Abstract
Approximately 10 to 15% of tuberculosis (TB) cases in India are estimated to have extrapulmonary disease, and due to a lack of diagnostic means, they often remain untreated. The early detection of Mycobacterium tuberculosis and multidrug resistance is a priority in TB diagnosis to improve the successful treatment rate of TB and reduce transmission. The Xpert MTB/RIF (Xpert) test, recently endorsed by the World Health Organization for the detection of pulmonary TB, was evaluated to test its utility in 547 patients with suspected extrapulmonary tuberculosis. Five hundred forty-seven extrapulmonary specimens were split and processed simultaneously for both culture (solid and liquid) and Xpert testing. For culture, the sensitivity was low, 53% (150/283 specimens). Xpert sensitivity and specificity results were assessed in comparison to a composite reference standard made up of smear and culture results and clinical, radiological, and histological findings. The sensitivity of the Xpert assay was 81% (228/283 specimens) (64% [89/138] for smear-negative cases and 96% [139/145] for smear-positive cases), with a specificity of 99.6%. The sensitivity was found to be high for the majority of specimen types (63 to 100%) except for cerebrospinal fluid, the sensitivity of which was 29% (2/7 specimens). The Xpert test correctly identified 98% of phenotypic rifampin (RIF)-resistant cases and 94% of phenotypic RIF-susceptible cases. Sequencing of the 6 discrepant samples resolved 3 of them, resulting in an increased specificity of 98%. In conclusion, the results of this study suggest that the Xpert test also shows good potential for the diagnosis of extrapulmonary TB and that its ease of use makes it applicable for countries where TB is endemic.
INTRODUCTION
India has the world's largest burden of tuberculosis (TB), accounting for one-fifth of the global TB incidence. The global annual incidence estimate is 9.4 million cases, of which 1.98 million cases are from India (10). TB remains the largest infectious killer disease affecting adults in developing countries (1). In India, TB disproportionately involves the young. Almost 50% of multidrug-resistant TB (MDR-TB) (resistant to at least rifampin [RIF] and isoniazid) cases worldwide are estimated to occur in China and India (21). TB manifests clinically as pulmonary or extrapulmonary tuberculosis (EPTB), with the former being more common. In India, 10 to 15% of TB cases are estimated to be cases of EPTB (which affects mainly the lymph nodes, meninges, kidney, spine, and growing ends of the bones), with a 25 to 50% case mortality rate within months. In this situation, not only rapid TB case detection but also the early determination of MDR status is important. The major challenge in the diagnosis of EPTB is the frequently atypical clinical presentation simulating other inflammatory and neoplastic conditions, which frequently results in a delay or deprivation of treatment. Therefore, a high index of suspicion is necessary to make an early diagnosis, and quite often, more than one procedure is necessary for the confirmation of the diagnosis. In lower-income countries, the lack of diagnostic infrastructure substantially aggravates the problem (1). Reports on biological tests such as enzyme-linked immunosorbent assays, slide agglutination techniques, and PCR are available for EPTB; however, the specificities and sensitivities of these tests are variable (5). Also, these tests require a number of manual steps, and some have a relatively long turnaround time.
The recently developed CE-marked Xpert MTB/RIF (Xpert) test (Cepheid Inc.), based on nested real-time PCR and molecular beacon technology, has been shown to be rapid, with a result for TB and RIF resistance in under 2 h (12); is not prone to cross-contamination; requires minimal biosafety facilities (4); can be performed by technicians with little training; and has a high sensitivity in smear-negative pulmonary TB (the last factor being particularly relevant for patients with HIV infection). These characteristics also make it a potentially attractive tool for extrapulmonary specimens. A series of meta-analyses has shown that nucleic acid amplification tests (NAATs) have high specificity and positive predictive value with highly variable sensitivity, especially in cases of EPTB (11, 16–18). In those studies, NAAT has usually been compared to culture, which is known to be a very suboptimal reference standard for EPTB. Therefore, we have also compared it to a composite reference standard (CRS) to evaluate the true diagnostic potential of the Xpert test for EPTB (3, 6, 14). The CRS for this study was composed of smear microscopy, culture (both liquid and solid), clinical findings, histology/cytology, site-specific computerized tomography scan/magnetic resonance imaging, and follow-up (FU) after 3 months from the date of enrollment. This study was carried out in accordance with previously reported recommendations on the design and conduct of diagnostic accuracy assessments (9).
