more-comprehensive identification schemes. The potential clinical significance of these infrequently isolated enterococcal species, especially when associated with relevant antimicrobial resistance markers, should draw attention to the need for the application of identification procedures of greater precision, allowing their proper recognition and characterization and improving our knowledge of their routes of transmission.
Acknowledgments
We thank Corey G. Franzen and Ifeoma Ezeoke for technical support.
L.M.T. was supported in part by grants from the Brazilian government agencies (FAPERJ and CNPq).
Footnotes
Published ahead of print on 4 May 2011.
REFERENCES
- 1. Brenner D. J., McWhorter A. C., Leete Knutson J. K., Steigerwalt A. G. 1982. Escherichia vulneris: a new species of Enterobacteriaceae associated with human wounds. J. Clin. Microbiol. 15:1133–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Carvalho M. G., et al. 2008. Designation of the provisional new Enterococcus species CDC PNS-E2 as Enterococcus sanguinicola sp. nov., isolated from human blood, and identification of a strain previously named Enterococcus CDC PNS-E1 as Enterococcus italicus Fortina, Ricci, Mora, and Manachini 2004. J. Clin. Microbiol. 46:3473–3476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Carvalho M. G. S., et al. 2004. Characterization of three new enterococcal species, Enterococcus sp. nov. CDC PNS-E1, Enterococcus sp. nov. CDC PNS-E2, and Enterococcus sp. nov. CDC PNS-E3, isolated from human clinical specimens. J. Clin. Microbiol. 42:1192–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Carvalho M. G., et al. 2006. Enterococcus caccae sp. nov., isolated from human stools. Int. J. Syst. Evol. Microbiol. 56:1505–1508 [DOI] [PubMed] [Google Scholar]
- 5. Drancourt M., Roux V., Fournier P.-E., Raoult D. 2004. rpoB gene sequence-based identification of aerobic gram-positive cocci of the genera Streptococcus, Enterococcus, Gemella, Abiotrophia, and Granulicatella. J. Clin. Microbiol. 42:497–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Euzéby J. P. 1997. List of bacterial names with standing in nomenclature. Int. J. Syst. Bacteriol. 47:590–592 http://www.bacterio.cict.fr [DOI] [PubMed] [Google Scholar]
- 7. Facklam R. R., Carvalho M. G. S., Teixeira L. M. 2002. History, taxonomy, biochemical characteristics, and antibiotic susceptibility testing of enterococci, p. 1–54 In Gilmore M. S., Clewell D. B., Courvalin P., Dunny G. M., Murray B. E., Rice L. B. (ed.), The enterococci: pathogenesis, molecular biology, and antibiotic resistance. American Society for Microbiology, Washington, DC [Google Scholar]
- 8. Kumaar S., Tamura K., Nei M. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150–163 [DOI] [PubMed] [Google Scholar]
- 9. Švec P., et al. 2006. Enterococcus silesiacus sp. nov. and Enterococcus termitis sp. nov. Int. J. Syst. Evol. Micobiol. 56:577–581 [DOI] [PubMed] [Google Scholar]
- 10. Tanasupawat S., Sukontasing S., Lee J.-S. 2008. Enterococcus thailandicus sp. nov. isolated from fermented sausage (′mum') in Thailand. Int. J. Syst. Evol. Microbiol. 58:1630–1634 [DOI] [PubMed] [Google Scholar]
- 11. Teixeira L. M., et al. 1995. Correlation between phenotypic characteristics and DNA relatedness within Enterococcus faecium strains. J. Clin. Microbiol. 33:1520–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Teixeira L. M., Carvalho M. G. S., Facklam R. R. 2007. Enterococcus, p. 430–442 In Murray P. R., Baron E. J., Jorgen J. H., Landry M. L., Pfaller M. A. (ed.), Manual of clinical microbiology, 9th ed. American Society for Microbiology, Washington, DC [Google Scholar]
- 13. Thompson J. D., Higgins D. G., Gibson T. J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wayne L. G., et al. 1987. International Committee on Systematic Bacteriology. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37:463–464 [Google Scholar]
- 15. Whiley R. A., Kilian M. 2003. International Committee on Systematics of Prokaryotes subcommittee on the taxonomy of staphylococci and streptococci: minutes of the closed meeting, 31 July 2002, Paris, France. Int. J. Syst. Evol. Microbiol. 53:915–917 [Google Scholar]