Abstract
Of 38 vancomycin-intermediate Staphylococcus aureus (VISA) clinical strains, 27 (71%) possessed a mutation(s) in rpoB encoding the β-subunit of RNA polymerase. Furthermore, 95.6% of the rifampin-resistant mutants obtained from 9 methicillin-resistant S. aureus (MRSA) clinical isolates showed decreased vancomycin susceptibilities. These data indicate the involvement of an rpoB mutation in VISA phenotype expression.
TEXT
Staphylococcus aureus is a leading cause of hospital-acquired infections (10, 17). Vancomycin has been the therapy of choice for treating serious infections caused by multidrug-resistant S. aureus since the 1950s. However, the emergence of vancomycin-intermediate S. aureus (VISA), hetero-VISA (hVISA) and vancomycin-resistant S. aureus (VRSA) has threatened the rank of vancomycin as the frontline antibiotic in methicillin-resistant S. aureus (MRSA) chemotherapy (1, 8, 9).
VISA and hVISA were first described in 1997. Since then the mechanism of resistance has been pursued, which is based on the accumulation of spontaneous chromosomal mutations (10, 11). Mutations identified in a couple of two-component regulatory systems, vraSR and graRS, were shown to be responsible for the VISA phenotype expression in the Mu3-Mu50 lineage of strains (MIC ≥ 4 μg/ml) (5, 19). Howden et al. also reported the contribution of a graRS mutation to vancomycin resistance (12). Recently, however, we noticed that introduction of vraS(I5N) and graR(N197S) mutations onto the chromosome of hVISA strain Mu3 was not sufficient to achieve the level of vancomycin resistance of VISA strain Mu50 (MIC = 8 μg/ml). We then found out that, besides the two regulator mutations, another mutation in rpoB, rpoB(H481Y), was additionally required for the complete acquisition of the VISA phenotype expressed by Mu50 (M. Matsuo et al., submitted for publication). We also demonstrated that introduction of another mutation, rpoB(A621E), into a vancomycin-susceptible S. aureus (VSSA) strain conferred vancomycin and daptomycin heteroresistance onto it (6). Therefore, it is likely that the rpoB mutation is an important contributor to the VISA phenotype. Although the way that rpoB mutation affects vancomycin resistance is currently unknown, a thickened cell wall was noted with the strains with rpoB(H481Y) mutations, as well as those with rpoB(A621E) mutations (6; M. Matsuo et al., submitted). In any case, if an rpoB mutation were a major contributor for VISA phenotype, it would be predictable for an rpoB mutation to be frequently found in VISA clinical strains throughout the world. It is also predicted that selection of clinical S. aureus strains by rifampin should yield rifampin-resistant rpoB mutants with reduced susceptibilities to vancomycin. This study was performed to test these predictions.
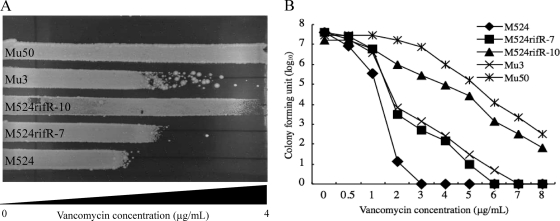
VISA strain Mu50, carrying an rpoB(H481Y) mutation, is highly rifampin resistant (MIC > 128 μg/ml). On the other hand, the rpoB mutation, rpoB(A621E), identified in the in vitro-derived hVISA strain, did not cause rifampin resistance (6). This indicates that there should be two groups among VISA or hVISA strains with rpoB mutations; one that is rifampin resistant, and another that is rifampin susceptible. With this in mind, we first generated a series of rifampin-resistant laboratory strains by selecting methicillin-resistant S. aureus (MRSA) clinical strains by using rifampin. We then evaluated the vancomycin susceptibilities of the mutants, along with those of their respective parent strains. A total of nine MRSA strains, isolated during 2005 and 2006 from bacteremia patients in Juntendo University Hospital, were used. All of the strains were susceptible to vancomycin and rifampin, with MICs of ≤2 and <0.25 μg/ml, respectively (Table 1). They belonged to the type IIA SCCmec-type II coagulase that corresponds to the most dominant Japanese hospital-associated MRSA (HA-MRSA) clone with the ST5 genotype (20, 23). A total of 107 CFU of MRSA cells were inoculated on brain heart infusion (BHI) agar plates containing 10 μg/ml rifampin, and