Abstract
Several studies have identified associations between single nucleotide polymorphisms (SNPs) occurring near the interleukin-28B (IL-28B) gene and response to antiviral treatment among hepatitis C virus (HCV) patients. Here, we describe a reliable melt-mismatch amplification mutation assay (melt-MAMA) PCR-based genotyping method for IL-28B which can be used in the management of HCV patients, helping to better define the course of therapy.
TEXT
Hepatitis C virus (HCV) is a positive-polarity, single-stranded RNA virus belonging to the genus Hepacivirus in the family Flaviviridae (12). Worldwide, an estimated of three million new infections occur annually and approximately 130 million people have been infected, the vast majority of infections becoming chronic infections (4). Moreover, a significant number of infected patients develop severe liver disease, including cirrhosis and hepatocellular carcinoma (7, 9, 17). Currently, the first line of HCV antiviral therapy is based on administration of pegylated alpha interferon (PEG-IFN-α) and ribavirin (RBV). Unfortunately, this therapeutic strategy is effective only in around 50% of patients infected with HCV genotype 1, although higher rates are reached in individuals infected with other viral genotypes (2, 20). There are a number of adverse effects to the PEG-IFN-α/RBV therapy such as depression, hematological “cytopenias,” thyroid dysfunction, and skin rash, making the treatment not well tolerated in many cases. Therefore, the ability to predict failures prior to treatment could save a great deal of pain and expense and lead to better clinical decisions.
Diverse predictor markers have been reported to influence the outcome of anti-HCV treatment such as virus genotype, viral load, complexity of viral population, and viral genome sequence (1, 5, 6, 10, 16). Recently, several genome-wide association studies (GWAS) have reported associations between different single nucleotide polymorphisms (SNPs) located near the interleukin-28B (IL-28B) gene and antiviral treatment, spontaneous viral clearance, and progression to chronicity (8, 14, 18, 19, 21). These findings suggest that these polymorphisms could be used as predictor factors to personalize antiviral therapy.
The goal of this work was to develop a rapid, highly specific and sensitive assay suitable for the identification of two IL-28B SNPs (rs12979860 and rs8099917) strongly associated with therapy outcome. For this, 20 patients with chronic HCV, 52 to 65 years old, and 30 healthy donors, age matched, were enrolled in this study. All patients had completed the corresponding antiviral treatment and were being seen as part of the follow-up standard protocol after completion of therapy.
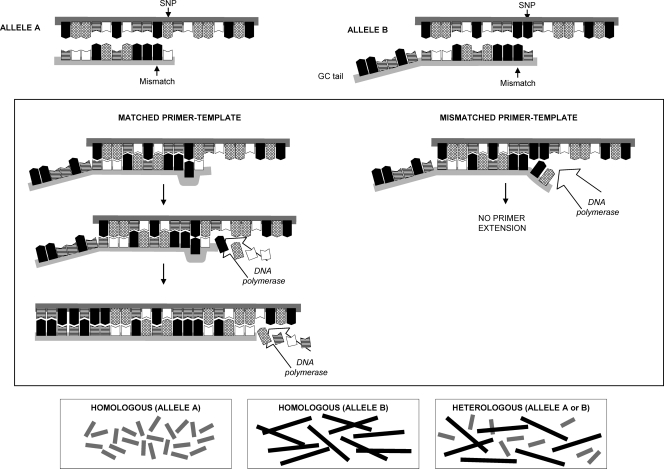
The genomic regions including SNPs rs12979860 and rs8099917 were used to design two different primer sets capable of differentiating between the two alleles for each polymorphism, based on their respective nucleotide patterns (Table 1). For this, the 3′ ends in both forward primers were designed to carry the nucleotide complementary to either allele. Additionally, one mismatch, located at the penultimate nucleotide position, was also incorporated into the allele-specific primer, increasing the primer specificity and enhancing the discrimination power of the two alleles (Fig. 1). The incorporation of a GC clamp (17-bp GC tail) at the 5′ end in one of the allele-specific primers was used to increase the melting temperature (Tm) (∼2.5°C) of that particular PCR fragment, thus facilitating visual identification (Fig. 2). Primers including one mismatch at the antepenultimate position and two mismatches, at both the penultimate and antepenultimate positions, were also tested to determine the optimal experimental mismatching combination.