MATERIALS AND METHODS
Study population and samples.
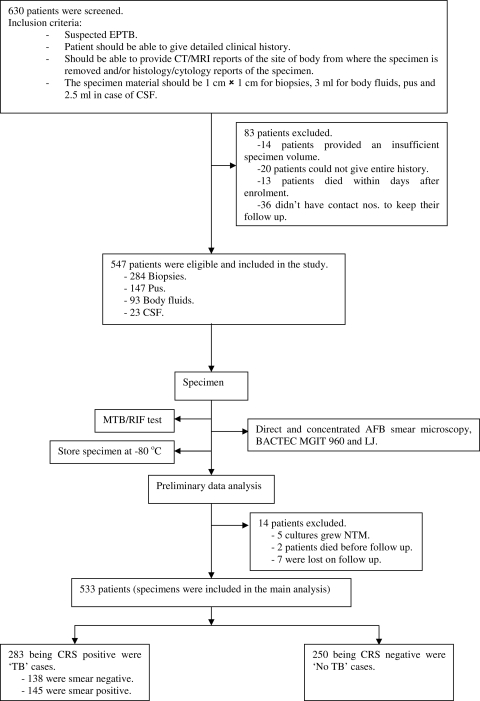
This study was conducted in a private tertiary care hospital, Mumbai, India, from January to August 2010. After the screening of 630 consecutively presenting patients with symptoms suggestive of EPTB, 547 patients met all inclusion criteria and were enrolled at the point of presentation to the consulting physician. Consenting patients were enrolled only if they could provide detailed clinical history and radiological and histology/cytology reports, along with an adequate amount of specimen material. The collected specimen types included 284 biopsy specimens (from tissues [n = 147], lymph nodes [n = 82], and fine-needle aspirates [n = 55]), 147 specimens of pus, 93 specimens of body fluids (synovial [n = 11], pericardial [n = 3], pleural [n = 66], and peritoneal [n = 13]), and 23 cerebrospinal fluid (CSF) specimens. The minimum volumes of sample required were as follows: 3 ml for any kind of body fluid, including pus; 2.5 ml for CSF; and 1 cm by 1 cm for biopsy specimens. Patients were excluded if they were initiated on antitubercular treatment (ATT) within the past 60 days. This study was approved by the Institutional Review Board of our hospital, and informed consent was obtained from each patient. The sample was divided equally into 3 parts, and each part was uniquely coded. Two parts were assigned to 2 different technologists, one in the mycobacteriology laboratory, where the technologist read smears, inoculated cultures, and performed drug susceptibility testing (DST), and the other in the research laboratory, where the technologist performed the Xpert assay, thus blinding the technologists to the results of other tests. The third part was stored at −80°C.
Methods.
The sample was divided equally into 3 parts: one part was used for the Xpert test, the second was stored at −80°C, and the third was tested by direct and concentrated acid-fast bacillus (AFB) microscopy (Ziehl-Neelsen [ZN] staining), followed by processing with N-acetyl-l-cysteine and sodium hydroxide (NALC-NaOH) (15) and centrifugation. The resuspended pellet was subjected to cultivation on both solid medium (egg-based Löwenstein-Jensen [LJ] medium [20]) and liquid medium (Bactec MGIT [mycobacteria growth indicator tube] 960 culture; BD Microbiology Systems). Culture-positive results were confirmed for Mycobacterium tuberculosis by a p-nitro-benzoic acid assay (20) and subjected to indirect drug susceptibility testing with MGIT SIRE (19).
Xpert.
The Xpert assay was performed as described previously (7, 8). A 2:1 volume of sample reagent (SR) buffer was added to biopsy specimens after they had been chopped into very small pieces with a sterile blade in a sterile petri dish. Care was taken to ensure that at least one piece entered the cartridge. Fluids were processed directly by the addition of a 2:1 volume of SR buffer, except for CSF (usually <1 ml), which was raised to 2 ml by the addition of SR buffer. The results obtained were in a simple text format which could be read easily. In case where results were reported as being “invalid,” “no result,” or “error,” the sample was reprocessed and rerun if sufficient material was available.
Patient categories.
