Abstract
Our study provides an alert regarding the transmission of rifampin-susceptible strains of Mycobacterium tuberculosis with a silent substitution in codon 514 of rpoB. Among 1,450 cases, we identified 12 isolates sharing this mutation and related restriction fragment length polymorphism (RFLP) types. The mutation impaired hybridization with the wild-type probes in three independent commercial assays, which could lead to misassignment of resistance.
TEXT
The demand for rapid detection of resistance in Mycobacterium tuberculosis has led to a switch from standard phenotyping assays to faster genotyping assays able to identify resistance mutations directly from clinical specimens.
Solid-phase hybridization tests use amplified DNA from the clinical isolate and a set of immobilized probes (containing wild-type [wt] and mutant alleles) and have proven to be efficient when applied prospectively (2, 4, 11). Resistance to rifampin (RIF) is assumed to be present when hybridization is observed with the probes including resistance mutations but not with the wt probe. However, as commercial designs do not include probes for all of the possible mutations in the RIF resistance-determining region (RRDR), the test instructions state that lack of hybridization with the wt probes allows the investigator to indirectly assume resistance, even in the absence of hybridization with the mutant probes.
Our primary objective was to evaluate the frequency of this pattern (no hybridization with the wt probe/no hybridization with the mutant probes), which leads to the indirect assumption of RIF resistance. We also analyzed whether this pattern could also be due to substitutions in the RRDR that impaired hybridization with the wt probe but were not associated with resistance. Therefore, we analyzed data from a reverse-hybridization assay (INNO-LiPA RIF.TB; Innogenetics NV, Ghent, Belgium) that was systematically performed between 2004 and 2011 on all M. tuberculosis isolates from independent tuberculosis (TB) cases received in the Mycobacteriology Reference Unit (1,084,341 inhabitants) of Hospital Universitario Central de Asturias (Oviedo, Spain). Both absence of hybridization with the wt probe S1 and absence of hybridization with the mutant probes were observed in 12 isolates of the 1,450 tested (all during 2006 to 2011; four were involved in a nosocomial cluster) (5) (Fig. 1). The reverse-hybridization test result was not reported to the clinician in any of the 12 cases, and the reference laboratory decided to wait for data from the antibiogram. All 12 isolates were pansusceptible by the agar proportion method (the RIF MICs were ≤1 μg/ml) according to an international standard (15) and by Etest (AB Biodisk, Solna, Sweden) (the RIF MICs were ≤0.125 μg/ml). Once we observed mutations impairing hybridization with the RIF wt probe but not associated with resistance, we evaluated other commercial genotyping tests applied to these isolates (Fig. 1). The GenoType MTBDRplus assay (Hain Lifescience GmbH, Nehren, Germany) showed no hybridization with the rpoB WT3 probe or with the rpoB mutant probes, and the GeneXpert MTB/RIF system (Cepheid, Sunnyvale, California) revealed no amplification detection by probe B.
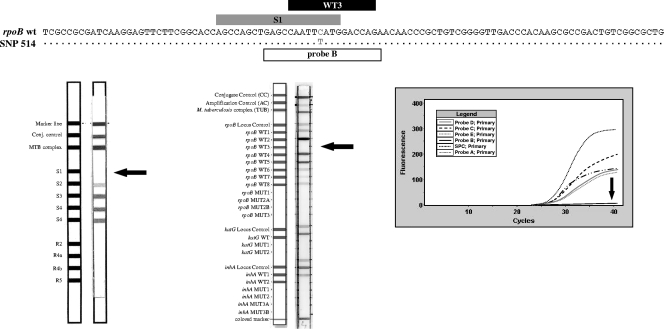
Fig. 1.
Top: reference RRDR wt sequence aligned with a representative sequence of the 12 isolates harboring the single nucleotide polymorphism (SNP) in codon 514. The regions covered by probes homologous with the wt sequence in INNO-LiPA, GenoType MTBDRplus, and GeneXpert are shown above the sequences (in gray, black, and white, respectively). Bottom: results for a representative isolate with the different genotyping techniques. From left to right, INNO-LiPA, GenoType MTBDRplus, and GeneXpert.
Finally, we sequenced the RRDR in order to define the specific mutations in the 12 isolates sharing these abnormal behaviors. Unexpectedly, all 12 isolates shared the same mutation, namely, TTC/TTT, a silent mutation in Phe514.