Table 1.
Primer sequences
| Primer | Sequencea of: |
|
|---|---|---|
| Forward primer | Reverse primer | |
| MAMA-PCR primers | ||
| rsl2979860 (allele C)b | GGAGCTCCCCGAAGGCCC | CCTATGTCAGCGCCCACAATTCC |
| rs12979860 (allele T)b | GCGCGCGCCGGCCGCGGGGGGAGCTCCCCGAAGGCCT | CCTATGTCAGCGCCCACAATTCC |
| rsl2979860 (allele C) | AGGGAGCTCCCCGAAGGAGC | CCTATGTCAGCGCCCACAATTCC |
| rsl2979860 (allele Τ) | GCGCGCGCCGGCCGCGGCAGGGAGCTCCCCGAAGGAGT | CCTATGTCAGCGCCCACAATTCC |
| rsl2979860 (allele C) | AGGGAGCTCCCCGAAGGACC | CCTATGTCAGCGCCCACAATTCC |
| rsl2979860 (allele T) | GCGCGCGCCGGCCGCGGCAGGGAGCTCCCCGAAGGACT | CCTATGTCAGCGCCCACAATTCC |
| rs8099917 (allele G)b | CTTTTGTTTTCCTTTCTGTGAGCAAGG | CCCAGGAGCTTGCACTAGCTCTTG |
| rs8099917 (allele T)b | GCGCGCGCCGGCCGCGGCCTTTTGTTTTCCTTTCTGTGAGCAACΤ | CCCAGGAGCTTGCACTAGCTCTTG |
| rs8099917 (allele G) | CTTTTGTTTTCCTTTCTGTGAGCATTG | CCCAGGAGCTTGCACTAGCTCTTG |
| rs8099917 (allele T) | GCGCGCGCCGGCCGCGGCCTTTTGTTTTCCTTTCTGTGAGCATTT | CCCAGGAGCTTGCACTAGCTCTTG |
| rs8099917 (allele G) | CGTTATGCATCCTTTCTGTGAGCATGG | CCCAGGAGCTTGCACTAGCTCTTG |
| rs8099917 (allele T) | GCGCGCGCCGGCCGCGGCCTTTTGTTTTCCTTTCTGTGAGCATCΤ | CCCAGGAGCTTGCACTAGCTCTTG |
| Sequencing primers | ||
| rsl2979860 sequencing | CGCTTATCGCATACGGCTAGGCC | CGCTACGTAAGTCACCGCCCAGC |
| rs8099917 sequencing | TGTTCCTTGTAAAAGATTCCATCCATACAA | CCCAGGAGCTTGCACTAGCTCTTG |
| Cloning primers | ||
| rsl2979860 cloning C | GCCTGTCGTGTACTGAACCAGGGAGCTCCCCGAAGGCGCGAACC | CTCGCCTGCTGCAGAAGCAGAG |
| rsl2979860 cloning T | GCCTGTCGTGTACTGAACCGGGAGCTCCCCGAAGGCGTGAACC | CTCGCCTGCTGCAGAAGCAGAG |
| rs8099917 cloning T | CAATTTGTCACTGTTCCTCCTTTTGTTTTCCTTTCTGTGAGCAATTTCACC | CCCAGGAGCTTGCACTAGCTCTTG |
| rs8099917 cloning G | CAATTTGTCACTGTTCCTCCTTTTGTTTTCCTTTCTGTGAGCAATGTCACC | CCCAGGAGCTTGCACTAGCTCTTG |
Polymorphic sites are shown in boldface. Mismatching nucleotides are in italics. GC tails are underlined.
Mismatches according to Li et al. (12a).
Fig. 1.
Melt-MAMA PCR principle. Two allele-specific primers were designed in such way that the last position at the 3′ end matches the SNP of interest, and therefore, if the SNP is not complementary, the DNA polymerase fails to extend the primers, preventing the synthesis of the nascent DNA strand. Mismatches purposely incorporated on the 3′ end of the MAMA primer improve allele discrimination by further destabilizing the nucleotide base pairing and avoiding amplification of the undesired allele. One of the allele-specific primers also differs at the 5′ end, where a GC tail was incorporated in order to increase the melting temperature of the corresponding amplicon to facilitate allele identification. Measurement of fluorescence (SYBR green) allows amplicon identification and allele discrimination by MC analysis.