Based on the CRS, patients were categorized into 4 groups: confirmed TB cases (culture positive, smear negative/culture positive, or smear positive/culture positive), probable TB cases (culture negative but showing clinical symptoms, radiological findings, and/or histology/cytology suggestive of TB), possible TB cases (negative culture and other tests and only clinical symptoms and/or signs suggestive of TB; in this group the patient follow-up indicated response to empirical ATT after 3 months), and not TB (culture and all other tests for TB were negative, and patient improved without receiving TB treatment). In cases of smear-positive, culture-negative patients, their LJ cultures were checked for 10 weeks before the samples were discarded, and all culture-negative patients were followed up after 3 months. All those patients whose cultures grew nontuberculous mycobacteria (NTM), who were lost to follow-up, or who died before follow-up were excluded from the study (see patient flow chart in Fig. 1). Based on clinical history, smear microscopy, culture reports, radiological reports and/or histology/cytology results, and follow-up, two experts in this field who were blinded to the Xpert test results categorized the patients into the four diagnostic groups. Table 1 shows the symptoms and signs taken into consideration according to the site of infection from where the specimen was obtained. Table 2 represents a detailed algorithm used for the categorization of patients into different categories of the composite reference standard.
Fig. 1.
Flowchart explaining the patient flow in this study. CT, computed tomography; MRI, magnetic resonance imaging.
Table 1.
Symptoms and signs taken into consideration based on site of infectiona
| Site of infection | Symptoms |
|---|---|
| Brain | Irritability, restlessness, neck stiffness, headache persistent for 2-3 wk, vomiting, seizures, changes in mental condition or behavior |
| Intestinal tract, abdomen | Abdominal pain, diarrhea |
| Lymph nodes | Enlargement of lymph nodes, mass formation in the neck |
| Cardiorespiratory | Shortness of breath, hypertension, chest pain, dyspnea |
| Endometrium | Pelvic pain, pelvic mass, irregular periods, infertility |
| Skin (cutaneous) | Visible presence of ulcers or lesions, tender nodules |
Weight loss, persistent cough, and fever for 2 to 3 weeks were also considered for all kinds of specimens.
Table 2.
Algorithm for patient categorization into different categories of the composite reference standard
| CRS category | Result |
|||||
|---|---|---|---|---|---|---|
| AFB smear | Culture | Symptoms/ signsa | Radiologyb | Histology/ cytologyc | Follow-up at 3 mod | |
| Confirmed TB | +/− | + | + | +/− | +/− | + |
| Probable TB | +/− | − | + | + | + | + |
| +/− | − | + | + | − | + | |
| +/− | − | + | − | + | + | |
| Possible TB | +/− | − | + | − | − | + |
| Not TB | − | − | + | − | − | − |
As described in Table 1.
For radiology, a specimen was positive if the presence of infiltrates or cavities, hilar lymph nodes, pleural effusions, or tuberculomas was noted.
For histology/cytology, a specimen was positive if the presence of caseation necrosis with epitheloid granulomas was reported irrespective of the visual presence or absence of acid-fast bacilli.
For follow-up at 3 months, a specimen was positive if the patient was on antitubercular treatment (ATT) and negative if the patient responded to non-ATT.
Analysis of RIF-discordant strains.
Bidirectional sequencing was carried out on the RIF resistance-determining region of the rpoB gene in all the RIF-discordant strains using forward (CGTTGATCAACATCCGGCCGGTG) and reverse (CCACCTTGCGGTACGGCGTT) primers and analyzed by using Chromas, version 2.33, software.
Statistical analysis.
The sensitivities and specificities of smear microscopy, culture, and Xpert methods were calculated against the CRS based on the single direct test run. Forest plots displaying sensitivity and specificity estimates and their 95% confidence intervals (CIs) for each specimen were created by using Meta-Disc software, version 1.4 (22). Wilson's binomial method was used to calculate 95% CIs (2). The indeterminate rate was the number of tests classified as “invalid,” “error,” or “no result” divided by the total number of tests performed. When results were indeterminate and a sufficient amount of the sample remained, the assay was repeated once, and the second result was used for analysis.
RESULTS
Patients.
