Abstract
Multilocus sequence typing (MLST) has been successfully applied to the epidemiology of Candida albicans isolates not only within the hospital setting but also in multiple locations nationwide. We performed MLST to investigate the genetic relatedness among bloodstream infection (BSI) isolates of C. albicans recovered from 10 Korean hospitals over a 12-month period. The 156 isolates yielded 112 unique diploid sequence types (DSTs). While 95 DSTs were each derived from a single isolate, 17 DSTs were shared by 61 isolates (39.1%). Interestingly, 111 (71.1%) isolates clustered within previously known clades, and 29 (18.6%) clustered within a new clade that includes strains of Asian origin previously typed as singletons. This MLST study was complemented by restriction endonuclease analysis of genomic DNA using BssHII (REAG-B) in order to evaluate whether strains with identical DSTs and originating from the same hospital corresponded to nosocomial clusters. Importantly, only those isolates with a strong epidemiological relationship showed ≥95% identical REAG-B types. Our results indicate that REAG-B typing can be complementary to MLST but should be limited to the investigation of isolates of identical DSTs and when interhuman transmission is suspected.
INTRODUCTION
Nosocomial bloodstream infection (BSI) due to Candida albicans is a significant cause of morbidity and mortality in hospital patients worldwide (8, 9). C. albicans BSI may be caused by either a commensal strain carried by the patient before hospitalization or a strain acquired nosocomially (15, 18, 20, 22). Pfaller et al. demonstrated previously that some C. albicans BSI strains were more highly concentrated in particular geographic locales and that established BSI strains were endemic in some hospitals (15). Because nosocomial BSIs due to C. albicans constitute an important issue, it has proven quite important to achieve a precise understanding of its epidemiology by use of a reliable typing system (2, 5, 13, 18, 20). Several molecular typing methods have been used to track endemic or epidemic C. albicans clones (3). These methods include Southern blot hybridization with discriminating probes, electrophoretic karyotyping (EK), restriction fragment length polymorphism (RFLP) analysis, randomly amplified polymorphic DNA (RAPD) analysis, and multilocus sequence typing (MLST).
MLST was developed to meet the increasing need for global surveillance and for the comparison of genotypes in a central database accessed via the Internet (2–5, 14). MLST is easy to perform and can group C. albicans isolates in a manner similar to that of Ca3 Southern hybridization (6). The results of MLST are unambiguous, and sequence data obtained from different hospitals can be shared and compared (3). However, because MLST analyzes the sequences of seven 300- to 400-bp loci only, isolates with identical diploid sequence types (DSTs) may differ substantially through large genomic rearrangements in regions that do not encompass the sequenced loci (loss of heterozygosity, aneuploidies, and chromosome size polymorphisms) as well as in the haplotype structures at the sequenced loci (3, 7, 16). Thus, isolates of identical MLST types may need further genotypic characterization to assess clonal relationships. In this respect, two previous studies used MLST analysis for the genotyping of C. albicans BSI isolates from different hospitals (2, 13). Both studies identified some strains of identical DSTs, but these were not further characterized.
The present study was performed to investigate the genetic relatedness among C. albicans BSI isolates from 10 Korean hospitals. We first analyzed MLST data for 156 C. albicans strains isolated from 10 hospitals over a 12-month period. Subsequently, we assessed the genotypic diversity among C. albicans isolates with the same DSTs by restriction digestion analysis of genomic DNA using BssHII (REAG-B) followed by pulsed-field gel electrophoresis (PFGE), which has proven useful for the discrimination of C. albicans isolates (17, 18, 21, 22). This is, to our knowledge, the first reported nationwide study analyzing genotypic relationships among C. albicans BSI isolates from South Korea by both MLST and REAG-B typing.
