Abstract
The large underestimations of HIV RNA quantification observed in 17 patients with the first version of Cobas TaqMan assay have been successfully corrected in the upgraded version 2.0. In comparison with the Abbott RealTime assay, the mean difference that was 1.18 log10 copies/ml is now zero. The discrepancies have disappeared.
TEXT
The quantification of plasma HIV viral load (VL) is an essential element in the management of antiretroviral therapy (7). New-generation real-time PCR assays are available for HIV-1 RNA quantification: the Abbott RealTime HIV-1 assay (ABB; Abbott, Wiesbaden, Germany) and the Cobas AmpliPrep/Cobas TaqMan HIV-1 assay (CTM; Roche Diagnostics, Mannheim, Germany) (1). These two tests use a cutting-edge molecular quantification technology but differ in all other respects: in the extraction system, the target of primers, the probe design, and the quantification method (the use of a calibration curve for the ABB assay versus that of an internal quantification standard for the CTM assay). They are increasingly replacing the older assays in the developed countries and seem to be the most reliable (2). All these reasons justify our choice to compare these two assays. However, possible primer or probe mismatches have been reported for the first version of the CTM assay (CTM1), and various studies have demonstrated that significant discrepancies exist between the ABB and CTM1 assays with possible clinical implications (3–5). Our team has observed large differences exceeding 1 log10 copies/ml between these two techniques (8). This involved about 3% of patients infected with HIV-1 group M, non-B subtypes, with the ABB assay always reporting the higher value. The unreliability of CTM1 for some HIV-1 strains led Roche Diagnostics to upgrade their test to CTM version 2.0 (CTM2). To address the genetic variability of HIV, CTM2 uses a dual-target strategy. In addition, the limit of quantification (LOQ) was reduced from 40 copies/ml for the CTM1 and ABB assays to a value of 20 copies/ml for CTM2. This new-version CTM2 has been used in our laboratory since 2009, and we sought to determine whether all the underestimations observed with CTM1 had been corrected by the CTM2 test.
Using the CTM2 and ABB assays, we tested multiple samples from all patients for whom underestimation with the CTM1 assay had repeatedly exceeded 0.5 log10 copies/ml relative to the ABB test. For each sample, we compared the difference between the CTM1 and ABB assay values (CTM1 − ABB = DIFF1) to the difference between the CTM2 and ABB assay values (CTM2 − ABB = DIFF2). We used the Wilcoxon signed-rank test for analysis, replacing, where necessary, VL below the working range by the respective LOQ. To be sure of the presence of HIV in plasma, we excluded all samples for which viremia was below the LOQ for each assay in the pairing. Because one aim of this study was to be clear about the consistency of the corrections observed, we chose to show the results for all samples from each patient. The medians of the assay differentials were also calculated for each patient.
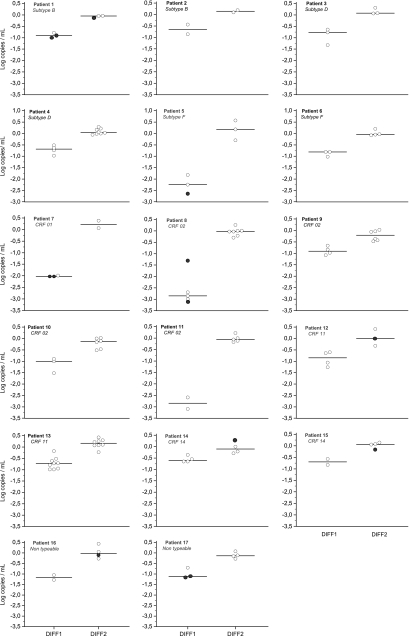
One hundred twenty-five samples from 17 patients were analyzed: 49 with CTM1 and ABB, 66 with CTM2 and ABB, and 10 with all three assays. The mean number of samples tested from each patient was 3.5 (range, 2 to 9) for the comparison between CTM1 and ABB and 4.5 (range, 2 to 8) for the comparison between CTM2 and ABB. The distribution of the various HIV subtypes and other circulating recombinant forms (CRF) found in the patients was as follows: B, 2; D, 2; F, 2; CRF01, 1; CRF02, 4; CRF11, 2; CRF14, 2; nontypeable, 2. For all samples taken together, the means for DIFF1 and DIFF2 were −1.18 log10 copies/ml (range, −3.10 to −0.18) and zero (−0.52 to +0.57), respectively. In detail, the median differences obtained for all 17 patients were between −2.85 and −0.60 log10 copies/ml for DIFF1 and between −0.21 and +0.22 log10 copies/ml for DIFF2 (Fig. 1) and were significantly different (P < 10−4). The median of DIFF1 exceeded 1 log10 copies/ml for 7 patients and 2 log10 copies/ml for 4 patients. Among the 59 values for DIFF1, 95%, 42%, and 17% exceeded 0.5, 1, and 2 log10 copies/ml, respectively, with the ABB assay always producing the higher value in each case. Conversely, only 2 of the 76 values for DIFF2 exceeded 0.5 log10 copies/ml: in one case (patient 10) the ABB result was lower (DIFF2 = −0.52 log10 copies/ml) and in the other (patient 5) the CTM2 result was lower (DIFF2 = +0.57 log10 copies/ml). However, these two discrepancies were not repeated in tests of other samples from these two patients.