A total of 547 patients were enrolled in the study (Fig. 1). A total of 14 (3%) patients were excluded from the study, since 5 (9%) were NTM positive, 7 (1%) were lost to follow-up, and 2 (0.5%) died; thus, the final sample size for the analysis was 533 patients. Of these patients, 150 (27%) were culture-positive “confirmed TB” cases (58 [11%] being smear negative and 92 [17%] being smear positive); 129 (24%) were clinically, radiologically, and/or histologically/cytologically positive, suggestive of “probable TB” cases; 4 (1%) were only clinically positive and responded to ATT, suggestive of “possible TB” cases; and 250 (46%) patients had no evidence of TB and were “not TB” cases. Of the total culture-positive cases, 50 (33%) patients were found to have MDR-TB upon phenotypic DST. Out of 547 patients, 16 patients (3%) were found to be HIV positive. The median age of the patients was 37 years (range, 8 months to 94 years). The male-to-female ratio was 0.85.
Sensitivity and specificity. (i) Case detection.
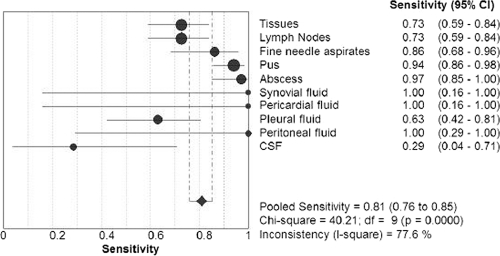
The sensitivities of smear microscopy were found to be 61% (91/150 specimens) among patients with positive cultures and 51% (145/283) among patients with a positive CRS. Upon comparison with a composite reference standard, the pooled sensitivity of culture was found to be 53% (150/283 specimens), with a 42% (59/138) sensitivity for smear-negative, CRS-positive (S−CRS+) cases and a 63% (91/145) sensitivity for smear-positive, CRS-positive (S+CRS+) cases. The sensitivity of the Xpert test against the CRS was found to be 81% (228/283 specimens), 64% (89/138) for S−CRS+ and 96% (139/145) for S+CRS+ cases, with a specificity of 99.6% (249/250). The Xpert test in comparison to culture showed a pooled sensitivity of 83% (125/150 specimens), with a 66% (38/58) sensitivity for smear-negative, culture-positive cases and a 95% (87/92) sensitivity for smear-positive, culture positive cases, with a specificity of 73% (277/382). Cultures had an average time to positivity (TTP) of 25 days with liquid culture and an average TTP of 5 weeks with solid media. Table 3 describes in detail the sensitivities and specificities of culture and Xpert methods with respect to different specimen groups in comparison with the CRS. Table 4 describes in detail the sensitivities and specificities of the Xpert test compared to culture among different specimen groups. Figure 2 gives details of the sensitivity of the Xpert test compared to CRS for each kind of specimen.
Table 3.
Sensitivities and specificities of culture and Xpert methods with respect to different specimen groups in comparison with a composite reference standard (CRS)
| Method compared to CRSa | Biopsy specimens |
Pus |
Body fluids |
CSF |
Total (pooled) |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| % positive specimens (no. of positive specimens/total no. of specimens) | 95% CI | % positive specimens (no. of positive specimens/total no. of specimens) | 95% CI | % positive specimens (no. of positive specimens/total no. of specimens) | 95% CI | % positive specimens (no. of positive specimens/total no. of specimens) | 95% CI | % positive specimens (no. of positive specimens/total no. of specimens) | 95% CI | |
| Culture sensitivity | 50 (70/139) | 42–59 | 64 (56/103) | 45–64 | 62 (21/34) | 45–76 | 43 (3/7) | 16–75 | 53 (150/283) | 47–59 |
| Xpert sensitivity | ||||||||||
| All CRS positive | 75 (105/139) | 68–82 | 95 (98/103) | 89–98 | 71 (24/34) | 54–83 | 29 (2/7) | 8–65 | 81 (228/283) | 76–85 |
| S−CRS+ | 62 (48/78) | 50–72 | 90 (26/29) | 73–97 | 57 (13/23) | 37–74 | 29 (2/7) | 8–65 | 64 (89/138) | 56–72 |
| S+CRS+ | 93 (56/60) | 84–98 | 97 (72/74) | 90–100 | 100 (11/11) | 77–100 | 0 (0/0) | 0 | 96 (139/145) | 91–98 |
| Xpert specificity | 100 (139/139) | 98–100 | 97 (37/38) | 85–100 | 100 (58/58) | 95–100 | 100 (15/15) | 82–100 | 99.6 (249/250) | 98–100 |
S-CRS+, smear negative CRS positive; S+CRS+, smear positive CRS positive.