MATERIALS AND METHODS
C. albicans BSI isolates.
A prospective surveillance study was conducted at 10 university hospitals (hospitals A to J) located throughout South Korea over a 12-month period to assess genotypic relationships among C. albicans BSI isolates. BSI isolates were collected between September 2006 and August 2007 from blood cultures by the method(s) in routine use in each laboratory. Duplicate isolates of C. albicans from the same patient were excluded. During the study period, nosocomial clusters were not reported by the hospital surveillance teams in any of the 10 hospitals.
In total, 156 C. albicans BSI isolates from 156 patients were submitted to the Chonnam National University Hospital for genotyping. The identification of isolates to the species level was based on colony morphology on CHROMagar Candida (BBL, Becton Dickinson, Sparks, MD) at 35°C, microscopic morphology on cornmeal-Tween 80 agar, and a commercially available biochemical identification system (API 20C [bioMérieux, Marcy L'Etoile, France] or the Vitek 2 system Vitek 2 ID-YST [bioMérieux]).
MLST analysis.
C. albicans isolates were typed by using an MLST scheme described previously (4). The internal regions of seven housekeeping genes (AAT1a, ACC1, ADP1, MPIb, SYA1, VPS13, and ZWF1b) were sequenced. Sequence analysis was performed on both strands by use of an ABI Prism 3130xl genetic analyzer (Applied Biosystems, Foster City, CA). Each strain was characterized by a DST resulting from the combination of the genotypes obtained at the seven loci. MLST data have been deposited at the MLST database (http://calbicans.mlst.net). Clades were defined and numbered as described previously (12, 14). Isolates that did not cluster with a known clade and clusters containing fewer than 10 isolates were labeled as singletons. A dendrogram was constructed by use of the unweighted-pair group method using average linkages (UPGMA) and the MEGA 4 software package, as described previously (19). Numbers at nodal points indicate bootstrap values (percent) for 1,000 replications.
REAG-B typing.
REAG-B typing was performed through PFGE of genomic DNA cleaved using the restriction endonuclease BssHII as described previously (17, 18). Isolates were considered identical when all bands observed by PFGE matched. Banding patterns with ≥95% but less than 100% of the bands matching were termed “similar” and grouped into subtypes (subtype a, b, or c) of a given REAG-B type. Isolates with less than 95% of bands matching were considered different (10, 18, 21). All analyses were performed at least twice.
RESULTS
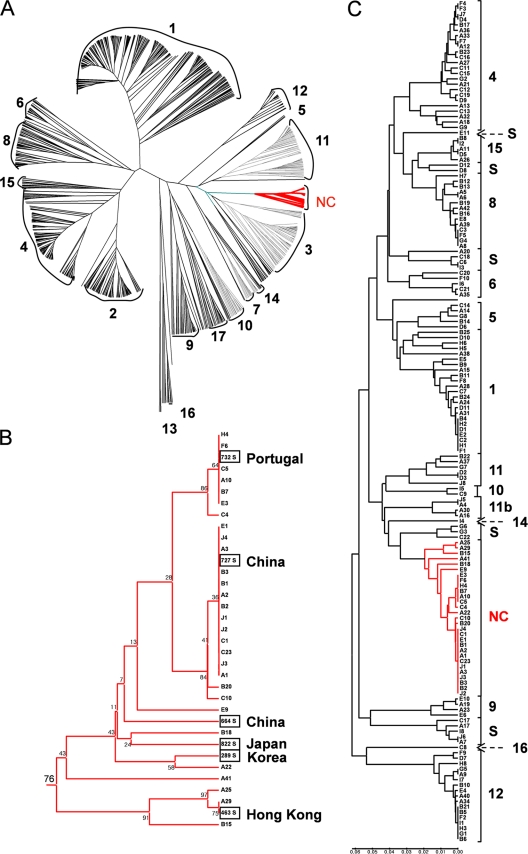
A prospective surveillance study was conducted at 10 university hospitals (hospitals A to J) (Table 1) located throughout South Korea over a 12-month period. In total, 156 C. albicans BSI isolates from 156 patients were obtained and typed by MLST using a combination of seven housekeeping genes. One hundred twelve unique DSTs were obtained from the 156 C. albicans BSI isolates. Sixty-five isolates (41.7%) belonged to previously described types (29 DSTs), whereas 91 (58.3%) belonged to 83 novel DSTs. In