rifampin-resistant colonies that appeared on the plates were picked after 24 h of incubation at 37°C. The picked colonies were cultivated in drug-free BHI medium and spread on drug-free BHI agar plates to carry out colony purification before they were established as strains. Rifampin resistance of the established mutant strains was confirmed by determining the rifampin MIC according to CLSI guidelines (2). Finally, a total of 90 rifampin-resistant mutant strains (10 from each MRSA strain) with rifampin MICs of >32 μg/ml were established, and their vancomycin susceptibilities were compared to those of the respective parent strains (Table 1). The change in vancomycin susceptibilities of these mutant strains was first determined by a vancomycin-gradient gel assay (V-GGA), which is convenient to operate and allows evaluation of a small range of susceptibility changes with a continuous value scale (7). As shown in Fig. 1A, all the mutant strains were streaked side by side, with their parent strains on the V-GGA plate using Mu3 and Mu50 as hVISA and VISA reference strains, respectively. The length (in cm) of cell growth was measured after 48 h of incubation at 37°C. The V-GGA was performed in duplicate, with reproducible results. Table 1 shows the results. Remarkably, as many as 86 of 90 rifampin-resistant mutant strains showed reduced susceptibilities to vancomycin in various degrees compared to those of their respective parent strains. Among the 86 mutant strains with increased V-GGA values, 5 (5.8%), 23 (26.7%), 43 (50%), and 15 (17.4%) strains exceeded those of the respective parent strains by 1, 5, 10, and >20%, respectively. The results demonstrated that the acquisition of rifampin resistance tends to decrease susceptibility to vancomycin. Figure 1A shows a representative result of a V-GGA. We then carried out MIC determination and population analysis with the mutant strains that had V-GGA results similar to or exceeding that of hVISA strain Mu3. By testing 21 such strains, 10 strains were found to have attained the level of vancomycin resistance equivalent either to VISA (MIC ≥ 4 μg/ml) or hVISA. hVISA was defined as having a subpopulation of cells capable of growth in the presence of 4 μg/ml vancomycin at a frequency of 1 × 10−6 or above, as judged by population analysis. As shown in Table 2, two strains, M524rifR-8 and M524rifR-10, turned out to be VISA (MIC = 4 μg/ml). The other 8 strains, M2184rifR-6, M2184rifR-10, M1894rifR-9, M1485rifR-8, M1156rifR-10, M1112rifR-5, M1112rifR-6, and M524rifR-7, had MIC values below 4 μg/ml but showed typical heterogeneous-type vancomycin resistance when tested by population analysis. Figure 1B shows a representative result of the population analysis of three independent experiments. The two VISA mutant strains were derived from the same MRSA strain, while the eight hVISA mutant strains were derived from six different MRSA strains (Table 2).
Table 1.
Vancomycin and rifampin susceptibilities of rifampin-selected mutantsa
| Parent strain (mutant strain [∼10]) | MIC of the parent strain (MIC of the mutants) (mg/liter) forb: |
V-GGA result of the parent (range; avg ± SD of mutants) (cm)c | |
|---|---|---|---|
| Vancomycin | Rifampin | ||
| M2184 (M2184rifR-1) | 1.5 (1.5–2) | <0.25 (>32) | 7.39 (7.70–9.67; 8.65 ± 0.54) |
| M1894 (M1894rifR-1) | 1 (1–2) | <0.25 (>32) | 5.74 (6.23–7.05; 6.57 ± 0.26) |
| M1485 (M1485rifR-1) | 0.5 (0.5–2) | <0.25 (>32) | 5.42 (5.74–7.87; 6.37 ± 0.57) |
| M1156 (M1156rifR-1) | 0.5 (0.5–1) | <0.25 (>32) | 5.51 (5.90–6.88; 6.29 ± 0.40) |
| M85 (M85rifR-1) | 1 (1–1) | <0.25 (>32) | 6.01 (6.00–6.95; 6.53 ± 0.28) |
| M1112 (M1112rifR-1) | 0.5 (0.5–1.5) | <0.25 (>32) | 5.45 (5.68–6.95; 6.17 ± 0.33) |
| M524 (M524rifR-1) | 1 (1–4) | <0.25 (>32) | 5.53 (5.53–11.53; 7.12 ± 1.94) |
| M1845 (M1845rifR-1) | 1 (1–1.5) | <0.25 (>32) | 6.01 (5.84–6.95; 6.48 ± 0.32) |
| M694 (M694rifR-1) | 0.5 (0.5–1) | <0.25 (>32) | 5.86 (6.16–7.11; 6.36 ± 0.30) |
Ten rifampin-resistant mutants were obtained by rifampin selection from each of the 9 clinical MRSA strains, and their data are shown in parentheses.