Table 4.
Sensitivities and specificities of the Xpert test with respect to different specimen groups upon comparison with culture results
| Method compared to culture | Biopsy specimens |
Pus |
Body fluids |
CSF |
Total (pooled) |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| % positive specimens (no. of positive specimens/total no. of specimens) | 95% CI | % positive specimens (no. of positive specimens/total no. of specimens) | 95% CI | % positive specimens (no. of positive specimens/total no. of specimens) | 95% CI | % positive specimens (no. of positive specimens/total no. of specimens) | 95% CI | % positive specimens (no. of positive specimens/total no. of specimens) | 95% CI | |
| Xpert sensitivity | ||||||||||
| All culture positive | 77 (54/70) | 66–86 | 96 (54/56) | 87–100 | 76 (16/21) | 55–90 | 33 (1/3) | 6–80 | 83 (125/150) | 77–89 |
| Smear negative, culture positive | 62 (21/34) | 45–76 | 89 (8/9) | 54–100 | 62 (8/13) | 35–82 | 33 (1/3) | 6–80 | 66 (38/58) | 53–76 |
| Smear positive, culture positive | 92 (33/36) | 77–98 | 98 (46/47) | 88–100 | 100 (8/8) | 71–100 | 0 (0/0) | 0 | 95 (87/92) | 88–98 |
| Xpert specificity | 75 (157/208) | 69–81 | 46 (40/85) | 37–58 | 89 (63/71) | 79–84 | 95 (18/19) | 74–100 | 73 (277/382) | 68–77 |
Fig. 2.
Forest plot giving details of the sensitivity of the Xpert test against the CRS for each kind of specimen.
(ii) Detection of RIF resistance.
The sensitivity and specificity of the Xpert test compared to phenotypic DST were found to be 97.5% (39/40) for correctly determining RIF resistance and 94% (80/85) for correctly determining RIF susceptibility. However, there were 6 patients whose phenotypic DST results for RIF were in discordance with the Xpert result. Five of these samples were RIF sensitive by phenotypic DST but RIF resistant by Xpert, and 1 sample was RIF resistant by phenotypic DST and RIF sensitive by Xpert. This discrepancy in results was resolved by bidirectional sequencing. Of the 5 phenotypically proven RIF-sensitive strains, 2 were found to have a wild-type sequence, while the other 3 showed the same point mutation at codon 533 (CTG to CCG). The correlation between the mutation at codon 533 and RIF resistance is controversial (13). The remaining sample showed the presence of a mutation at codon 531 (TCG to TTG) upon sequencing. Additionally, results of 4 randomly sequenced samples were in concordance with the Xpert results. Considering the phenotypic DST and sequencing results together, the sensitivity using Xpert was found to be 98% (42/43), and the specificity was found to be 98% (80/82) (Table 5).
Table 5.
Sensitivities and specificities of the Xpert test for detection of rifampin resistance compared with phenotypic DST alone and in combination with sequencing for discrepant samples
| Xpert comparison | % positive specimens (no. of positive specimens/total no. of specimens) | 95% CI |
|---|---|---|
| Phenotypic DSTa | ||
| Sensitivity | 98 (39/40) | 86–100 |
| Specificity | 94 (80/85) | 87–98 |
| Phenotypic DST + sequencing for discrepant samplesa | ||
| Sensitivity | 98 (42/43) | 87–100 |
| Specificity | 98 (80/82) | 91–100 |
This is the reference standard for the Xpert test.
(iii) Indeterminate rate.
The indeterminate rate was the number of tests classified as invalid, error, or no result divided by the total number of tests performed. The Xpert result was indeterminate for 0.7% (4/547) of tests performed, a rate lower than the overall contamination rate (2.1%) for 11/547 cultures, both liquid and solid. Allowing for one repeat test, the indeterminate result rate dropped to 0% (0/547), with 100% (4/4) valid results.