order to assign the Korean isolates to existing clades, we performed an analysis of these isolates together with 1,003 DSTs retrieved from the MLST database (55% of all DSTs currently available in the MLST database [http://calbicans.mlst.net]). Nine hundred fifty-nine of the 1,003 DSTs had been previously assigned to one of 17 clades, and 44 had been classified as singletons. Of 156 Korean BSI isolates, 111 (71.1%) isolates clustered within known clades (clades 1 to 17), while 45 (28.9%) did not cluster within these clades. Noticeably, among the unclassified isolates, 29 (18.6%) clustered within a single, highly structured group (Fig. 1A), which corresponded to a new clade, as it contained more than 10 different DSTs that clustered within a 0.035 threshold, which we used to define clades (see Discussion; also see Fig. S1 in the supplemental material). As shown in Fig. 1B, this new clade included 35 of the 1,159 isolates and had a high level of bootstrap support (0.76). Strikingly, 5 of 13 singletons corresponding to isolates originating from Asia clustered within this new clade, while only 1 of 31 singletons with a non-Asian origin clustered within the new clade (Fig. 1B). These data suggested that the new clade identified in our study is highly enriched for isolates of Asian origin if not specific to Asia.
Table 1.
MLST results for 156 C. albicans bloodstream isolates from 156 patients from 10 Korean hospitals
| Clade | No. (%) of isolates | No. of isolates in hospital: |
No. of DSTs | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | H | I | J | |||
| 1 | 23 (14.7) | 5 | 5 | 2 | 3 | 2 | 2 | 4 | 17 | |||
| 4 | 24 (15.4) | 8 | 2 | 6 | 2 | 3 | 2 | 1 | 21 | |||
| 5 | 5 (3.2) | 1 | 1 | 1 | 1 | 1 | 5 | |||||
| 6 | 5 (3.2) | 1 | 2 | 1 | 1 | 5 | ||||||
| 8 | 13 (8.3) | 5 | 3 | 1 | 1 | 1 | 1 | 1 | 9 | |||
| 9 | 4 (2.6) | 2 | 2 | 4 | ||||||||
| 10 | 2 (1.3) | 1 | 1 | 2 | ||||||||
| 11 | 6 (3.8) | 1 | 1 | 2 | 1 | 1 | 5 | |||||
| 11b | 4 (2.6) | 3 | 1 | 2 | ||||||||
| 12 | 18 (11.5) | 3 | 4 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 12 | |
| 14 | 1 (0.6) | 1 | 1 | |||||||||
| 15 | 5 (3.2) | 2 | 1 | 1 | 1 | 3 | ||||||
| 16 | 1 (0.6) | 1 | 1 | |||||||||
| New clade | 29 (18.6) | 8 | 7 | 5 | 3 | 1 | 1 | 4 | 11 | |||
| Singletons | 16 (10.3) | 3 | 4 | 2 | 2 | 2 | 2 | 1 | 14 | |||
| Total | 156 (100.0) | 42 | 25 | 23 | 12 | 11 | 10 | 9 | 8 | 8 | 8 | 112 |
Fig. 1.
MLST analysis of Korean C. albicans isolates identifies a novel clade. (A) UPGMA dendrogram for 1,159 C. albicans isolates. The 17 clades (clades 1 to 17) and the new Asia-specific clade (NC) are indicated. (B) Details of the UPGMA dendrogram of A showing relationships between 35 isolates of the new Asia-specific clade. Boxed isolates identify singletons retrieved from the MLST database, and their origins are indicated. Numbers at nodal points indicate bootstrap values (percent) for 1,000 replications. (C) UPGMA dendrogram showing the genetic relatedness among 156 BSI isolates of C. albicans by use of MLST typing. The vertical bar to the right of each cluster indicates the clade or singleton designation. The scale indicates p-distances.