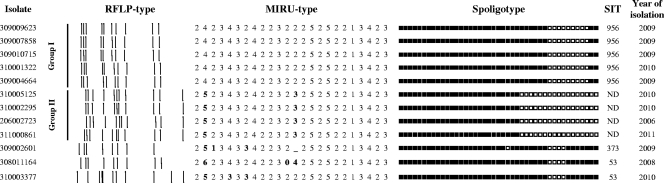
This mutation has only been anecdotally reported in the literature, accompanying another substitution associated with resistance (in codon 531) (10), and it has recently been found in several isolates in the United States (Grace Lin, California Department of Public Health, personal communication). The fact that it was shared by all 12 isolates in our study suggested that they, and not only those involved in the nosocomial cluster, could be clonally related. We applied IS6110 restriction fragment length polymorphism (RFLP), spoligotyping, and mycobacterial interspersed repetitive-unit–variable-number tandem-repeat (MIRU-VNTR) analysis (24-locus format). The four isolates from the nosocomial cluster, together with another unrelated isolate (310001322), shared identical RFLP types, MIRU types, and spoligotypes (Fig. 2, group I). Another group of four isolates (Fig. 2, group II) shared identical patterns by all three techniques; these patterns resembled those of group I, although with subtle differences in mobility and the presence/absence of certain bands and allelic variants at some MIRU loci. Spoligotyping revealed marked differences between the group I pattern and the group II pattern, with eight additional absent spacers adjacent to those that were absent in the group I pattern. Differences in adjacent spacers between the spoligotypes of epidemiologically linked cases can arise as a result of single independent rearrangements in the direct repeat (DR) region of clonally related isolates (13). The remaining three isolates shared identical or highly similar spoligotypes but showed a certain degree of difference from the group I and II patterns (Fig. 2).
Fig. 2.
IS6110 RFLP, MIRU-VNTR, and spoligotyping results for all the isolates. MIRU alleles with differences are indicated in boldface. The isolates were grouped according to similarity (groups I and II). The SIT number and the year of isolation are included. SIT, spoligo-international-type number according to the designation of the SpolDB4 database.
The finding of 12 cases sharing this silent substitution in the RRDR, which may be responsible for misassignment of resistance, could be due to one of the following two scenarios.
Scenarios. (i) Convergent scenario.
The silent mutation in codon 514 of rpoB was acquired by independent unrelated strains. Thus, we would expect completely different genotypic features for each of the strains. Instead, apart from the identity of the isolates involved in the nosocomial cluster, a certain global similarity between the genotyping patterns was observed for most of the remaining cases.
(ii) Divergent scenario.
The strains sharing the silent mutation at codon 514 come from a common ancestor, since very few isolates harbored this substitution (only 12 out of 1,450). Other singular genotypic features found in the patterns for the strains sharing the 514 mutation were allele 4 in the MIRU16 locus, which was identified in only 135 strains (7.6%) of the 1,779 MIRU types obtained in our laboratory, and the fact that some of the spoligotypes detected were specific for these isolates and either not registered or underrepresented in the Asturias regional spoligotype database (2004 to 2011) and in the international SpolDB4 database (3). It could be considered that our observations are region specific; additional studies in other contexts are needed to clarify this possibility. The hypothesis of a common ancestor in our study allows for a certain degree of microevolution, probably during transmission from one to another of the 12 hosts over a 6-year period. Partial modification of the genotyping patterns during transmission chains has been reported (1, 6, 9, 12, 14), and the same may have occurred in our cases. Nevertheless, we recognize that the heterogeneity observed was higher than expected. With respect to the host, bacilli could have the opportunity to microevolve faster due to diagnostic delay, substandard therapy, and poor adherence (14). No evidence of failed therapy or adherence was found for the cases analyzed, although diagnostic delay was found in five cases (1, 2, 4, 6, and 8 months). In addition, certain bacterial mechanisms have been found to be responsible for the faster generation of variability in specific M. tuberculosis strains (7, 8).
Our study provides an alert regarding the existence of silent mutations in the RRDR that are able to impair binding with wt probes in hybridization-based commercial genotyping assays, a pattern that can be interpreted as an indicator of resistance. The credibility gained by commercial genotyping tests for detecting resistance in M. tuberculosis means that they play an important role in therapeutic decision making. New probes must be designed and included in the commercial tests to rule out misassignment of resistance caused by phenomena such as that described here. However, hybridization failures with wt probe(s) in the absence of hybridization with mutant probes should be investigated by direct sequencing before assigning resistance. Analysis of isolates showing this abnormal hybridization pattern revealed the accumulation of related isolates sharing an identical silent mutation whose relatedness had not been suspected using standard genotyping methods applied to M. tuberculosis.
Acknowledgments
We are indebted to Martínez Lirola, who shared with us the original idea on which this study is based. We are grateful for the participation of Ainhoa Simón Zárate and Milagros González in the sequencing analysis.