DISCUSSION
A recent study by Boehme et al. (8) successfully showed the use of the Xpert test for point-of-care treatment in low-income countries for the detection of RIF resistance in pulmonary TB cases. Along with high specificity, the study showed a sensitivity of 90% for smear-negative pulmonary TB cases. Since we were one of the sites evaluating the test, we decided to also evaluate its utility for paucibacillary extrapulmonary specimens. The test identified 83% (125/150 specimens) of all “confirmed TB” cases, including 64% (38/59) of smear-negative TB cases. It was also observed that the Xpert test detected TB in 80% (103/129) of the samples from “probable TB” cases, whose cultures were negative but who had positive radiological tests and/or positive histology/cytology reports, while some of the patients were already on antitubercular treatment at the point of enrollment in the study.
The specificity of the Xpert test (99.6%) was found to be similar to that reported by Boehme et al. (8). In the case of extrapulmonary specimens, the sensitivities of smear (51%) and culture (53%), though comparable, were found to be low in comparison with that of the Xpert test (81%); and the average TTP of culture is 25 days for MGIT only. Culture is seen to have a low sensitivity in cases of smear-positive patients (n = 57, smear positive, culture negative), because 80% (45/57) of the patients were on ATT for various periods of time ranging from 4 to 6 months when enrolled in the study, and 16% (9/57) of the patients had completed their treatment regimen. Thus, both groups of patients were expected to become culture negative.
The low of sensitivity of culture (53%) in comparison with that of the Xpert test (81%) against the CRS can be explained as follows: (i) 78% (104/133) of culture negative, Xpert-positive patients were on antitubercular treatment for various periods of time when enrolled in the study; (ii) the paucibacillary nature of extrapulmonary specimens with a tendency of M. tuberculosis to form clumps leads to an uneven distribution of the bacilli; (iii) there is loss of viable bacilli during NALC-NaOH processing (due to decanting supernatant steps), unlike Xpert processing, wherein the entire volume of the processed specimen is used; and (iv) the Xpert sample reagent has a better homogenization and liquefaction efficiency than NALC-NaOH processing.
The study shows that the Xpert test has true diagnostic potential with good sensitivity (86 to 100%) for specimens such as synovial, pericardial, and peritoneal fluids; pus; and fine-needle aspirates and moderate sensitivity (63 to 73%) for tissues, lymph nodes, and pleural fluid but poor sensitivity (29%) in the case of CSF, at least in this small number of samples. A preprocessing step (concentrating the specimen by centrifuging it at high speed and then using the pellet for processing) might be required to increase the sensitivity for paucibacillary specimen types such as CSF. There is a need to evaluate and confirm the utility of this tool on a large sample size with specimens such as CSF, other body fluids, and urine, which are easier to obtain.
Finally, not only M. tuberculosis detection but also rapidly determining the patient's MDR status is of prime importance in bringing to an end the spread of MDR-TB and decreasing mortality. Conventional DST results take at least 2 months from the time when the culture is inoculated. Faster methods that allow MDR regimens to be started early are urgently needed. Conventional procedures are laborious and require high-infrastructure laboratories and trained personnel, a luxury that is available only in a few reference centers and not in resource-limited settings or decentralized laboratory settings, where they are most required.
The high cost of this sophisticated technology is offset to an extent by the rapid turnaround time, similar to that of smear microscopy (<2 h), with less biohazard risk and only minimal training needed (22).
In conclusion, the GeneXpert MTB/RIF test not only has good sensitivity and specificity for the diagnosis of TB and detection of RIF resistance in EPTB but also perfectly fits the requirements of the Indian health care setting.
Acknowledgements
We thank National Health and Education Society, P. D. Hinduja National Hospital and Medical Research Centre, Mumbai, for their encouragement and support. David Alland was supported by U.S. National Institutes of Health grant AI52523, and David Alland is among a group of coinvestigators who invented molecular beacons and receive income from licensees, including to Cepheid, for M. tuberculosis detection.
Footnotes
Published ahead of print on 18 May 2011.