Figure 1C illustrates the UPGMA dendrogram based on MLST data for the 156 C. albicans isolates and their clade assignments, and Table 1 presents the clade distribution of the 156 C. albicans BSI isolates from the 10 hospitals. The novel clade contained the greatest proportion of isolates (18.6%), followed by clade 4 (15.4%), clade 1 (14.7%), clade 12 (11.5%), and clade 8 (8.3%). Within each hospital, isolates from 4 to 11 different clades were found.
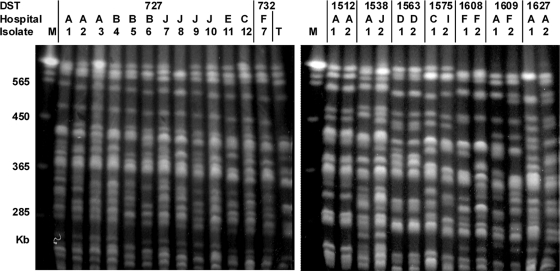
Of 112 DSTs, 95 (60.9%) were unique to a single isolate, and 17 (39.1%) were shared among 61 isolates. DSTs, hospital and ward of origin, and date of isolation for each of these 61 isolates are shown in Table 2. DST 727 was shared by 13 isolates, DST 732 and DST 69 were shared by 7 isolates, DST 601 was shared by 6 isolates, and DST 365 was shared by 4 isolates (Table 2). The remaining 12 DSTs accounted for two isolates each (Table 2). Overall, 32.8% of these 61 isolates accounted for DSTs 727 and 732, which belonged to the same (novel) clade. Noticeably, 26 of these isolates formed groups of two, three, or four isolates each originating from a single hospital, representing 11 potential nosocomial clusters (DST 69, isolates 1 and 2; DST 601, isolates 1 and 2; DST 727, isolates 1 to 3, 4 to 6, 7 to 10, 12, and 13; DST 732, isolates 1 and 2; DST 1512, isolates 1 and 2; DST 1563, isolates 1 and 2; DST 1608, isolates 1 and 2; and DST 1627, isolates 1 and 2) (Table 2). These 26 isolates, as well as the other 35 isolates with identical DSTs, were further characterized by REAG-B typing, yielding 54 different REAG-B patterns (Fig. 2 and Table 2). Of the 11 groups of strains with identical DSTs and each originating from a single hospital, only 1 had isolates with identical REAG-B patterns (DST 1512, isolates 1 and 2), and 1 had isolates with similar REAG-B patterns (DST 1563, isolates 1 and 2). The other nine groups contained two, three, or four isolates with different REAG-B patterns. Interestingly, while DST 1512 or 1563 isolates had been recovered from two pairs of patients who were concurrently hospitalized in the same ward, other isolates with identical DSTs and from the same hospitals had been isolated in different wards, most at an interval of more than 2 months (Table 2). Finally, it should be noted that isolates with identical DSTs and identical or closely related REAG-B patterns and originating from different hospitals were also observed (DST 69, isolates 2, 3, and 6; DST 69, isolates 5 and 7; DST 659, isolates 1 and 2; DST 732, isolates 2 and 3) (Table 2).
Table 2.