Vancomycin concentrations in 0.5-mg/liter increments were used to determine MICs by using the MH agar dilution method according to CLSI guidelines (2).
The vancomycin gradient gel assay (V-GGA) evaluates subtle differences in vancomycin resistance between the parent and its mutant strains. It measures the length of bacterial growth on the vancomycin gradient gel plate. The length of growth of the parent strains and the ranges, averages, and SD of lengths of growth of the rifampin-resistant mutant strains are shown.
Fig. 1.
Evaluation of vancomycin susceptibility change for rifampin-selected mutant strains. Subtle changes in the vancomycin susceptibility were detected by using the vancomycin gradient gel assay (V-GGA) (A), and heterogeneous vancomycin resistance was evaluated by analyzing the vancomycin-resistant subpopulations as previously described (8) (B). The V-GGA was carried out using BHI agar plates containing increasing concentrations of vancomycin (from left to right), up to 4 μg/ml. The population analysis was performed using BHI agar plates with various concentrations of vancomycin, and the colonies formed on the agar plates were enumerated after 48 h of incubation at 37°C.
Table 2.
Characteristics of rifampin-selected mutant strains with confirmed VISA or hVISA phenotype
| Strain | Description | MIC (mg/liter) for: |
Phenotypea | V-GGA result (cm) | RpoB aa substitutionb | |
|---|---|---|---|---|---|---|
| Vancomycin | Rifampin | |||||
| M2184 | Clinical isolate | 1.5 | <0.25 | VSSA | 7.39 | |
| M2184rifR-6 | Derivative | 2 | >32 | hVISA | 9.34 | A477D |
| M2184rifR-l0 | Derivative | 2 | >32 | hVISA | 9.67 | R484C, E520G |
| M1894 | Clinical isolate | 1 | <0.25 | VSSA | 5.74 | |
| M1894rifR-9 | Derivative | 2 | >32 | hVISA | 7.05 | H481Y |
| M1485 | Clinical isolate | 0.5 | <0.25 | VSSA | 5.42 | |
| M1485rifR-8 | Derivative | 2 | >32 | hVISA | 7.87 | R484C, S529P |
| M1156 | Clinical isolate | 0.5 | <0.25 | VSSA | 5.51 | |
| M1156rifR-10 | Derivative | 1 | >32 | hVISA | 6.88 | R484H |
| M1112 | Clinical isolate | 0.5 | <0.25 | VSSA | 5.45 | |
| M1112rifR-5 | Derivative | 1 | >32 | hVISA | 6.32 | H481D |
| MI1112ifR-6 | Derivative | 1.5 | >32 | hVISA | 6.95 | A477V, H481Y |
| M524 | Clinical isolate | 1 | <0.25 | VSSA | 5.65 | |
| M524rifR-7 | Derivative | 1 | >32 | hVISA | 6.79 | H481Y |
| M524rifR-8 | Derivative | 4 | >32 | VISA | 10.25 | A477V, S529L |
| M524rifR-l0 | Derivative | 4 | >32 | VISA | 11.53 | R484C, L887F |
| Mu3c | Clinical isolate | 2 | <0.25 | hVISA | 8.34 | |
| Mu50c | Clinical isolate | 8 | >32 | VISA | >13.5 | H481Y |
VSSA, vancomycin-susceptible S. aureus with a vancomycin MIC of <4 mg/liter; VISA, vancomycin-intermediate S. aureus with a vancomycin MIC of ≥4 mg/liter; hVISA, hetero-VISA with a vancomycin MIC of <4 mg/liter, but containing a subpopulation of cells that grow on the BHI agar plate that contained ≥4 mg of vancomycin/liter at a frequency of 1 in 106 or greater (8).