María Alonso is supported by a contract (REF CA09/00054) from the Instituto de Salud Carlos III (Fondo de Investigaciones Sanitarias) and provides technical support in the Unidad Central de Análisis Molecular of Hospital General Universitario Gregorio Marañón. The study was partially financed by Fondo de Investigaciones Sanitarias (grant S09/02205). Ainhoa Simón Zárate, who participated in the sequencing analysis, holds a grant from the Fondo de Investigaciones Sanitarias (Línea Instrumental Secuenciación). The 3130xl Genetic Analyzer was partially financed by grants from Fondo de Investigaciones Sanitarias (grants IF01-3624 and IF08-36173).
We thank the Microbiology Laboratories of Servicio de Salud del Principado de Asturias SESPA (María Isabel Blanco, Lucía Barreiro, Henar Villar, Aurora Torreblanca, Elisa Hidalgo, Julio Díaz, Flor Hidalgo, and Soledad Calzón) for providing strains. We thank Dolores Pérez Hernández from Servicio de Vigilancia y Alertas Epidemiológicas (Dirección General de Salud Pública y Participación del Principado de Asturias) for the clinical and epidemiological information provided.
We thank Thomas O'Boyle for proofreading the manuscript.
Footnotes
Published ahead of print on 11 May 2011.
REFERENCES
- 1. Alito A., et al. 1999. The IS6110 restriction fragment length polymorphism in particular multidrug-resistant Mycobacterium tuberculosis strains may evolve too fast for reliable use in outbreak investigation. J. Clin. Microbiol. 37:788–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barnard M., Albert H., Coetzee G., O'Brien R., Bosman M. E. 2008. Rapid molecular screening for multidrug-resistant tuberculosis in a high-volume public health laboratory in South Africa. Am. J. Respir. Crit. Care Med. 177:787–792 [DOI] [PubMed] [Google Scholar]
- 3. Brudey K., et al. 2006. Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiol. 6:23–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bwanga F., Hoffner S., Haile M., Joloba M. L. 2009. Direct susceptibility testing for multi drug resistant tuberculosis: a meta-analysis. BMC Infect. Dis. 9:67–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Casas-Fisher R., Penedo-Pallares A., Palacios-Gutiérrez J. J., Moreno-Torrico A. 30 May 2011, posting date. Outbreak or coincidental cases of tuberculosis? Genotyping gives the clue. Am. J. Infect. Control. doi: 10.1016/j.ajic.2011.03.003 [DOI] [PubMed] [Google Scholar]
- 6. Cave M. D., et al. 2005. Epidemiologic import of tuberculosis cases whose isolates have similar but not identical IS6110 restriction fragment length polymorphism patterns. J. Clin. Microbiol. 43:1228–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dos Vultos T., et al. 2008. Evolution and diversity of clonal bacteria: the paradigm of Mycobacterium tuberculosis. PLoS One 3:e1538–e1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ebrahimi-Rad M., et al. 2003. Mutations in putative mutator genes of Mycobacterium tuberculosis strains of the W-Beijing family. Emerg. Infect. Dis. 9:838–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Glynn J. R., et al. 2004. DNA fingerprint changes in tuberculosis: reinfection, evolution, or laboratory error? J. Infect. Dis. 190:1158–1166 [DOI] [PubMed] [Google Scholar]
- 10. Kapur V., et al. 1994. Characterization by automated DNA sequencing of mutations in the gene (rpoB) encoding the RNA polymerase beta subunit in rifampin-resistant Mycobacterium tuberculosis strains from New York City and Texas. J. Clin. Microbiol. 32:1095–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Morgan M., Kalantri S., Flores L., Pai M. 2005. A commercial line probe assay for the rapid detection of rifampicin resistance in Mycobacterium tuberculosis: a systematic review and meta-analysis. BMC Infect. Dis. 5:62–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Perri B. R., et al. 2011. Mycobacterium tuberculosis cluster with developing drug resistance, New York, New York, U.S.A., 2003-2009. Emerg. Infect. Dis. 17:372–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schurch A. C., et al. 2011. Preferential deletion events in the direct repeat locus of Mycobacterium tuberculosis. J. Clin. Microbiol. 49:1318–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schurch A. C., et al. 2010. The tempo and mode of molecular evolution of Mycobacterium tuberculosis at patient-to-patient scale. Infect. Genet. Evol. 10:108–114 [DOI] [PubMed] [Google Scholar]
- 15. Woods G. L. 2003. Susceptibility testing of mycobacteria, nocardiae and other aerobic actinomycetes: approved standard. National Committee for Clinical Laboratory Standards, Wayne, PA: [PubMed] [Google Scholar]