REAG-B results for 61 bloodstream isolates sharing 17 DSTsa
| DST | Hospital | Isolate | Department/ward | Isolation date (mo/day/yr) | REAG-B type |
|---|---|---|---|---|---|
| 69 | H | 1 | NS/SICU | 07/01/2007 | R1 |
| H | 2b | PUL/32W | 08/25/2007 | R2a | |
| B | 3b | IM/80W | 12/09/2006 | R2b | |
| C | 4 | IM/MICU | 04/08/2007 | R3 | |
| D | 5b | IM/80W | 01/12/2007 | R4a | |
| E | 6b | IM/ICU | 07/02/2007 | R2c | |
| F | 7b | HEM/50W | 08/22/2007 | R4b | |
| 365 | A | 1 | HEM/36W | 06/10/2007 | R5 |
| G | 2 | IM/10W | 12/28/2006 | R6 | |
| F | 3 | HEM/16W | 05/09/2007 | R7 | |
| C | 4 | ONC/MICU | 08/10/2007 | R8 | |
| 461 | A | 1 | IM/23W | 08/30/2007 | R9 |
| J | 2 | PUL/MICU | 06/14/2007 | R10 | |
| 601 | B | 1 | NR/109W | 09/01/2006 | R11 |
| B | 2 | CV/MICU | 09/21/2006 | R12 | |
| F | 3 | GE/5W | 06/06/2007 | R13 | |
| G | 4 | GS/3W | 06/05/2007 | R14 | |
| H | 5 | GE/ER | 04/11/2007 | R15 | |
| I | 6 | GS/ICU | 04/02/2007 | R16 | |
| 659 | D | 1b | IM/80W | 11/13/1007 | R17a |
| J | 2b | NEP/9W | 08/11/2007 | R17b | |
| 719 | A | 1 | HEM/45W | 04/30/2007 | R18 |
| G | 2 | IM/6W | 10/06/2006 | R19 | |
| 727 | A | 1 | GS/191W | 04/17/2007 | R20 |
| A | 2 | ONC/45W | 08/14/2007 | R21 | |
| A | 3 | PED/78W | 08/17/2007 | R22 | |
| B | 4 | GS/SICU | 09/15/2006 | R23 | |
| B | 5 | CV/87W | 05/30/2007 | R24 | |
| B | 6 | GE/89W | 08/04/2007 | R25 | |
| J | 7 | PED/SICU | 09/25/2006 | R26 | |
| J | 8 | GS/52W | 01/09/2007 | R27 | |
| J | 9 | NEP/MICU | 03/10/2007 | R28 | |
| J | 10 | PUL/102W | 07/23/2007 | R29 | |
| E | 11 | IM/61W | 08/28/2006 | R30 | |
| C | 12 | ONC/72W | 11/11/2006 | R31 | |
| C | 13 | GE/184W | 07/02/2007 | R32 | |
| 732 | C | 1 | PUL/83W | 08/02/2007 | R33 |
| C | 2b | ONC/MICU | 07/05/2007 | R34a | |
| A | 3b | HEM/24W | 09/02/2006 | R34b | |
| B | 4 | PUL/MICU | 02/16/2007 | R35 | |
| H | 5 | HEM/SICU | 12/17/2006 | R36 | |
| E | 6 | GS/51W | 10/30/2006 | R37 | |
| F | 7 | GS/70W | 06/11/2006 | R38 | |
| 929 | B | 1 | NEP/ICU | 04/05/2007 | R39 |
| I | 2 | PUL/ICU | 07/24/2007 | R40 | |
| 1512 | A | 1b,c | NEP/ICU B | 10/07/2006 | R41 |
| A | 2b,c | PUL/ICU B | 10/07/2006 | R41 | |
| 1538 | A | 1 | IM/23W | 08/07/2007 | R42 |
| J | 2 | ONC/MICU | 03/31/2007 | R43 | |
| 1563 | D | 1b,c | GS/90W | 09/13/2006 | R44a |
| D | 2b,c | GS/90W | 09/22/2006 | R44b | |
| 1575 | C | 1 | PED/62W | 05/29/2007 | R45 |
| I | 2 | NS/9W | 02/20/2007 | R46 | |
| 1608 | F | 1 | GS/69W | 11/26/2006 | R47 |
| F | 2 | HEM/17W | 01/26/2007 | R48 | |
| 1609 | F | 1 | HEM/16W | 04/05/2007 | R49 |
| A | 2 | ONC/35W | 06/30/2007 | R50 | |
| 1614 | A | 1 | ONC/36W | 10/15/2006 | R51 |
| D | 2 | IM/80W | 12/18/2006 | R52 | |
| 1627 | A | 1 | PUL/ICU | 01/07/2007 | R53 |
| A | 2 | IM/101W | 06/20/2007 | R54 |
REAG-B, restriction endonuclease analysis of genomic DNA using BssHII; NS, neurosurgery; SICU, surgical intensive care unit; PUL, pulmonology; IM, internal medicine; MICU, medical intensive care unit; HEM, hematology; ONC, oncology; NR, neurology; CV, cardiology; GE, gastroenterology; GS, general surgery; PED, pediatrics; NEP, nephrology. Two- or three-number designations prefixed to the letter “W” represent ward numbers.