The substituted RpoB amino acid (aa) in the mutated strain is shown as a one-letter alphabetical notation after the numeral, indicating the position of the substituted aa. The first letter denotes the substituted aa found at the corresponding position of the RpoB amino acid of strain N315. Bold font indicates an aa substitution located beyond the rifampin resistance determining region (RRDR).
Control strain.
Next, we determined the rpoB gene sequences of all the VISA and hVISA mutant strains and compared them with those of the parent strains. The sequence of entire rpoB gene, including its promoter region, was determined with the forward and reverse primers as described previously (6). All of the rifampin-selected strains tested harbored one or two mutations with amino acid (aa) substitutions. All of the mutations, except for one in M524rifR-10, were located within the rifampin resistance determining region (RRDR) that spans amino acid residues 463 to 550 (Table 2). Mutations in RRDR were reported to decrease the binding affinity of rifampin to RNA polymerase holoenzyme by lowering the hydrophobic interaction between RpoB and rifampin (21). It is interesting in this regard that a rifampin-selected mutant strain, M524rifR-10, possessed two rpoB mutations: one, R484C, within the RRDR, and the other, L887F, located outside the RRDR. Four rpoB mutations causing aa substitutions A477D, H481Y, R484H, and H481D were found as solitary mutations, indicating that they are directly associated with the reduced vancomycin susceptibility in their respective strains. In fact, the rpoB mutation with H481Y amino acid substitution is able to confer vancomycin resistance when introduced into an S. aureus cell (see below). There were only four rifampin-resistant mutants derived from three parent MRSA strains that did not reduce vancomycin susceptibility. They carried rpoB mutations with aa substitutions S464P, Q468K, D471Y, and S486L. It is possible that certain types of rpoB mutations may not raise vancomycin resistance. In agreement with this hypothesis, none of the four mutations was found in the list of in vitro-generated VISA or hVISA strains (Table 2). The former three mutations were not found in VISA clinical strains, either (Table 3). Although the fourth aa substitution, S486L, was found in VISA strain HIP09662, it was not the solitary change, and another aa substitution, D471N, was found in RpoB of the strain HIP09662 (Table 3).
Table 3.
List of rpoB nonsynonymous mutations found in the worldwide clinical VISA strains
| Strain | Description |
MIC (mg/liter) for: |
Predicted RpoB aa substitutionb | ||||
|---|---|---|---|---|---|---|---|
| NARSA strain IDa | Yr | Country | Reference | Vancomycin | Rifampin | ||
| Mu50 | NRS1 | 1996 | Japan | 4 | 8 | >32 | H481Y |
| MI (HIP5827) | NRS3 | 1997 | USA | 4 | 8 | <0.25 | R140S |
| NJ (HIP5836) | NRS4 | 1997 | USA | 4 | 8 | >32 | H481Y |
| PC (HIP06297) | NRS17 | 1998 | USA | 4 | 8 | >32 | Q468L |
| IL | NRS79 | 2001 | USA | 4 | 8 | >32 | H481R |
| AMC11094 | NRS49 | 1997 | Korea | 4 | 8 | <0.25 | |
| 99/3759-V | NRS39 | 1999 | UK | 4 | 8 | >32 | H481N, S529L |
| 99/3700-W | 1999 | UK | 4 | 8 | <0.25 | ||
| LIM2 | NRS36 | 1995 | France | 4 | 8 | >32 | H481N, S529L |