Fig. 1.
Differences between the first version of Cobas TaqMan (CTM1) and Abbott RealTime (ABB) assay values (CTM1 − ABB = DIFF1) and differences between the CTM version 2 (CTM2) and ABB assay values (CTM2 − ABB = DIFF2) observed for each sample and for each patient. The medians are illustrated by the dark lines. The black circles indicate that one of the viral loads was undetectable and was replaced by the limit of quantification: 9 among the DIFF1 values (always with the CTM1 assay) and 5 among the DIFF2 values (always with the ABB assay).
The study results clearly show that the underestimation of VL that we observed by comparing the first version of CTM with the ABB assay was corrected successfully in the improved CTM version 2.0. The large number of samples analyzed in this study and the detailed results illustrated in Fig. 1 show that there was not a subgroup of samples for which CTM2 did not perform better than the previous version. In fact, corrections were observed for each sample and for each patient, regardless of HIV subtype, and even in the 4 patients where DIFF1 exceeded 2 log10 copies/ml. Although 17 patients infected with various subtypes and CRF were included in the present study, it is not possible to conclude that the unreliability of CTM1 has been completely corrected for all HIV strains in circulation. For this reason, it is necessary to remain vigilant with respect to the accuracy of these assays, especially in cases where there is an apparent discord between a patient's HIV viral load and the patient's immune or clinical status. This is also valid for the ABB assay, since Sire et al. have recently reported that some strains showed higher values with CTM2, with differences sometimes exceeding 1 log10 copies/ml (6). Our previous study already showed higher values with CTM1, but the differences did not exceed 1 log10 copies/ml (8).
In conclusion, the results of the present study show that all underestimations of VL observed with the first version of CTM have been successfully and clearly corrected in the upgraded CTM version 2.0. Although not all HIV strains in circulation were studied, these results demonstrate good reliability of the Cobas TaqMan v2.0 technique.
Footnotes
Published ahead of print on 4 May 2011.
REFERENCES
- 1. Braun P., et al. 2007. Comparison of four commercial quantitative HIV-1 assays for viral load monitoring in clinical daily routine. Clin. Chem. Lab. Med. 45:93–99 [DOI] [PubMed] [Google Scholar]
- 2. Church D., et al. 2011. Comparison of the RealTime HIV-1, COBAS TaqMan 48 v1.0, Easy Q. v1.2, and Versant v3.0 assays for determination of HIV-1 viral loads in a cohort of Canadian patients with diverse HIV subtype infections. J. Clin. Microbiol. 49:118–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Delaugerre C., et al. 2009. Clinical and resistance consequences of misquantification of plasma and cerebrospinal fluid human immunodeficiency virus type 1 (HIV-1) RNA in samples from an HIV-1 subtype G-infected patient. J. Clin. Microbiol. 47:3763–3764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gueudin M., et al. 2007. Evaluation of the Roche Cobas TaqMan and Abbott RealTime extraction-quantification systems for HIV-1 subtypes. J. Acquir. Immune Defic. Syndr. 44:500–505 [DOI] [PubMed] [Google Scholar]
- 5. Holguin A., Lopez M., Molinero M., Soriano V. 2008. Performance of three commercial viral load assays, Versant human immunodeficiency virus type 1 (HIV-1) RNA bDNA v3.0, Cobas AmpliPrep/Cobas TaqMan HIV-1, and NucliSens HIV-1 EasyQ v1.2, testing HIV-1 non-B subtypes and recombinant variants. J. Clin. Microbiol. 46:2918–2923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sire J. M., et al. 2011. Comparative RNA quantification of HIV-1 group M and non-M with the Roche Cobas AmpliPrep/Cobas TaqMan HIV-1 v2.0 and Abbott Real Time HIV-1 PCR assays. J. Acquir. Immune Defic. Syndr. 56:239–243 [DOI] [PubMed] [Google Scholar]
- 7. Thompson M. A., et al. 2010. Antiretroviral treatment of adult HIV infection: 2010 recommendations of the International AIDS Society-USA panel. JAMA 304:321–333 [DOI] [PubMed] [Google Scholar]
- 8. Wirden M., et al. 2009. Impact of discrepancies between the Abbott RealTime and Cobas TaqMan assays for quantification of human immunodeficiency virus type 1 group M non-B subtypes. J. Clin. Microbiol. 47:1543–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]