REFERENCES
- 1. Agarwal S. P., Chauhan L. S. 2005. Tuberculosis control in India. Directorate General of Health Services/Ministry of Health and Family Welfare, New Delhi, India [Google Scholar]
- 2. Agresti A., Coull B. 1998. Approximate is better than “exact” for interval estimation of binomial proportions. Am. Stat. 52:119–126 [Google Scholar]
- 3. Alonzo T. A., Pepe M. S. 1999. Using a combination of reference tests to assess the accuracy of a new diagnostic test. Stat. Med. 18:2987–3003 [DOI] [PubMed] [Google Scholar]
- 4. Banada P. P., et al. 18 August 2010. Containment of bioaerosol infection risk by the Xpert MTB/RIF assay and its applicability to point-of-care settings. J. Clin. Microbiol. doi:10.1128/JCM.01053-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barnes P. F. 1997. Rapid diagnostic tests for tuberculosis: progress but no gold standard. Am. J. Respir. Crit. Care Med. 165:1497–1498 [DOI] [PubMed] [Google Scholar]
- 6. Baughman A. L., et al. 2008. Utility of composite reference standards and latent class analysis in evaluating the clinical accuracy of diagnostic tests for pertussis. Clin. Vaccine Immunol. 15:106–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Blakemore R., et al. 2010. Evaluation of the analytical performance of the Xpert MTB/RIF Assay. J. Clin. Microbiol. 48:2495–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boehme C. C., et al. 1 September 2010, posting date. Rapid molecular detection of tuberculosis and rifampin resistance. N. Engl. J. Med. doi:10.1056/NEJMoa0907847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bossuyt P. M., et al. 2003. Towards complete and accurate reporting of studies of diagnostic accuracy. The STARD initiative: Standards for Reporting of Diagnostic Accuracy. Clin. Chem. 49:1–6 [DOI] [PubMed] [Google Scholar]
- 10. Central TB Division 2010. TB India 2010: RNTCP status report. Ministry of Health and Family Welfare, New Delhi, India [Google Scholar]
- 11. Daley P., Thomas S., Pai M. 2007. Nucleic acid amplification tests for the diagnosis of tuberculous lymphadenitis: a systematic review. Int. J. Tuberc. Lung Dis. 11:1166–1176 [PubMed] [Google Scholar]
- 12. Helb D., et al. 2010. Rapid detection of Mycobacterium tuberculosis and rifampin resistance by use of on-demand, near-patient technology. J. Clin. Microbiol. 48:229–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hillemann D., Sabine R. G., Richter E. 2007. Evaluation of the GenoType MTBDRplus assay for rifampin and isoniazid susceptibility testing of Mycobacterium tuberculosis strains and clinical specimens. J. Clin. Microbiol. 45:2635–2640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Johnson R. E., et al. 2000. Evaluation of nucleic acid amplification tests as reference tests for Chlamydia trachomatis infections in asymptomatic men. J. Clin. Microbiol. 38:4382–4386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kent P. T., Kubica G. P. 1985. Public health mycobacteriology: a guide for the level III laboratory. Centers for Disease Control, Atlanta, GA [Google Scholar]
- 16. Pai M., Ramsay A., Brien R. 2008. Evidence-based tuberculosis diagnosis. PLoS Med. 5:e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pai M., Ling D. I. 2008. Rapid diagnosis of extrapulmonary tuberculosis using nucleic acid amplification tests: what is the evidence? Future Microbiol. 3:1–4 [DOI] [PubMed] [Google Scholar]
- 18. Pai M., Flores L. L., Hubbard A., Riley L. W., Colford J. M. 2004. Nucleic acid amplification tests in the diagnosis of tuberculous pleuritis: a systematic review and meta-analysis. BMC Infect. Dis. 4:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Siddiqi S. H., Ruesch-Gerdes S. 2006. MGIT procedure manual for BACTEC MGIT 960 TB system. Becton Dickinson, Franklin Lakes, NJ [Google Scholar]
- 20. Weyer K. 1998. Laboratory services in tuberculosis control. Part III: culture. World Health Organization, Geneva, Switzerland [Google Scholar]
- 21. World Health Organization 2010. Multidrug and extensively drug-resistant TB (M/XDR-TB): 2010 global report on surveillance and response. WHO/HTM/TB/2010.3. World Health Organization, Geneva, Switzerland [Google Scholar]
- 22. Zamora J., Abraira V., Muriel A., Khan K., Coomarasamy A. 2006. Meta-DiSc: a software for meta-analysis of test accuracy data. BMC Med. Res. Methodol. 6:31. [DOI] [PMC free article] [PubMed] [Google Scholar]