| 98141 | 1998 | France | 4 | 8 | >32 | H481N, S529L | |
| 28160 | 1998 | South Africa | 4 | 8 | >32 | H481N, S529L | |
| BR1 | 1998 | Brazil | 4 | 8 | >32 | H481N, 1527M | |
| SA MER-S6 | NRS12 | 1999 | France | NAc | 8 | <0.25 | |
| HIP06854 | NRS18 | 1998 | USA | NA | 4 | <0.25 | |
| HIP07920 | NRS21 | 1998 | USA | NA | 4 | >32 | R484H |
| HIP07930 | NRS22 | 1999 | USA | NA | 4 | <0.25 | D320N |
| HIP08926 | NRS23 | 2000 | USA | NA | 4 | <0.25 | |
| HIP09143 | NRS24 | 2000 | USA | NA | 4 | <0.25 | |
| HIP09313 | NRS26 | 2000 | USA | NA | 4 | <0.25 | |
| HIP09433 | NRS27 | 2000 | USA | NA | 4 | <0.25 | D320N |
| HIP09662 | NRS28 | 2000 | USA | NA | 4 | >32 | D471N, S486L |
| HIP09735 | NRS29 | 2000 | USA | NA | 4 | <0.25 | Y737F |
| HIP09740 | NRS51 | 2000 | USA | NA | 6 | >32 | H481D |
| HIP09737 | NRS52 | 2000 | USA | NA | 4 | >32 | H481D |
| LY-1999-01 | NRS63 | 1998 | Oman | NA | 4 | <0.25 | R406S |
| LY-1999-03 | NRS65 | 1998 | Oman | NA | 4 | <0.25 | |
| HIP10540 | NRS73 | 2000 | USA | NA | 8 | >32 | V135A, A477V |
| HIP10267 | NRS74 | 2000 | USA | NA | 4 | 16 | D471V, A473S, A477S, E478D |
| C2000001227 | NRS76 | 2000 | USA | NA | 8 | <0.25 | |
| NRS118 | NRS118 | 2002 | USA | NA | 4 | >32 | H481N, S529L |
| NRS126 | NRS126 | 2000 | USA | NA | 4 | 4 | H481N |
| P1V44 | NRS272 | 1999 | Belgium | NA | 16 | >32 | H481N, S529L |
| H1P12864 | NRS402 | 2003 | USA | NA | 8 | 2 | P519L |
| HIP13057 | NRS403 | 2004 | USA | NA | 8 | >32 | H481Y |
| HIP13036 | NRS404 | 2004 | USA | NA | 8 | <0.25 | |
| JCSC7193 | 2007 | Thailand | 18 | 4 | >32 | H481N, S529L | |
| JCSC7195 | 2007 | Thailand | 18 | 4 | <0.25 | ||
| JCSC7203 | 2007 | Thailand | 18 | 4 | >32 | H481N, S529L | |
“NARSA strain ID” refers to strain identification according to the Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA) (http://www.narsa.net).
Bold font indicates amino acid substitutions located beyond the rifampin resistance determining region (RRDR). Those underlined are those found in the VISA and hVISA mutant strains listed in Table 2.
NA, not applicable.
VSSA-to-VISA conversion is a multistep genetic event that involves at least two sequential mutations (8). In this regard, the case of M524 was impressive in that two VISA mutant strains were directly generated from it (increments of vancomycin MICs from 1 to 4 μg/ml) by using only one-step rifampin selection. We thought that M524 was ready to generate VISA by having gained another mutation that promotes vancomycin resistance. To test this hypothesis, we determined sequences of vraSR and graRS of M524 and other strains, since vraSR and graRS mutations are known to raise vancomycin resistance in S. aureus (3, 5, 13, 19). Interestingly, we found that the parent strain M524 harbored two mutations in the vraS gene, with amino acid substitutions of I5N and L67F. The vraS(I5N) mutation has been shown to confer the hVISA phenotype on a VSSA strain, N315 (13). However, M524 did not express the heteroresistance phenotype (data not shown). We do not know at the moment if the doubly mutated vraS contributes to the rise of vancomycin resistance or not. It is also possible that the M524 genome contains a mutation(s) in other genes that promotes vancomycin resistance in collaboration with the rpoB mutation.