Fig. 2.
Primer annealing sites. Sequences of both plasmids rs12979860 (A) and rs8099917 (B), along with the allele-specific primers, are depicted. The conserved nucleotide positions throughout the alignment are indicated by dots. The target SNP position is designated by asterisks. Solid arrows represent the positions for the melt-MAMA (forward) primer, and the dashed arrows depict the locations of the corresponding reverse primers.
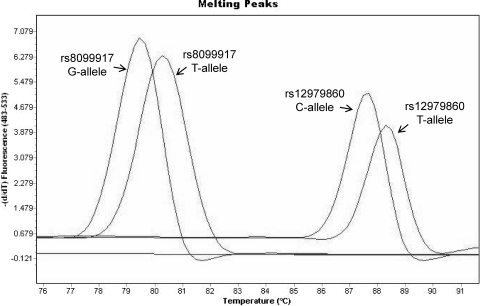
Four different constructions bearing the corresponding nucleotide substitutions for each target SNP were generated. All plasmid constructions were subjected to melt-mismatch amplification mutation assay (melt-MAMA) PCR using the corresponding primers. Melting curve (MC) analysis showed that SNP rs12979860-C exhibited a melting peak at 87.5°C while rs12979860-T exhibited a peak at 88.5°C. For SNP rs8099917, the G allele showed a melting peak at 79.3°C and the T allele displayed a melting peak at 80.5°C (Fig. 3). Primers including only one mismatch at the penultimate nucleotide position exhibited good discrimination between both alleles (Fig. 3). On the other hand, neither primer set containing a unique mismatch at the antepenultimate position achieved the desired degree of discrimination for either SNP. PCR efficiency was significantly reduced when primers including two mismatches were used, resulting in poor amplification.
Fig. 3.
IL-28B genotyping by melt-MAMA PCR. (A) Primer performance for both SNPs was assessed by melt-MAMA PCR. Primers bearing one single mismatch exhibited good discrimination between alleles. The Tm for SNP rs12979860 corresponding to the C allele amplicon was 87.5°C, while the T allele exhibited a Tm of 88.5°C. For SNP rs8099917, the experimental Tms observed for the G and T alleles were 79.3 and 80.5°C, respectively.
Blood samples from 30 healthy donors were simultaneously genotyped by melt-MAMA PCR and Sanger sequencing. Concordance between the two methods was 100%. For SNP rs12979860, 25 subjects were homozygous (3 for the T allele and 22 for the C allele) and five were heterozygous. For SNP rs8099917, 15 individuals exhibited a homozygous genotype (5 for the G allele and 10 for the T-allele) and 15 individuals were heterozygous (Table 2).
Table 2.
IL28B genotyping and patients' demographic characteristics
| Subject type | Characteristic | Value for SNP: |
|||||
|---|---|---|---|---|---|---|---|
| rsl2979S60 with genotype: |
rs8099917 with genotype: |
||||||
| TT | TC | CC | GG | TG | TT | ||
| Donors | No. | 3 | 5 | 22 | 5 | 15 | 10 |
| Mean age (yr) (SD) | 56.6 (6.4) | 57.6 (4.7) | 57.8 (3.6) | 56.6 (4.8) | 58.4 (4.5) | 57 (2.4) | |
| No. female | 1 | 4 | 12 | 3 | 14 | —a | |
| No. male | 2 | 1 | 10 | 2 | 1 | 10 | |
| Patients | No. | 16 | — | 4 | 5 | 3 | 12 |
| Mean age (yr) (SD) | 59.1 (4.3) | — | 60.2 (4.2) | 59 (4.8) | 59.6 (4.1) | 59.4 (4.3) | |
| No. female | 9 | — | 3 | 4 | 1 | 7 | |
| No. male | 7 | — | 1 | 1 | 2 | 5 | |
| No. with SVR | 2 | — | 4 | — | 1 | 5 | |
| No. with NVRb | 14 | — | — | 5 | 2 | 7 | |
—, not applicable.