Six sets of isolates (DST 69, isolates 2, 3, and 6; DST 69, isolates 5 and 7; DST 659, isolates 1 and 2; DST 732, isolates 2 and 3; DST 1512, isolates 1 and 2; DST 1563, isolates 1 and 2) with the same DSTs were similar by REAG-B typing (≥95% of the bands matched by REAG-B).
The same DSTs and indistinguishable REAG-B patterns in two patient isolates from the same hospital suggest a nosocomial cluster.
Fig. 2.
Representative PFGE patterns of C. albicans obtained by restriction endonuclease analysis of genomic DNA using BssHII (REAG-B). See Table 2 for detailed information on each isolate. Two isolates belonging to the same DST (DST 1512 and DST 1563), which were from the same hospital, showed indistinguishable REAG-B patterns. However, other isolates belonging to the same DST displayed patient-specific PFGE genotypes (less than 95% of the bands matching). T, C. albicans ATCC 90028; M, Saccharomyces cerevisiae DNA concatemers as a molecular size marker.
DISCUSSION
C. albicans isolates can be assigned by MLST to subsets of closely related strain types, referred to as clades (12, 14). Clades have been defined as clusters of at least 10 isolates with DSTs that have a p-distance below 0.04, while their p-distance with DSTs outside the clade is above 0.04. In this respect, a previously reported analysis of MLST data for 1,391 C. albicans isolates corresponding to 1,005 unique DSTs by Odds et al. (14) allowed the assignment of 97% of these isolates to one of 17 clades, with the remaining 3% being defined as singletons. Clade 1 accounted for the largest proportion of isolates (33.6%). As shown in Fig. S1 in the supplemental material, when a panel of 1,005 DSTs already assigned to clades (14) was combined with the 112 DSTs identified in our study, the p-distance threshold of 0.04 used by Odds et al. to define clades could not resolve clades 2 and 4, whereas a threshold of 0.035 slit clades 2 and 4. This suggested that the threshold used for clade assignments was somewhat reliant on the sample studied and that p-distances between 0.04 and 0.035 should be used to separate clades, provided that they include at least 10 DSTs. On these grounds, only 71.1% of 156 BSI isolates of Korean origin clustered within 12 of the 17 known clades. Significantly, a minority of the 156 isolates (29 isolates; 18.6%) belonged to the new MLST clade. It is noteworthy that 6 DSTs previously labeled as singletons belong to this new clade and that 5 of these 6 DSTs correspond to isolates of Asian origin (China, South Korea, Japan, and Hong Kong). As the sample of DSTs that we have analyzed contained 44 singletons, 13 of which originated from Asia, we hypothesize that the new clade revealed by our analysis of Korean isolates corresponds to an Asia-specific clade. If this hypothesis holds true, it is, to our knowledge, the first time that a clade with such a geographical specificity would have been identified within the C. albicans population. Indeed, while clades show some enrichment for isolates from a specific geographical origin, most of them include isolates from a multitude of geographical origins (14). Therefore, the identification of this new clade supports the hypothesis that C. albicans clades have a geographical origin that is being blurred by the increased traveling of human populations over recent years (11). Surprisingly, none of the 156 isolates characterized in this study could be assigned to clade 3, one of the major clades identified by all other studies (11, 14). However, Fig. 1A shows that the new clade is closely related to clade 3. Therefore, it cannot be excluded that the new clade, despite meeting the criteria for clade definition, constitutes a subgroup of clade 3 including mainly Asian strains.