If the rpoB mutation has a significant role in raising vancomycin resistance, the mutation is expected to have occurred at a considerable frequency in VISA clinical strains throughout the world. To test this hypothesis, we performed a prevalence study of an rpoB nonsynonymous mutation with a total of 38 VISA strains isolated from 10 countries, including 23 provided from the Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA) (http://www.narsa.net). Vancomycin and rifampin MICs were determined by the Mueller-Hinton (MH) agar dilution method, and the whole rpoB gene sequence was determined for all the strains. Table 3 shows the results. It was reconfirmed that all the strains satisfied the VISA criterion (vancomycin MIC ≥ 4 μg/ml). Among the 38 VISA strains, as many as 21 strains (55%) were resistant to rifampin (MIC ≥ 4 μg/ml); one was intermediate (MIC = 2 μg/ml), and 16 were susceptible (MIC < 1 μg/ml). The rpoB mutations with amino acid substitutions were found in all of the 22 rifampin-resistant and intermediate strains, but also in 5 of the 16 rifampin-susceptible strains. All mutations found in rifampin-resistant or intermediate strains were located within the RRDR, whereas those identified in rifampin-susceptible strains were found outside the RRDR (Table 3). In total, 27 of 38 VISA strains (71%) were found to carry the rpoB mutation. This frequency is significantly high compared to that reported with MRSA clinical strains (see below). It is also noticed that, among the 27 strains with the rpoB mutation, 12 strains had multiple mutations; the most mutations were four, found in strain HIP10267. At this moment we do not know whether all the rpoB mutations found in Table 3 contribute to the VISA phenotype of each strain except for the H481Y mutation found in Mu50. The rpoB(H481Y) mutation has been proven by a gene replacement experiment to raise vancomycin resistance (M. Matsuo et al., submitted). It is also noted that a considerable number of the amino acid substitutions underlined in Table 3 correspond to those correlated with the raised vancomycin resistance, as shown in Table 2.
Rifampin has been used in combination with other antibiotics to treat MRSA infection, because resistant strains of S. aureus are rapidly observed when rifampin is used as a single agent (22). Rifampin-resistant S. aureus is prevalent, and its frequency was reported to be from approximately 1.9 to 4.5 to 18% (14–16, 22). However, the rate of rifampin resistance in VISA strains revealed in this study was extremely high (55%), supporting the immanent link between the resistance of vancomycin and rifampin in S. aureus. However, it should be noted that rpoB mutations were observed even in the rifampin-susceptible VISA strains at a high frequency of 5 of 16 (31%). Evidently, these mutations are not the outcome of clinical rifampin therapy. We recently published a paper on the role of the rpoB(A621E) mutation to confer vancomycin and daptomycin heteroresistance on a VSSA strain without raising rifampin resistance (6). Therefore, it is very likely that the 5 rpoB mutations were selected not by rifampin but by vancomycin or other related antibiotics. Finally, 11 of 38 VISA strains (29%) were rifampin susceptible and free from rpoB mutation. These VISA strains should have a different resistance mechanism(s).
Taken altogether, the results from our study indicate that the rpoB mutation, although not exclusive, is one of the major contributors to vancomycin resistance in S. aureus. The use of rifampin in the treatment of MRSA infections would be better if reevaluated to prevent further increase of hVISA and VISA in clinical settings.
Acknowledgments
This work was supported by a Grant-in-Aid for the 21st Century COE Program and a Grant-in-Aid for Scientific Research (18590438) to L. Cui from the Ministry of Education, Science, Sports, Culture and Technology of Japan.
Footnotes
Published ahead of print on 27 April 2011.