NVR, nonvirological response.
Twenty HCV cases with known therapy outcome were genotyped. The most common genotype for SNP rs12979860 (16 patients) among the HCV cases was homozygous for the T allele. Four patients were homozygous for the C allele, and no individuals heterozygous for this particular SNP were identified in this cohort. All four C allele carriers (100%) successfully achieved a sustained virological response (SVR), while only 2 individuals (12.5%) of 16 carrying the T allele attained SVR. In general, patients carrying the T allele (87.5%) were prone to fail the antiviral treatment (Table 2).
For SNP rs8099917, 17 cases showed a homozygous genotype (5 for the G allele and 12 for the T allele) and 3 heterozygous cases were also identified (Table 2). Seven individuals (87.5%) carrying the G allele did not achieve SVR, and only 1 subject (12.5%) successfully eliminated the virus. Subjects homozygous for the T allele achieved SVR in 41.6% of the cases, while 58.3% showed a null viral response.
Until very recently, there were no reliable baseline markers that could predict the outcome of anti-HCV therapy. Findings showing associations between several SNPs located in the proximity of the IL-28B gene and viral response are promising results, suggesting that the associations can be used for decision making before start of treatment, preventing patients from undergoing a great deal of unnecessary distress. Thus, this might potentially represent a major step toward customization of medical care for HCV patients. Remarkable attention has been given to IL-28B SNPs rs12979860 and rs8099917, which have shown strong association with therapy outcome and therefore are leading candidates for predictor markers.
The predictive value of the SNPs located near the IL-28B gene needs to be compared with those of other predictor factors such as the infecting genotype, viral load, viral population complexity, nucleotide substitutions, etc., to identify possible complementation between predictor factors and thus improve their capacity to predict therapy outcome. The importance of IL-28B SNPs as predictor markers is crucial even in the case of future therapies based on viral enzyme inhibitors since IFN-α will remain the backbone of the anti-HCV treatment.
Despite the capacity of both IL-28B SNPs to predict SVR among HCV cases, these markers seem to lack the ability to forecast when a patient could relapse, and therefore the IL-28B genotype should not be used exclusively to determine treatment outcome. Moreover, IL-28B genotyping does not provide any guidance regarding treatment duration and should always be supplemented with a response-guided, personalized approach based on viral titer monitoring during the patient's follow-up.
Development of reliable methods for the correct identification of IL-28B SNPs is of high importance for the management of HCV cases. MAMA-PCR has been largely used for timely nucleotide substitution identification in diverse settings (3, 11, 15). The combination of MC analysis resolution and the simplicity of MAMA-PCR-based assays provides a powerful tool for the accurate identification of SNPs without the need for extra steps after DNA amplification (13).
In summary, we present a reproducible, inexpensive, and accurate method that allows rapid genotyping of IL-28B polymorphisms in either clinical or research laboratories with various throughputs. This methodological approach is especially convenient in clinical scenarios, where rather quick decisions should be made regarding medical attention of chronic HCV cases.
Acknowledgments
We thank Leopoldo Flores-Romo for critical reading of the manuscript.
S. Fonseca-Coronado received funding from Programa de Formación e Incorporación de Profesores de Carrera en Facultades y Escuelas para el Fortalecimiento de la Investigación (PROFIP), DGAPA, UNAM.
Footnotes
Published ahead of print on 25 May 2011.