An analysis limited to European isolates, thereby minimizing geographical effects, suggested that the proportion of isolates from blood, commensal carriage, and superficial infections differed between the most populous clades (14). Interestingly, 79.5% of the 327 BSI isolates characterized previously by Odds et al. (14) fell into one of the five major clades (clades 1 to 4 and 11). This is in contrast with our observation that only 34% of the 156 Korean BSI isolates belong to these major clades. This might represent differences in the clade distribution of C. albicans isolates circulating in South Korea and Europe. Further characterization of non-BSI isolates in South Korea will be required to assess whether this difference is also influenced by the origins of our BSI isolates.
In this study, we have performed REAG-B typing for the further characterization of isolates indistinguishable by MLST. Our data show that most isolates with identical DSTs and originating from different hospitals have different REAG-B types, demonstrating that REAG-B typing can be used to further differentiate C. albicans BSI isolates of identical DSTs. However, four sets of two or three isolates from different hospitals remained indistinguishable by MLST and REAG-B typing. Although this may suggest the interhospital transfer of C. albicans strains, it is more likely to reflect the predominance of some DSTs in the global C. albicans population. Indeed, DST 69 and DST 659 have been shown to occur at a higher frequency than any other DST worldwide (14), and DST 732 was one of the two predominant DSTs in the novel Korean-specific clade. These data are consistent with data from a study reported previously by Odds et al. (13), who performed MLST typing on C. albicans BSI isolates (n = 152) in Scotland and suggested that the occurrence of BSI isolates with identical DSTs obtained from hospitals in different cities was unlikely to be the result of interhospital strain transmission.
The characterization by REAG-B typing of putative nosocomial clusters, i.e., a group of two or more isolates originating from the same hospital, showed that 9 of these clusters contained isolates with different REAG-B types and that 2 contained isolates with ≥95% identical REAG-B types. Importantly, the former 9 clusters included isolates collected from different wards and/or more than 2 months apart, while the latter 2 clusters included isolates sampled in the same ward and at the same time. Therefore, it is likely that the former 9 clusters do not represent nosocomial clusters and include strains with identical DSTs but different epidemiological origins, while the latter 2 clusters represent possible nosocomial clusters. However, this interpretation should be taken with caution, as REAG-B patterns are likely to reflect fast-evolving genetic markers, and it cannot be excluded that isolates with identical DSTs and different REAG-B types could nevertheless be derived from a common recent ancestor. Therefore, REAG-B typing of isolates with identical DSTs should be used preferentially for isolates most likely to belong to a nosocomial cluster, i.e., those identified in a same ward or at about the same time in the same hospital.
These data suggested that nosocomial clusters were relatively rare in our sample. This is in contrast with data from a recent study from Iceland, where as many as 23% of all cases of C. albicans BSI were attributable to nosocomial clusters (1). However, it should be noted that that study used RAPD typing, which has lower discriminatory power and reproducibility than MLST and REAG-B typing and may consequently overestimate the incidence of identical isolates.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by the Korea Research Foundation Grant funded by the Korean Government (MOEHRD) (KRF-2008-313-E00510).
Footnotes
Supplemental material for this article may be found at http://jcm.asm.org/.
Published ahead of print on 11 May 2011.