REFERENCES
- 1. Chang S., et al. 2003. Infection with vancomycin-resistant Staphylococcus aureus containing the vanA resistance gene. N. Engl. J. Med. 348:1342–1347 [DOI] [PubMed] [Google Scholar]
- 2. Clinical Laboratory Standards Institute 2009. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 8th ed., vol. 29, no. 2 Approved standard M07-A8. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 3. Cui L., Lian J., Neoh H., Ethel R., Hiramatsu K. 2005. DNA microarray-based identification of genes associated with glycopeptide resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 49:3404–3413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cui L., et al. 2003. Cell wall thickening is a common feature of vancomycin resistance in Staphylococcus aureus. J. Clin. Microbiol. 41:5–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cui L., Neoh H. M., Shoji M., Hiramatsu K. 2009. Contribution of vraSR and graSR point mutations to vancomycin resistance in vancomycin-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 53:1231–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cui L., Neoh H. M., Shoji M., Hiramatsu K. 2010. An RpoB mutation confers dual heteroresistance to daptomycin and vancomycin in Staphylococcus aureus. Antimicrob. Agents Chemother. 54:5222–5233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cui L., Tominaga E., Neoh H. M., Hiramatsu K. 2006. Correlation between reduced daptomycin susceptibility and vancomycin resistance in vancomycin-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 50:1079–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hiramatsu K., et al. 1997. Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin. Lancet 350:1670–1673 [DOI] [PubMed] [Google Scholar]
- 9. Hiramatsu K., et al. 1997. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J. Antimicrob. Chemother. 40:135–136 [DOI] [PubMed] [Google Scholar]
- 10. Hiramatsu K., et al. 2004. Advance in vancomycin-resistance research in Staphylococcus aureus, p. 289–298 In Alekshun M., McDermott P., White D. (ed.), Frontiers in antibiotic resistance: a tribute to Stuart B. Levy. ASM Press, Washington, DC [Google Scholar]
- 11. Howden B. P., Davies J. K., Johnson P. D., Stinear T. P., Grayson M. L. 2010. Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin-intermediate strains: resistance mechanisms, laboratory detection, and clinical implications. Clin. Microbiol. Rev. 23:99–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Howden B. P., et al. 2008. Genomic analysis reveals a point mutation in the two-component sensor gene graS that leads to intermediate vancomycin resistance in clinical Staphylococcus aureus. Antimicrob. Agents Chemother. 52:3755–3762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Katayama Y., Murakami-Kuroda H., Cui L., Hiramatsu K. 2009. Selection of heterogeneous vancomycin-intermediate Staphylococcus aureus by imipenem. Antimicrob. Agents Chemother. 53:3190–3196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kilic A., Li H., Stratton C. W., Tang Y. W. 2006. Antimicrobial susceptibility patterns and staphylococcal cassette chromosome mec types of, as well as Panton-Valentine leukocidin occurrence among, methicillin-resistant Staphylococcus aureus isolates from children and adults in middle Tennessee. J. Clin. Microbiol. 44:4436–4440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim H. B., et al. 2004. In vitro activities of 28 antimicrobial agents against Staphylococcus aureus isolates from tertiary-care hospitals in Korea: a nationwide survey. Antimicrob. Agents Chemother. 48:1124–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Limbago B., et al. 2009. Characterization of methicillin-resistant Staphylococcus aureus isolates collected in 2005 and 2006 from patients with invasive disease: a population-based analysis. J. Clin. Microbiol. 47:1344–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lowy F. D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520–532 [DOI] [PubMed] [Google Scholar]
- 18. Lulitanond A., et al. 2009. The first vancomycin-intermediate Staphylococcus aureus strains isolated from patients in Thailand. J. Clin. Microbiol. 47:2311–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Neoh H. M., et al. 2008. Mutated response regulator graR is responsible for phenotypic conversion of Staphylococcus aureus from heterogeneous vancomycin-intermediate resistance to vancomycin-intermediate resistance. Antimicrob. Agents Chemother. 52:45–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Okuma K., et al. 2002. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J. Clin. Microbiol. 40:4289–4294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. O'Neill A. J., Huovinen T., Fishwick C. W., Chopra I. 2006. Molecular genetic and structural modeling studies of Staphylococcus aureus RNA polymerase and the fitness of rifampin resistance genotypes in relation to clinical prevalence. Antimicrob. Agents Chemother. 50:298–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Strausbaugh L. J., Jacobson C., Sewell D. L., Potter S., Ward T. T. 1992. Antimicrobial therapy for methicillin-resistant Staphylococcus aureus colonization in residents and staff of a Veterans Affairs nursing home care unit. Infect. Control Hosp. Epidemiol. 13:151–159 [DOI] [PubMed] [Google Scholar]
- 23. Watanabe S., et al. 2009. Genetic diversity of staphylocoagulase genes (coa): insight into the evolution of variable chromosomal virulence factors in Staphylococcus aureus. PLoS One 4:e5714. [DOI] [PMC free article] [PubMed] [Google Scholar]