REFERENCES
- 1. Akuta N., et al. 2009. A matched case-controlled study of 48 and 72 weeks of peginterferon plus ribavirin combination therapy in patients infected with HCV genotype 1b in Japan: amino acid substitutions in HCV core region as predictor of sustained virological response. J. Med. Virol. 81:452–458 [DOI] [PubMed] [Google Scholar]
- 2. Alavian S. M., Behnava B., Tabatabaei S. V. 2010. Comparative efficacy and overall safety of different doses of consensus interferon for treatment of chronic HCV infection: a systematic review and meta-analysis. Eur. J. Clin. Pharmacol. 66:1071–1079 [DOI] [PubMed] [Google Scholar]
- 3. Alonso R., et al. 2005. MAMA-PCR assay for the detection of point mutations associated with high-level erythromycin resistance in Campylobacter jejuni and Campylobacter coli strains. J. Microbiol. Methods 63:99–103 [DOI] [PubMed] [Google Scholar]
- 4. Alter M. J. 2007. Epidemiology of hepatitis C virus infection. World J. Gastroenterol. 13:2436–2441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Aurora R., Donlin M. J., Cannon N. A., Tavis J. E. 2009. Genome-wide hepatitis C virus amino acid covariance networks can predict response to antiviral therapy in humans. J. Clin. Invest. 119:225–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Donlin M. J., et al. 2010. Contribution of genome-wide HCV genetic differences to outcome of interferon-based therapy in Caucasian American and African American patients. PLoS One 5:e9032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ferlay J., et al. 2010. GLOBOCAN 2008, cancer incidence and mortality worldwide: IARC CancerBase no. 10. International Agency for Research on Cancer, Lyon, France: http://globocan.iarc.fr/ [Google Scholar]
- 8. Ge D., et al. 2009. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature 461:399–401 [DOI] [PubMed] [Google Scholar]
- 9. Global Burden of Hepatitis C Working Group 2004. Global burden of disease (GBD) for hepatitis C. J. Clin. Pharmacol. 44:20–29 [DOI] [PubMed] [Google Scholar]
- 10. Hoofnagle J. H., Wahed A. S., Brown R. S., Jr., Howell C. D., Belle S. H. 2009. Early changes in hepatitis C virus (HCV) levels in response to peginterferon and ribavirin treatment in patients with chronic HCV genotype 1 infection. J. Infect. Dis. 199:1112–1120 [DOI] [PubMed] [Google Scholar]
- 11. Jinneman K. C., Hill W. E. 2001. Listeria monocytogenes lineage group classification by MAMA-PCR of the listeriolysin gene. Curr. Microbiol. 43:129–133 [DOI] [PubMed] [Google Scholar]
- 12. Lemon S. M., Walker C. M., Alter M. J., Yi M. 2007. Hepatitis C virus, p. 1253–1304In Knipe D. M., Howley P. M. (ed.), Fields virology, vol. 1 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 12a. Li B., Kodura I., Fu D. J., Watson D. E. 2004. Genotyping with Taq MAMA. Genomics 83:311–320 [DOI] [PubMed] [Google Scholar]
- 13. Papp A. C., Pinsonneault J. K., Cooke G., Sadee W. 2003. Single nucleotide polymorphism genotyping using allele-specific PCR and fluorescence melting curves. Biotechniques 34:1068–1072 [DOI] [PubMed] [Google Scholar]
- 14. Rauch A., et al. 2010. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology 138:1338–1345, 1345 e1–e7 [DOI] [PubMed] [Google Scholar]
- 15. Rodriguez-Castillo A., Vaughan G., Ramirez-Gonzalez J. E., Escobar-Gutierrez A. 2010. Simultaneous cocirculation of both European varicella-zoster virus genotypes (E1 and E2) in Mexico City. J. Clin. Microbiol. 48:1712–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Saludes V., et al. 2010. Baseline prediction of combination therapy outcome in hepatitis C virus 1b infected patients by discriminant analysis using viral and host factors. PLoS One 5:e14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Seeff L. B. 2002. Natural history of chronic hepatitis C. Hepatology 36:S35–S46 [DOI] [PubMed] [Google Scholar]
- 18. Suppiah V., et al. 2009. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat. Genet. 41:1100–1104 [DOI] [PubMed] [Google Scholar]
- 19. Tanaka Y., et al. 2009. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat. Genet. 41:1105–1109 [DOI] [PubMed] [Google Scholar]
- 20. Teoh N. C., Farrell G. C., Chan H. L. 2010. Individualisation of antiviral therapy for chronic hepatitis C. J. Gastroenterol. Hepatol. 25:1206–1216 [DOI] [PubMed] [Google Scholar]
- 21. Thomas D. L., et al. 2009. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature 461:798–801 [DOI] [PMC free article] [PubMed] [Google Scholar]