REFERENCES
- 1. Asmundsdóttir L. R., et al. 2008. Molecular epidemiology of candidemia: evidence of clusters of smoldering nosocomial infections. Clin. Infect. Dis. 47:e17–e24 [DOI] [PubMed] [Google Scholar]
- 2. Bougnoux M. E., et al. 2008. Candidemia and candiduria in critically ill patients admitted to intensive care units in France: incidence, molecular diversity, management and outcome. Intensive Care Med. 34:292–299 [DOI] [PubMed] [Google Scholar]
- 3. Bougnoux M. E., et al. 2004. Multilocus sequence typing of Candida albicans: strategies, data exchange and applications. Infect. Genet. Evol. 4:243–252 [DOI] [PubMed] [Google Scholar]
- 4. Bougnoux M. E., et al. 2003. Collaborative consensus for optimized multilocus sequence typing of Candida albicans. J. Clin. Microbiol. 41:5265–5266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen K. W., et al. 2006. Multilocus sequence typing for analyses of clonality of Candida albicans strains in Taiwan. J. Clin. Microbiol. 44:2172–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chowdhary A., et al. 2006. Comparison of multilocus sequence typing and Ca3 fingerprinting for molecular subtyping epidemiologically-related clinical isolates of Candida albicans. Med. Mycol. 44:405–417 [DOI] [PubMed] [Google Scholar]
- 7. Diogo D., Bouchier C., d'Enfert C., Bougnoux M. E. 2009. Loss of heterozygosity in commensal isolates of the asexual diploid yeast Candida albicans. Fungal Genet. Biol. 46:159–168 [DOI] [PubMed] [Google Scholar]
- 8. Gudlaugsson O., et al. 2003. Attributable mortality of nosocomial candidemia, revisited. Clin. Infect. Dis. 37:1172–1177 [DOI] [PubMed] [Google Scholar]
- 9. Horn D. L., et al. 2009. Epidemiology and outcomes of candidemia in 2019 patients: data from the prospective antifungal therapy alliance registry. Clin. Infect. Dis. 48:1695–1703 [DOI] [PubMed] [Google Scholar]
- 10. Lee J. S., et al. 2007. Kodamaea ohmeri isolates from patients in a university hospital: identification, antifungal susceptibility, and pulsed-field gel electrophoresis analysis. J. Clin. Microbiol. 45:1005–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Odds F. C. 2010. Molecular phylogenetics and epidemiology of Candida albicans. Future Microbiol. 5:67–79 [DOI] [PubMed] [Google Scholar]
- 12. Odds F. C., Jacobsen M. D. 2008. Multilocus sequence typing of pathogenic Candida species. Eukaryot. Cell 7:1075–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Odds F. C., et al. 2007. One year prospective survey of Candida bloodstream infections in Scotland. J. Med. Microbiol. 56:1066–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Odds F. C., et al. 2007. Molecular phylogenetics of Candida albicans. Eukaryot. Cell 6:1041–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pfaller M. A., et al. 1998. Hospital specificity, region specificity, and fluconazole resistance of Candida albicans bloodstream isolates. J. Clin. Microbiol. 36:1518–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Selmecki A. M., Dulmage K., Cowen L. E., Anderson J. B., Berman J. 2009. Acquisition of aneuploidy provides increased fitness during the evolution of antifungal drug resistance. PLoS Genet. 5:e1000705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shin J. H., et al. 2004. Microevolution of Candida albicans strains during catheter-related candidemia. J. Clin. Microbiol. 42:4025–4031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shin J. H., et al. 2005. Molecular epidemiological analysis of bloodstream isolates of Candida albicans from a university hospital over a five-year period. J. Microbiol. 43:546–554 [PubMed] [Google Scholar]
- 19. Tamura K., Dudley J., Nei M., Kumar S. 2007. Mega 4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 20. Viviani M. A., Cogliati M., Esposto M. C., Prigitano A., Tortorano A. M. 2006. Four-year persistence of a single Candida albicans genotype causing bloodstream infections in a surgical ward proven by multilocus sequence typing. J. Clin. Microbiol. 44:218–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Voss A., Pfaller M. A., Hollis R. J., Rhine-Chalberg J., Doebbeling B. N. 1995. Investigation of Candida albicans transmission in a surgical intensive care unit cluster by using genomic DNA typing methods. J. Clin. Microbiol. 33:576–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Waggoner-Fountain L. A., et al. 1996. Vertical and horizontal transmission of unique Candida species to premature newborns. Clin. Infect. Dis. 22:803–808 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.